Submitted:
26 June 2023
Posted:
27 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and methods
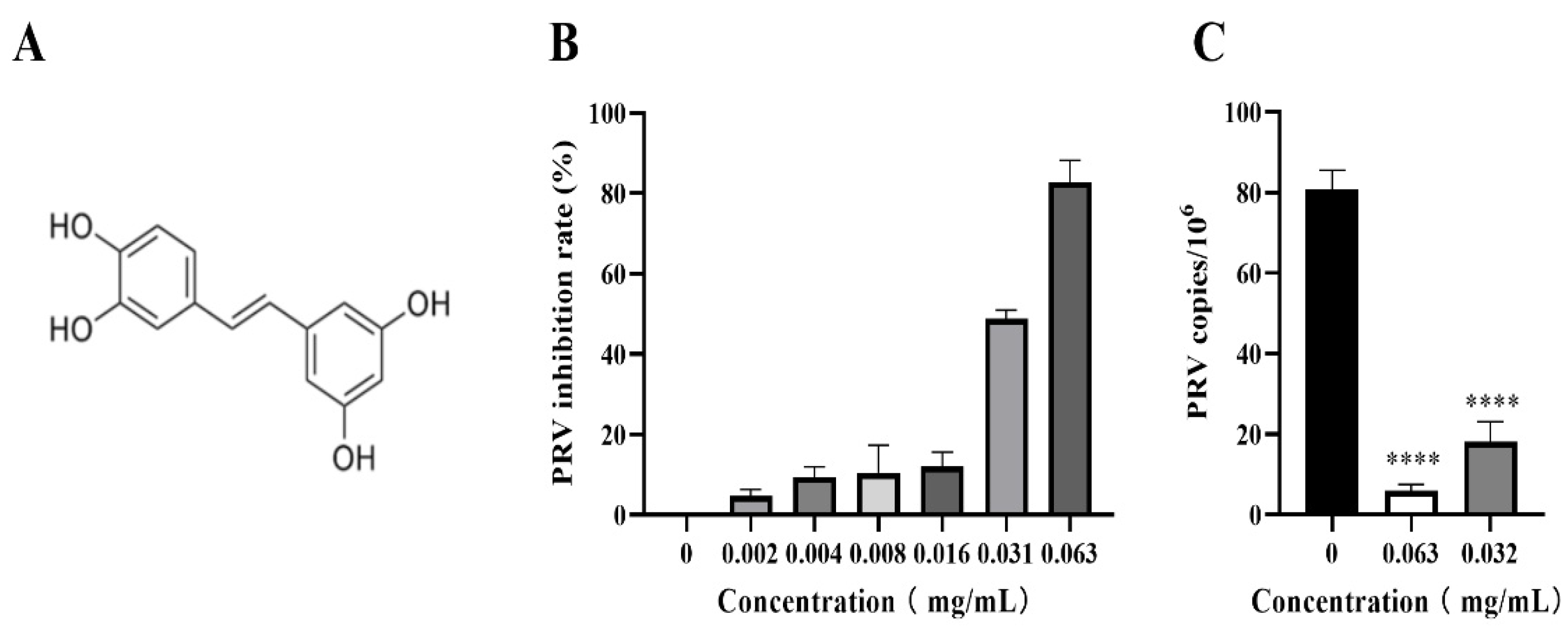
2.1. Cells, materials, and piceatannol
2.2. Determination of cytotoxicity and inhibitory activity
2.3. Effect of piceatannol on the PRV life cycle
2.4. Effects of piceatannol on PRV gene expression
2.5. Effect of piceatannol on PRV protein expression
