Human KRAS gene is a known oncogene that activates downstream cancer-causing pathway resulting in clinically detectable cancer and tumour formation [
1,
2]. Originally from the rat sarcoma virus, detection of KRAS can be construed as a sign of food contamination, most probably from rat or mouse infestation as they are the natural animal host for the rat sarcoma virus. Typically, KRAS can be detected clinically via the gold standard quantitative polymerase chain reaction (qPCR) assay or enzyme-linked immunosorbent assay (ELISA) [
3,
4]. Comparing the two approaches, qPCR is faster, easier and more widely used.
The work describes some results on the observed ineffective use of FAM probe for detecting the amplicons of human KRAS gene in qPCR of CloTest positive and negative samples. Highly specific primers of the human KRAS gene were designed based on the conserved regions of the gene. In a similar qPCR assay with SYBR green detection chemistry, KRAS gene was detected with good efficiency. Thus, it appears perplexing as to why a highly specific FAM probe for the KRAS gene did not yield good amplification curves for most of the CloTest positive and negative samples.
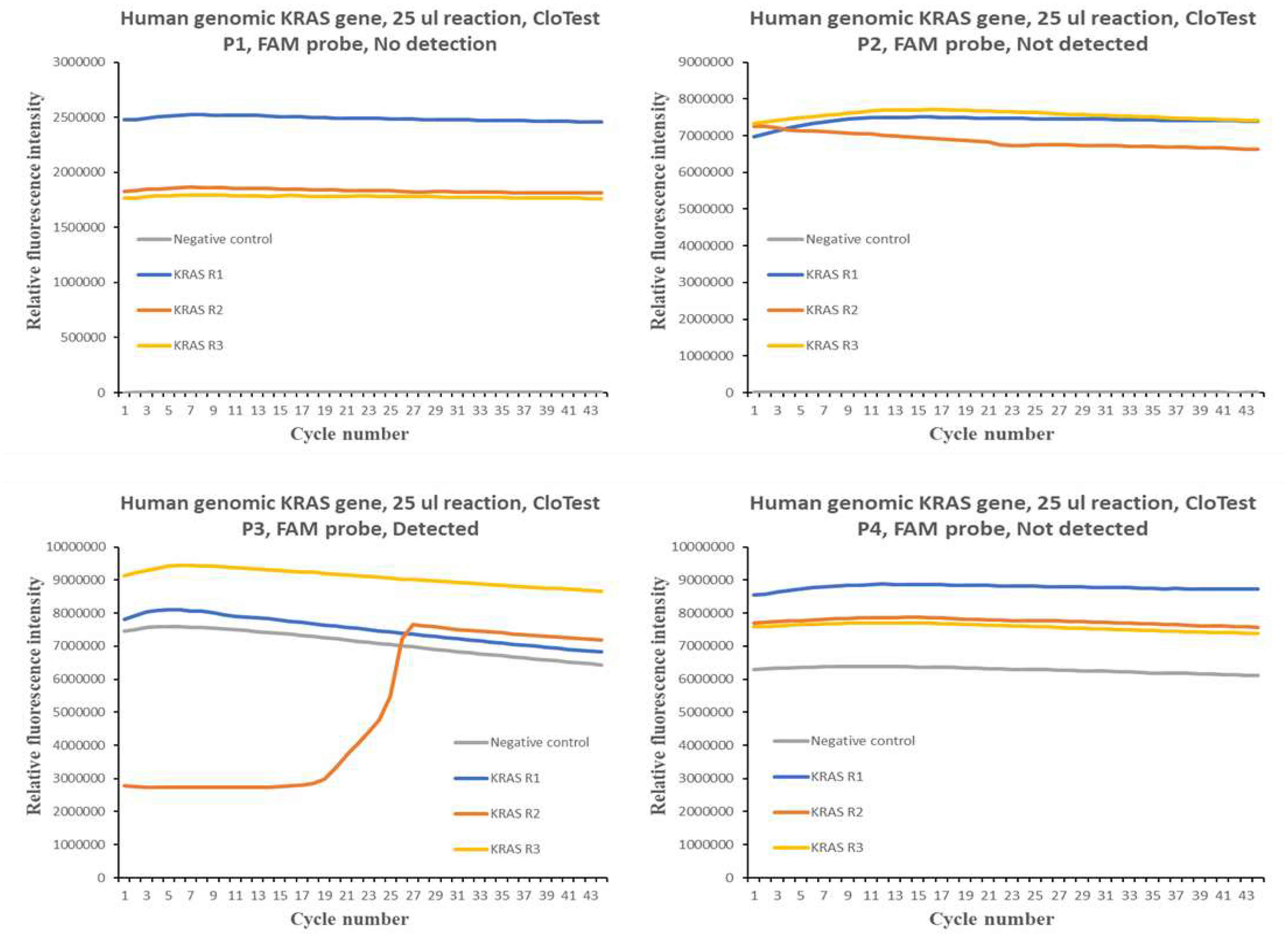
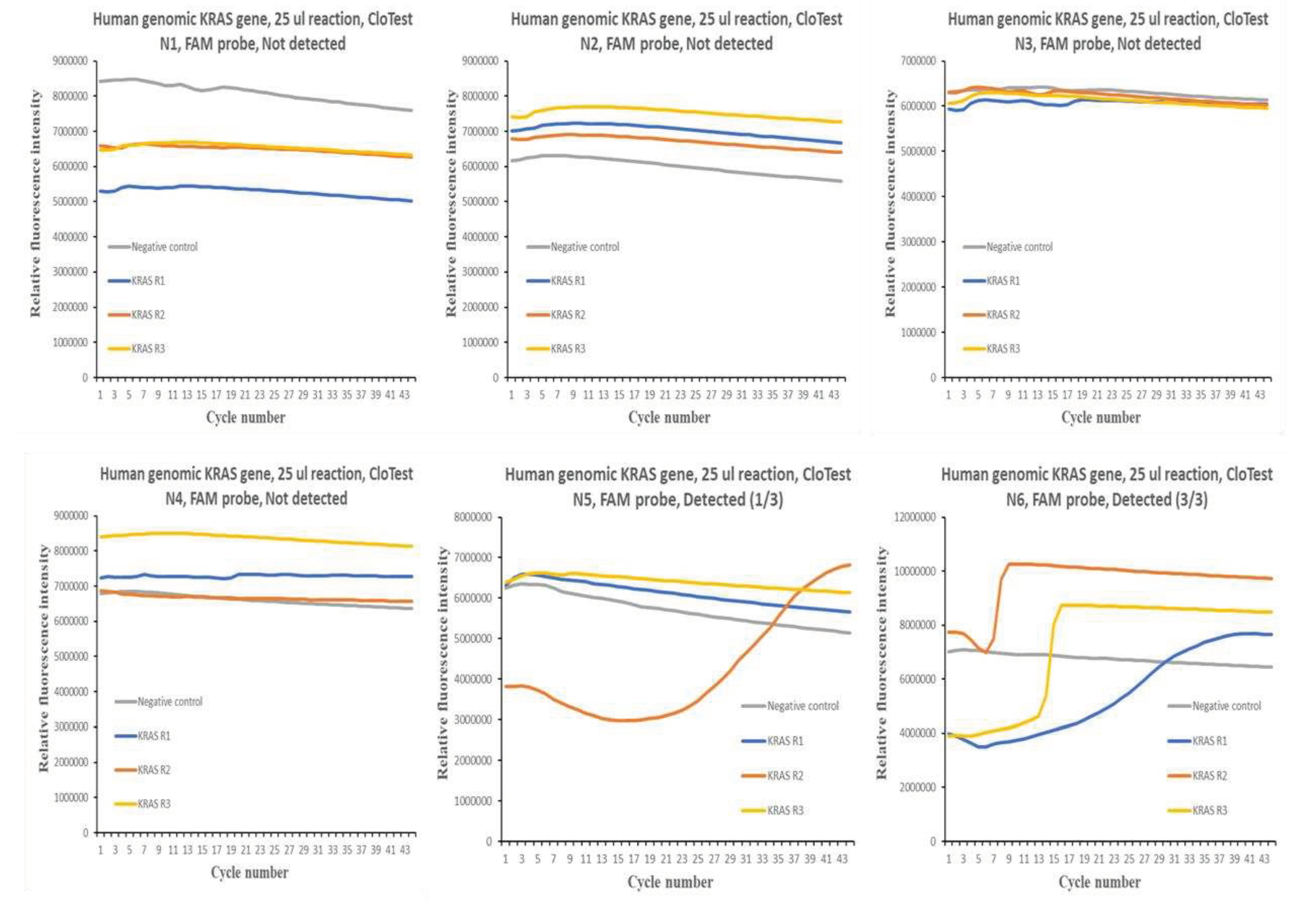
Figure 1 and
Figure 2 show the qPCR amplification plots for the different CloTest positive and negative samples. In the case of CloTest positive samples, there was only one amplification curve out of a possible 12. On the other hand, there were also very few instances of successful amplification of the human KRAS gene in six CloTest negative samples.