2.6. Apoptosis analysis
2.7. Assay of the antiviral activity of piceatannol against PRV in vivo
2.8. Statistical methods
3. Results
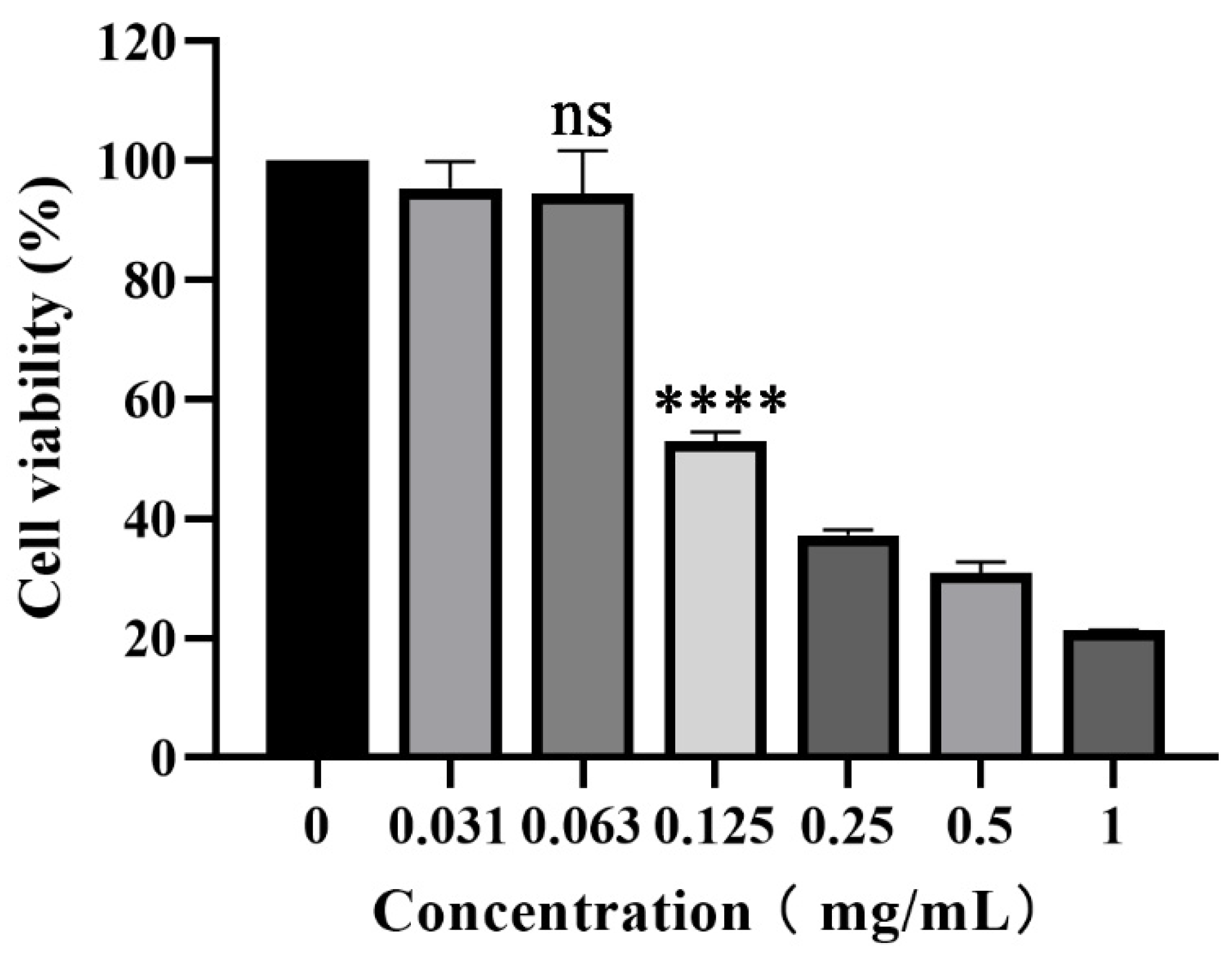
3.1. Cytotoxicity of piceatannol on PK-15 cells
3.2. Piceatannol inhibited PRV proliferation in PK-15 cells
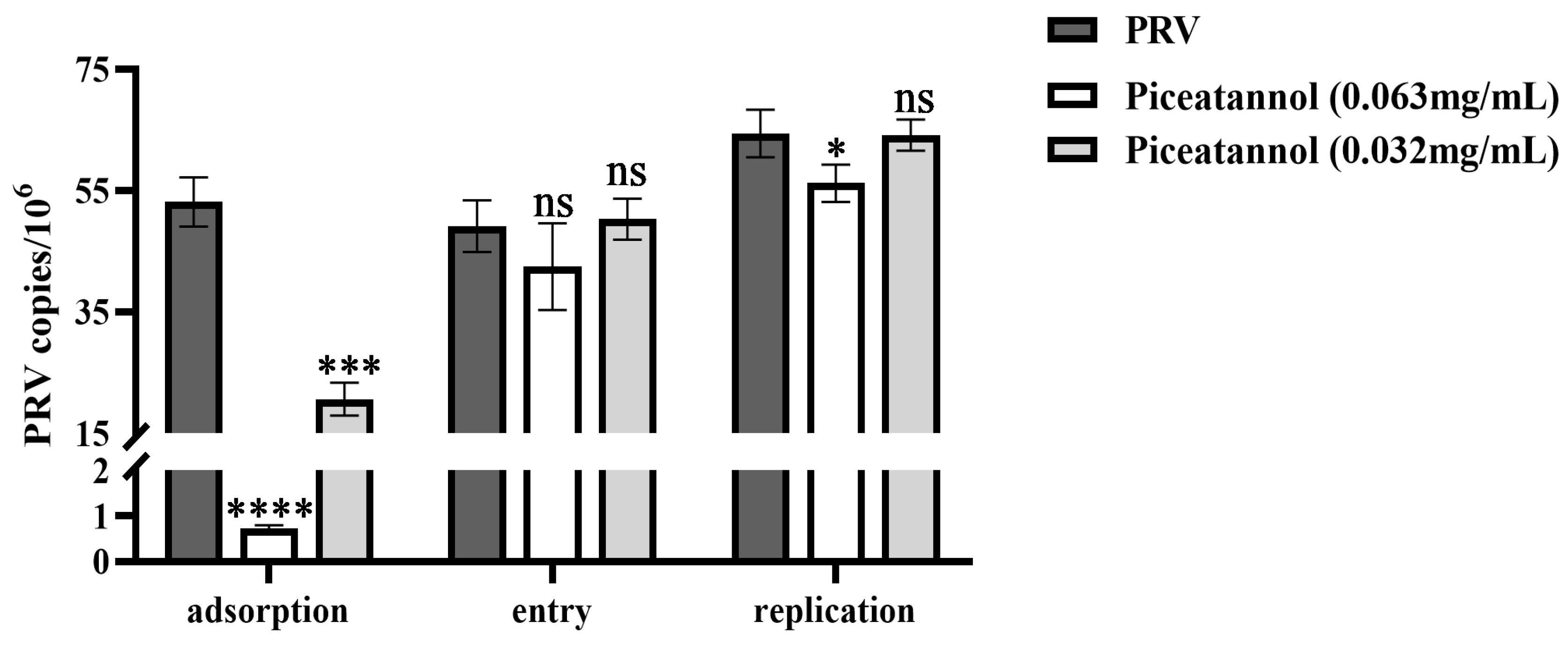
3.3. Effect of piceatannol on the replication cycle of PRV
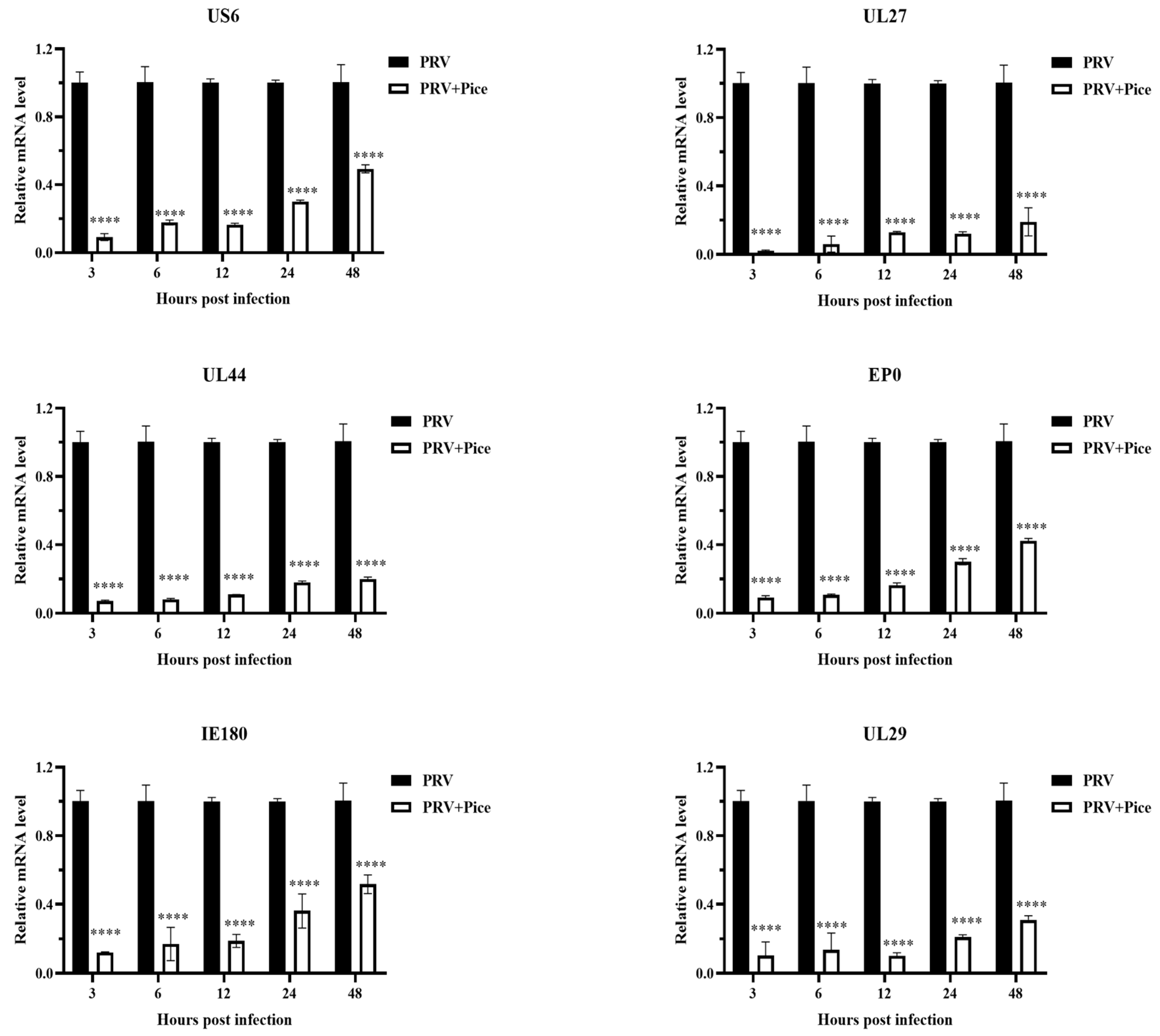
3.4. Inhibitory effect of piceatannol on PRV gene expression
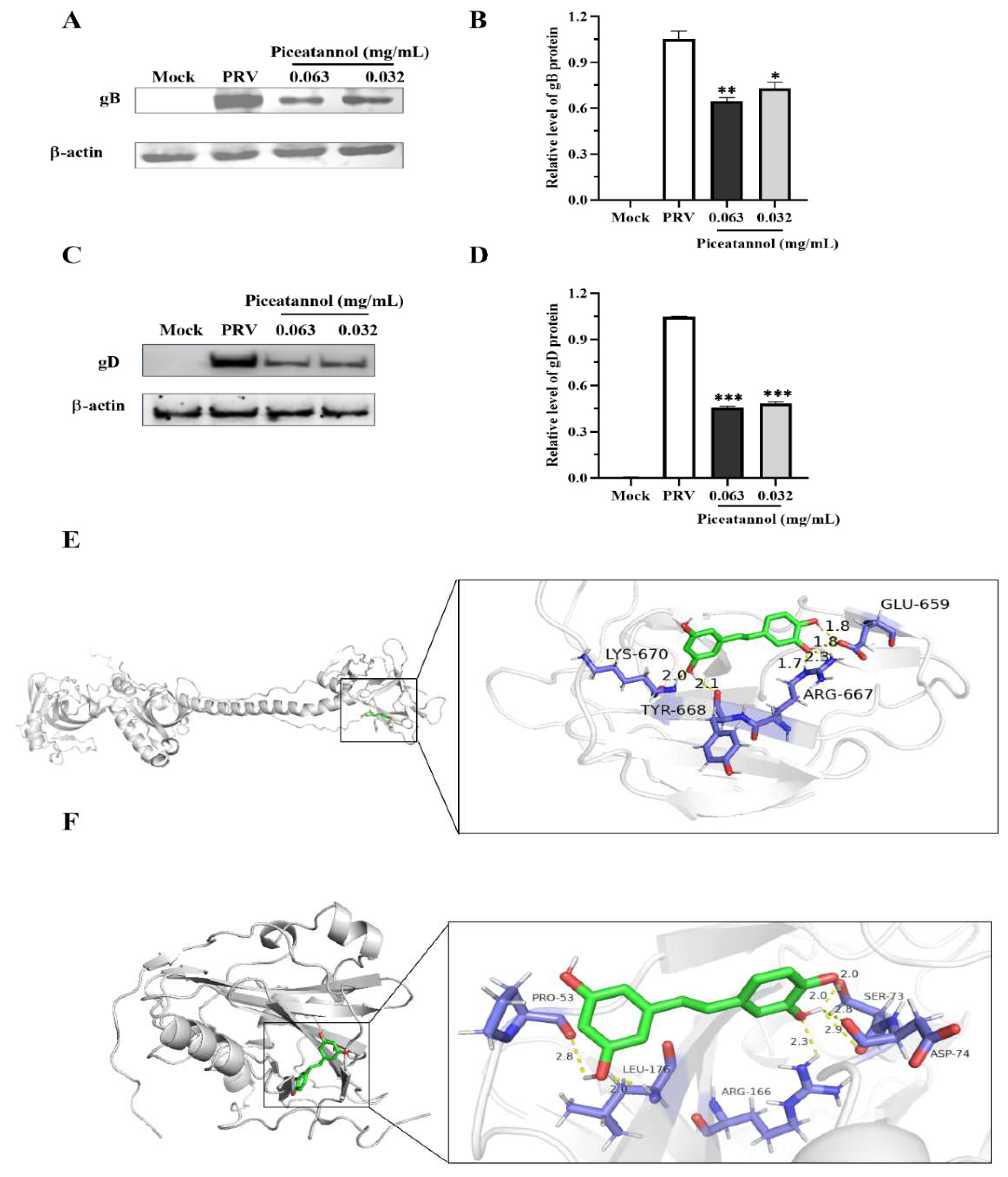
3.5. Piceatannol inhibited the expression of the PRV gB and gD proteins
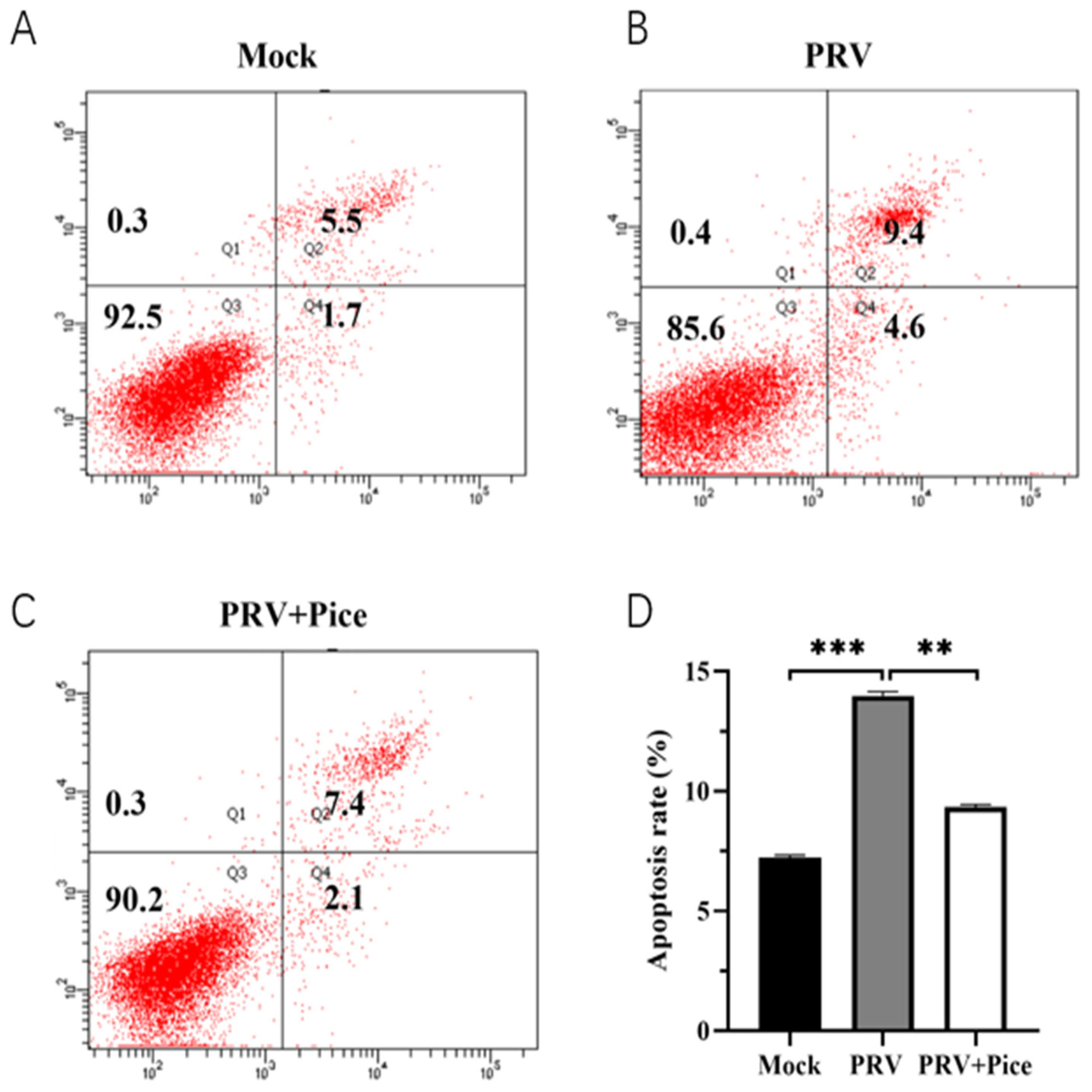
3.6. Piceatannol alleviates PRV-induced apoptosis
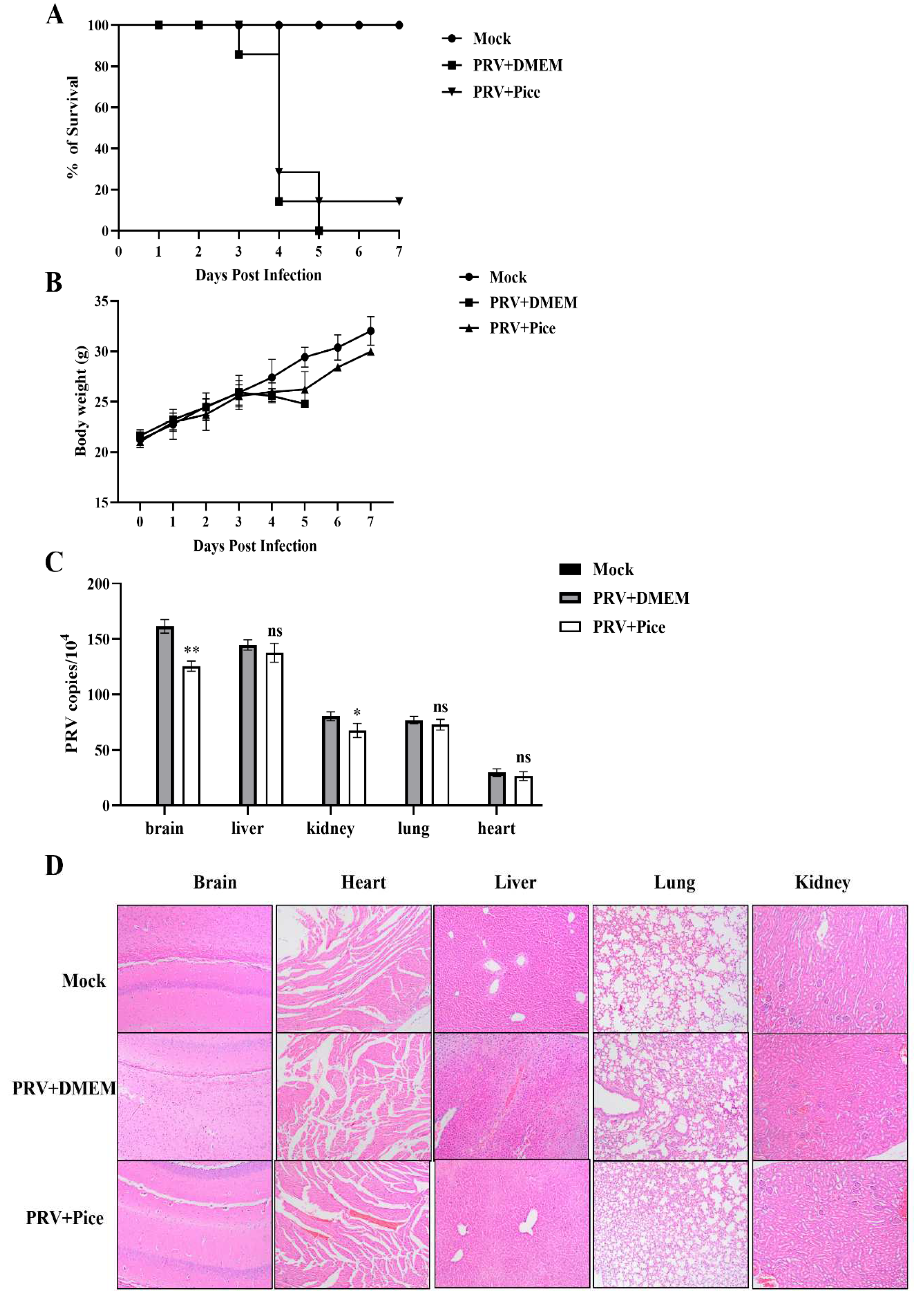
3.7. Piceatannol inhibits PRV infection in mice
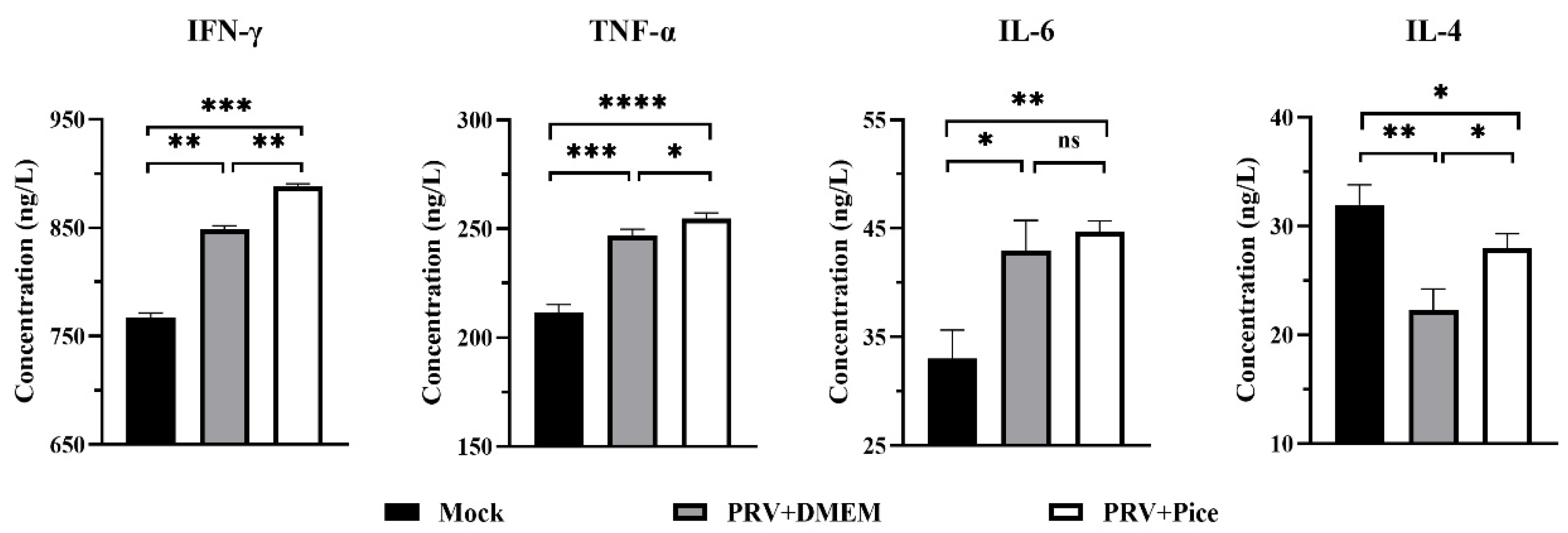
3.8. Changes in cytokine levels in the serum of PRV-infected mice treated with piceatannol
4. Discussion
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Ethics Statement
Sample Availability
References
- Masot:, A.J.; Gil, M.; Risco, D.; Jimenez, O.M.; Nunez, J.I.; Redondo, E. Pseudorabies virus infection (Aujeszky's disease) in an Iberian lynx (Lynx pardinus) in Spain: a case report. Bmc Vet. Res. 2017, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Okada, N.; Ohgitani, T.; Kokubu, T.; Shimizu, Y. Influence of cell surface glycoprotein gC produced by pseudorabies virus on cytopathic effect. J. Vet. Med. Sci. 1998, 60, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clinical Infectious Diseases : An Official Publication of the Infectious Diseases Society of America. 2020, 73.