Overall, the results obtained suggest significant difficulty in using FAM probe detection chemistry to profile for human KRAS gene in human gastric CloTest positive and negative samples, where a similar qPCR test using SYBR Green detection chemistry was efficient in generating good amplification curves. Such a result suggests that the FAM probe could not bind to the qPCR amplicons, which, in turn, indicates that the targeted KRAS gene region may be undergoing rapid genomic evolution, and thus, escape detection via probe chemistry. This region may be a candidate for close surveillance by the public health community as it may be a region important for molecular recognition or structural function in the human KRAS gene.
Funding
The author thank the National University of Singapore for financial support.
Conflict of Interest
The author declares no conflict of interest.
References
- S. Jančík, J. Drábek, D. Radzioch, and M. Hajdúch, “Clinical Relevance of KRAS in Human Cancers,” BioMed Res. Int., vol. 2010, p. e150960, Jun. 2010. [CrossRef]
- L. Huang, Z. Guo, F. Wang, and L. Fu, “KRAS mutation: from undruggable to druggable in cancer,” Signal Transduct. Target. Ther., vol. 6, no. 1, Art. no. 1, Nov. 2021. [CrossRef]
- Orue and, M. Rieber, “Optimized Multiplex Detection of 7 KRAS Mutations by Taqman Allele-Specific qPCR,” PLOS ONE, vol. 11, no. 9, p. e0163070, Sep. 2016. [CrossRef]
- J. Ugorcakova, T. Hlavaty, T. Novotna, and G. Bukovska, “Detection of point mutations in KRAS oncogene by real-time PCR-based genotyping assay in GIT diseases,” Bratisl. Med. J., vol. 113, no. 02, pp. 73–79, 2012. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).