- Wang, D.; Tao, X.; Fei, M.; Chen, J.; Guo, W.; Li, P.; Wang, J. Human encephalitis caused by pseudorabies virus infection: a case report. J. Neurovirol. 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Ghidoli, M.; Colombo, F.; Sangiorgio, S.; Landoni, M.; Giupponi, L.; Nielsen, E.; Pilu, R. Food Containing Bioactive Flavonoids and Other Phenolic or Sulfur Phytochemicals With Antiviral Effect: Can We Design a Promising Diet Against COVID-19? Front. Nutr. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Casado, A. The Health Potential of Fruits and Vegetables Phytochemicals: Notable Examples. Crit. Rev. Food Sci. Nutr. 2016, 56. [Google Scholar] [CrossRef]
- Maria, R.; Stefania, M.; Carmela, S.; Idolo, T.; Gian, L.R. Roles of flavonoids against coronavirus infection. Chem.-Biol. Interact. 2020, 328.
- Julia, S.; Johannes-Paul, F. Flavonoids: A complementary approach to conventional therapy of COVID-19? Phytochemistry Reviews : Proceedings of the Phytochemical Society of Europe. 2020.
- Mohammadi, P.P.; Fakhri, S.; Asgary, S.; Farzaei, M.H.; Echeverría, J. The Signaling Pathways, and Therapeutic Targets of Antiviral Agents: Focusing on the Antiviral Approaches and Clinical Perspectives of Anthocyanins in the Management of Viral Diseases. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Harazem, R.; Rahman, S.; Kenawy, A. Evaluation of Antiviral Activity of Allium Cepa and Allium Sativum Extracts Against Newcastle Disease Virus. Alexandria Journal of Veterinary Sciences. 2019, 61. [Google Scholar] [CrossRef]
- Alex, H.; Andrea, T.; Berlin, L.; Tonya, M.C.; Curt, H. Garlic Organosulfur Compounds Reduce Inflammation and Oxidative Stress during Dengue Virus Infection. Viruses-Basel. 2017, 9.
- Josling, P. Preventing the common cold with a garlic supplement: a double-blind, placebo-controlled survey. Adv. Ther. 2001, 18. [Google Scholar] [CrossRef]
- Bayan, L.; Koulivand, P.H.; Gorji, A. Garlic: a review of potential therapeutic effects. Avicenna J. Phytomedicine. 2014, 4. [Google Scholar]
- Ueda, K.; Kawabata, R.; Irie, T.; Nakai, Y.; Tohya, Y.; Sakaguchi, T. Inactivation of pathogenic viruses by plant-derived tannins: strong effects of extracts from persimmon (Diospyros kaki) on a broad range of viruses. Plos One. 2013, 8. [Google Scholar] [CrossRef]
- J., L.S.G.; R., C.L.; D., B.O.; S., S.C.; M., M.F.S.M.; M., T.V.R.; N., S.O.S.; M., D.W. In vitro anti-rotavirus activity of some medicinal plants used in Brazil against diarrhea. J. Ethnopharmacol. 2005, 99.
- Zhi-Yong, J.; Wen-Feng, L.; Xue-Mei, Z.; Jie, L.; Yun-Bao, M.; Ji-Jun, C. Anti-HBV active constituents from Piper longum. Bioorg. Med. Chem. Lett. 2013, 23. [Google Scholar]
- Nongluk, S.; Syuichi, F.; Kenji, K.; Hiroaki, H.; Takato, O.; Masato, T.; Yasuo, S. Antiviral effects of Psidium guajava Linn. (guava) tea on the growth of clinical isolated H1N1 viruses: Its role in viral hemagglutination and neuraminidase inhibition. Antiviral Res. 2012, 94.
- Sumra, N.; Shabbir, H.; Naureen, N.; Muhammad, P.; Madiha, R. The phytochemistry and medicinal value of Psidium guajava (guava). Clinical Phytoscience. 2018, 4. [Google Scholar]
- Prochazkova, D.; Bousova, I.; Wilhelmova, N. Antioxidant and prooxidant properties of flavonoids. Fitoterapia. 2011, 82, 513–523. [Google Scholar] [CrossRef]
- Matsui, Y.; Sugiyama, K.; Kamei, M.; Takahashi, T.; Suzuki, T.; Katagata, Y.; Ito, T. Extract of passion fruit (Passiflora edulis) seed containing high amounts of piceatannol inhibits melanogenesis and promotes collagen synthesis. J. Agric. Food Chem. 2010, 58. [Google Scholar] [CrossRef] [PubMed]
- Hanna, P.; Malgorzata, K.; Marek, M. Biological activity of piceatannol: Leaving the shadow of resveratrol. Mutat. Res.-Rev. Mutat. Res. 2011, 750.
- Efstathios, P.V.; Athanassios, K.; Ioannis, G.; Dimitrios, S.; Marilena, E.L. Screening of mushrooms bioactivity: piceatannol was identified as a bioactive ingredient in the order Cantharellales. Eur. Food Res. Technol. 2018, 244. [Google Scholar]
- Shah, U.; Shah, R.; Acharya, S.; Acharya, N. Novel anticancer agents from plant sources:Novel anticancer agents from plant sources. Chin. J. Nat. Med. 2014, 11. [Google Scholar]
- Rimando, A.M.; Kalt, W.; Magee, J.B.; Dewey, J.; Ballington, J.R. Resveratrol, pterostilbene, and piceatannol in vaccinium berries. J. Agric. Food Chem. 2004, 52, 4713–4719. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Smith, W.; Yan, L.; Kong, L. Overview of Cellular Mechanisms and Signaling Pathways of Piceatannol. Curr. Stem Cell Res. Ther. 2020, 15. [Google Scholar]
- Wang, S.Y.; Zhang, J.; Xu, X.G.; Su, H.L.; Xing, W.M.; Zhang, Z.S.; Jin, W.H.; Dai, J.H.; Wang, Y.Z.; He, X.Y.; et al. Inhibitory effects of piceatannol on human cytomegalovirus (hCMV) in vitro. J. Microbiol. 2020, 58, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Pflieger, A.; Waffo, T.P.; Papastamoulis, Y.; Chaignepain, S.; Subra, F.; Munir, S.; Delelis, O.; Lesbats, P.; Calmels, C.; Andreola, M.L.; et al. Natural stilbenoids isolated from grapevine exhibiting inhibitory effects against HIV-1 integrase and eukaryote MOS1 transposase in vitro activities. Plos One. 2013, 8, e81184. [Google Scholar] [CrossRef]
- Li, A.; Lu, G.; Qi, J.; Wu, L.; Tian, K.; Luo, T.; Shi, Y.; Yan, J.; Gao, G.F. Structural basis of nectin-1 recognition by pseudorabies virus glycoprotein D. Plos Pathog. 2017, 13, e1006314. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.H.; Fu, P.F.; Chen, H.Y.; Wang, Z.Y. Pseudorabies Virus: From Pathogenesis to Prevention Strategies. Viruses-Basel. 2022, 14.
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Dey, R.; Samadder, A.; Nandi, S. Exploring the Targets of Novel Corona Virus and Docking-based Screening of Potential Natural Inhibitors to Combat COVID-19. Curr. Top. Med. Chem. 2022, 22, 2410–2434. [Google Scholar] [CrossRef]
- Chen, Z.; Ye, S. Research progress on antiviral constituents in traditional Chinese medicines and their mechanisms of action. Pharm. Biol. 2022, 60. [Google Scholar] [CrossRef]
- Rossi, M.; Caruso, F.; Opazo, C.; Salciccioli, J. Crystal and molecular structure of piceatannol; scavenging features of resveratrol and piceatannol on hydroxyl and peroxyl radicals and docking with transthyretin. J. Agric. Food Chem. 2008, 56, 10557–10566. [Google Scholar] [CrossRef]
- Zakaria, M.Y.; Abd, E.S.M.; Beshay, B.Y.; Zaki, I.; Abourehab, M.A.S. 'Poly phenolic phytoceutical loaded nano-bilosomes for enhanced caco-2 cell permeability and SARS-CoV 2 antiviral activity': in-vitro and insilico studies. Drug Deliv. 2023, 30. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Xu, W.; Guo, T.; Pan, H.; Zou, H.; Jiang, L.; Li, C.; Gao, S. (-)-Epigallocatechin-3-Gallate Inhibits the Life Cycle of Pseudorabies Virus In Vitro and Protects Mice Against Fatal Infection. Front. Cell. Infect. Microbiol. 2020, 10, 616895. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.W.; Fu, P.F.; Zeng, L.; Qi, Y.L.; Li, X.Q.; Wang, Q.; Yang, G.Y.; Li, H.W.; Wang, J.; Chu, B.B.; et al. EGCG Restricts PRRSV Proliferation by Disturbing Lipid Metabolism. Microbiol. Spectr. 2022, 10, e227621. [Google Scholar] [CrossRef]
- Mou, Q.; Jiang, Y.; Zhu, L.; Zhu, Z.; Ren, T. EGCG induces beta-defensin 3 against influenza A virus H1N1 by the MAPK signaling pathway. Exp. Ther. Med. 2020, 20, 3017–3024. [Google Scholar]
- Stamos, J.D.; Lee, L.H.; Taylor, C.; Elias, T.; Adams, S.D. In Vitro and In Silico Analysis of the Inhibitory Activity of EGCG-Stearate against Herpes Simplex Virus-2. Microorganisms. 2022, 10. [Google Scholar] [CrossRef]
- Carneiro, B.M.; Batista, M.N.; Braga, A.; Nogueira, M.L.; Rahal, P. The green tea molecule EGCG inhibits Zika virus entry. Virology. 2016, 496, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Luo, G.; Zhang, C.; Feng, L.; Luo, X.; Gan, L. Curcumin protects rat hippocampal neurons against pseudorabies virus by regulating the BDNF/TrkB pathway. Sci Rep. 2020, 10, 22204. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hu, J.H.; Liang, X.D.; Chen, J.; Liu, C.C.; Liu, Y.Y.; Cheng, Y.; Go, Y.Y.; Zhou, B. Curcumin inhibits classical swine fever virus replication by interfering with lipid metabolism. Vet. Microbiol. 2021, 259, 109152. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Liu, Y.; Luo, X.; Lei, W.; Xie, L. Antiviral and virucidal effects of curcumin on transmissible gastroenteritis virus in vitro. The Journal of General Virology. 2020, 101. [Google Scholar] [CrossRef]
- Wei, Z.; Zhang, Y.; Ke, C.; Chen, H.; Ren, P.; He, Y.; Hu, P.; Ma, D.; Luo, J.; Meng, Z. Curcumin inhibits hepatitis B virus infection by down-regulating cccDNA-bound histone acetylation. World J. Gastroenterol. 2017, 23. [Google Scholar] [CrossRef]
- Padilla-S, L.; Rodriguez, A.; Gonzales, M.M.; Gallego-G, J.C.; Castano-O, J.C. Inhibitory effects of curcumin on dengue virus type 2-infected cells in vitro. Arch. Virol. 2014, 159, 573–579. [Google Scholar] [CrossRef]
- Zhao, X.; Tong, W.; Song, X.; Jia, R.; Li, L.; Zou, Y.; He, C.; Liang, X.; Lv, C.; Jing, B.; et al. Antiviral Effect of Resveratrol in Piglets Infected with Virulent Pseudorabies Virus. Viruses-Basel. 2018, 10.
- Zhao, X.; Xu, J.; Song, X.; Jia, R.; Yin, Z.; Cheng, A.; Jia, R.; Zou, Y.; Li, L.; Yin, L.; et al. Antiviral effect of resveratrol in ducklings infected with virulent duck enteritis virus. Antiviral Res. 2016, 130, 93–100. [Google Scholar] [CrossRef]
- Krishnan, V.; Zeichner, S.L. Host cell gene expression during human immunodeficiency virus type 1 latency and reactivation and effects of targeting genes that are differentially expressed in viral latency. J. Virol. 2004, 78, 9458–9473. [Google Scholar] [CrossRef]
- Zhao, X.; Cui, Q.; Fu, Q.; Song, X.; Jia, R.; Yang, Y.; Zou, Y.; Li, L.; He, C.; Liang, X.; et al. Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci Rep. 2017, 7. [Google Scholar] [CrossRef]
- Jiao, X.; Zhongqiong, Y.; Li, L.; Anchun, C.; Renyong, J.; Xu, S.; Hongke, L.; Shujun, D.; Cheng, L.; Xiaoxia, L.; et al. Inhibitory effect of resveratrol against duck enteritis virus in vitro. Plos One. 2017, 8. [Google Scholar]
- Huan, C.; Zhou, Z.; Yao, J.; Ni, B.; Gao, S. The Antiviral Effect of Panax Notoginseng Polysaccharides by Inhibiting PRV Adsorption and Replication In Vitro. Molecules. 2022, 27. [Google Scholar] [CrossRef]
- Johnson, D.C.; Baines, J.D. Herpesviruses remodel host membranes for virus egress. Nature Reviews. Microbiology. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiology and Molecular Biology Reviews : Mmbr. 2005, 69.
- Böhm, S.W.; Backovic, M.; Klupp, B.G.; Rey, F.A.; Mettenleiter, T.C.; Fuchs, W. Functional Characterization of Glycoprotein H Chimeras Composed of Conserved Domains of the Pseudorabies Virus and Herpes Simplex Virus 1 Homologs. J. Virol. 2016, 90. [Google Scholar] [CrossRef] [PubMed]
- Men, X.; Li, S.; Cai, X.; Fu, L.; Shao, Y.; Zhu, Y. Antiviral Activity of Luteolin against Pseudorabies Virus In Vitro and In Vivo. Animals. 2023, 13. [Google Scholar] [CrossRef]
- Ye, G.; Liu, H.; Zhou, Q.; Liu, X.; Huang, L.; Weng, C. A Tug of War: Pseudorabies Virus and Host Antiviral Innate Immunity. Viruses-Basel. 2022, 14.
- Liu, L. Fields Virology, 6th Edition. Clin. Infect. Dis. 2014, 59. [Google Scholar] [CrossRef]
- Teodoro, J.G.; Branton, P.E. Regulation of apoptosis by viral gene products. J. Virol. 1997, 71, 1739–1746. [Google Scholar] [CrossRef]
- Yang, B.; Luo, G.; Zhang, C.; Feng, L.; Luo, X.; Gan, L. Curcumin protects rat hippocampal neurons against pseudorabies virus by regulating the BDNF/TrkB pathway. Sci Rep. 2020, 10. [Google Scholar] [CrossRef]
- Hu, H.; Hu, Z.; Zhang, Y.; Wan, H.; Yin, Z.; Li, L.; Liang, X.; Zhao, X.; Yin, L.; Ye, G.; et al. Myricetin inhibits pseudorabies virus infection through direct inactivation and activating host antiviral defense13. Front. Microbiol. 2022, 13. [Google Scholar]
- Maresch, C.; Lange, E.; Teifke, J.P.; Fuchs, W.; Klupp, B.; Muller, T.; Mettenleiter, T.C.; Vahlenkamp, T.W. Oral immunization of wild boar and domestic pigs with attenuated live vaccine protects against Pseudorabies virus infection. Vet. Microbiol. 2012, 161, 20–25. [Google Scholar] [CrossRef]
- Cai, X.; Shao, Y.; Wang, Z.; Xu, Y.; Ren, Z.; Fu, L.; Zhu, Y. Antiviral activity of dandelion aqueous extract against pseudorabies virus both in vitro and in vivo. Front. Vet. Sci. 2022, 9, 1090398. [Google Scholar] [CrossRef]
- Brittle, E.E.; Reynolds, A.E.; Enquist, L.W. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J. Virol. 2004, 78, 12951–12963. [Google Scholar] [CrossRef]
- Zhang, S.; Ye, J.; Wang, L.; Zhong, X.; Zou, X.; Qiu, F.; Huang, Z. Piceatannol protects rat neuron cells from oxygen-glucose deprivation reperfusion injury via regulation of GSK-3beta/Nrf2 signaling pathway. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022, 51, 552–562. [Google Scholar] [PubMed]
- Jingyun, W.; Yanmei, M.; Long, W.; Xiaojuan, C.; Ruoxiang, Y.; Song, W.; Xinxin, L.; Xiaoyong, C.; Wenhan, S.; Ji-Long, C. Alpha/beta interferon receptor deficiency in mice significantly enhances susceptibility of the animals to pseudorabies virus infection. Vet. Microbiol. 2017, 203. [Google Scholar]
- Wang, F.; Zhang, S.; Jeon, R.; Vuckovic, I.; Jiang, X.; Lerman, A.; Folmes, C.D.; Dzeja, P.D.; Herrmann, J. Interferon Gamma Induces Reversible Metabolic Reprogramming of M1 Macrophages to Sustain Cell Viability and Pro-Inflammatory Activity. Ebiomedicine. 2018, 30, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Wolcott, R.M.; Chervenak, R.; Jennings, S.R. Control of acute cutaneous herpes simplex virus infection: T cell-mediated viral clearance is dependent upon interferon-gamma (IFN-gamma). Virology. 1994, 202, 76–88. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspect. Biol. 2014, 6, a16295. [Google Scholar] [CrossRef]
| Gene Name | Sequence (5’-3’) |
|---|---|
| UL44-F | CGTCAGGAATCGCATCA |
| UL44-R | CGCGTCACGTTCACCAC |
| IE180-F | CGCTCCACCAACAACC |
| IE180-R | TCGTCCTCGTCCCAGA |
| UL29-F | AGAAGCCGCACGCCATCACC |
| UL29-R | GGGAACCCGCAGACGGACAA |
| EP0-F | GGGCGTGGGTGTTT |
| EP0-R | GCTTTATGGGCAGGT |
| US6-F | AACATCCTCACCGACTTCA |
| US6-R | CGTCAGGAATCGCATCA |
| UL27-F | TCGTCCACGTCGTCCTCTTCG |
| UL27-R | CGGCATCGCCAACTTCTTCC |
| β-actin-F | TGCGGGACATCAAGGAGAA |
| β-actin-R | AGGAAGGAGGGCTGGAAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





