Submitted:
28 June 2023
Posted:
29 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction:
2. The interplay between the genome and Epigenome in KSHV infections:
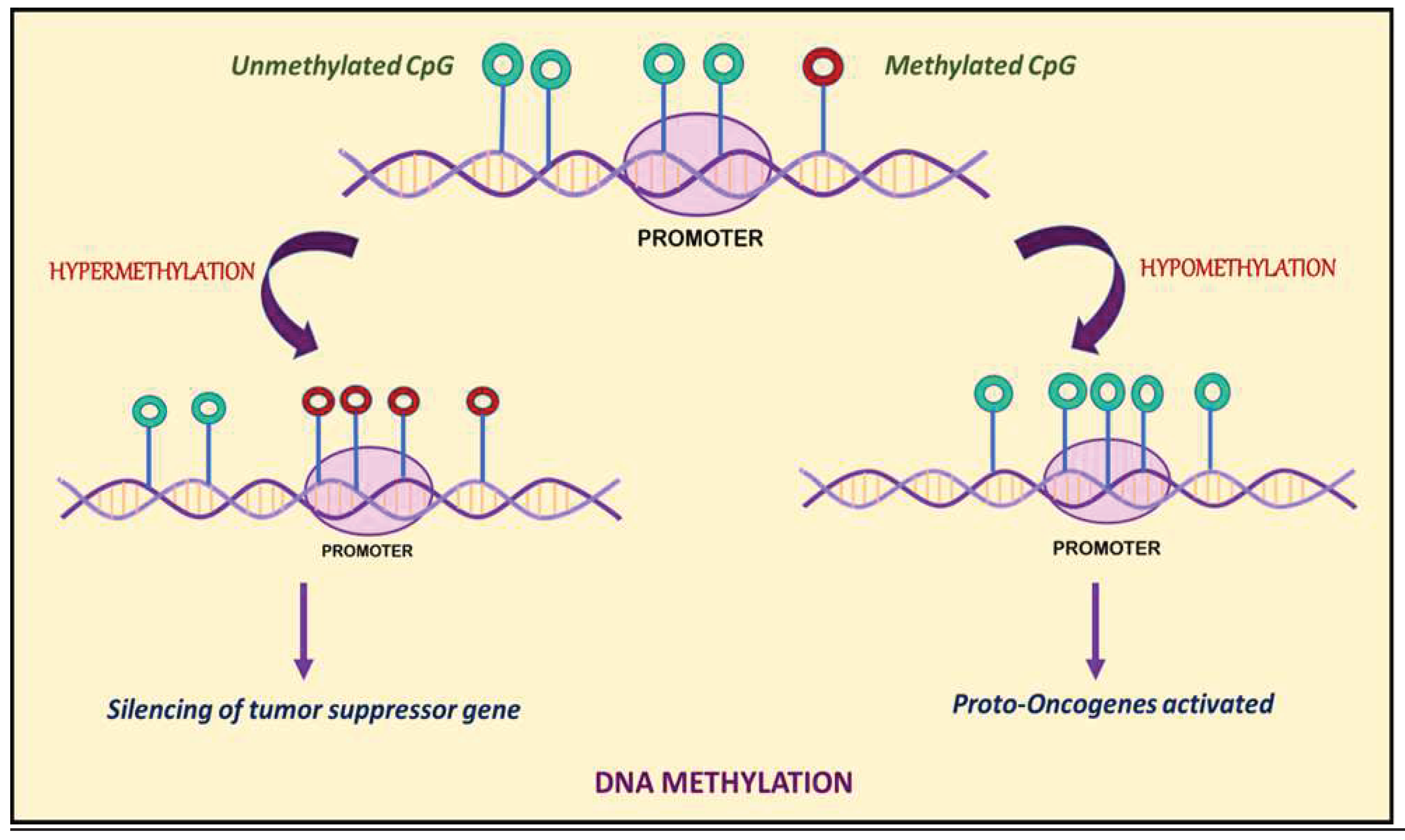
3. DNA Methylation:
4. DNA Methylation in KSHV infection:
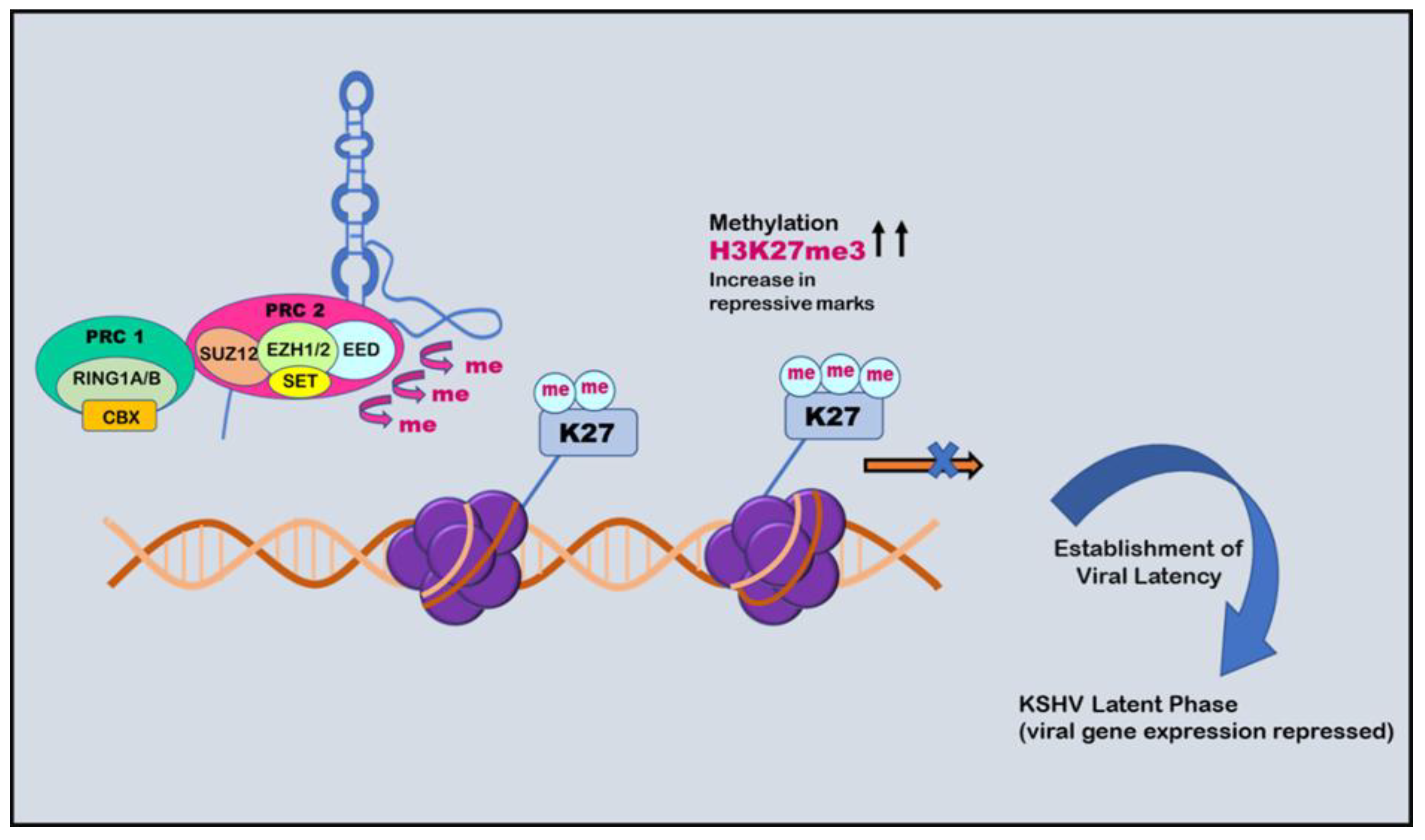
5. Histone Modification
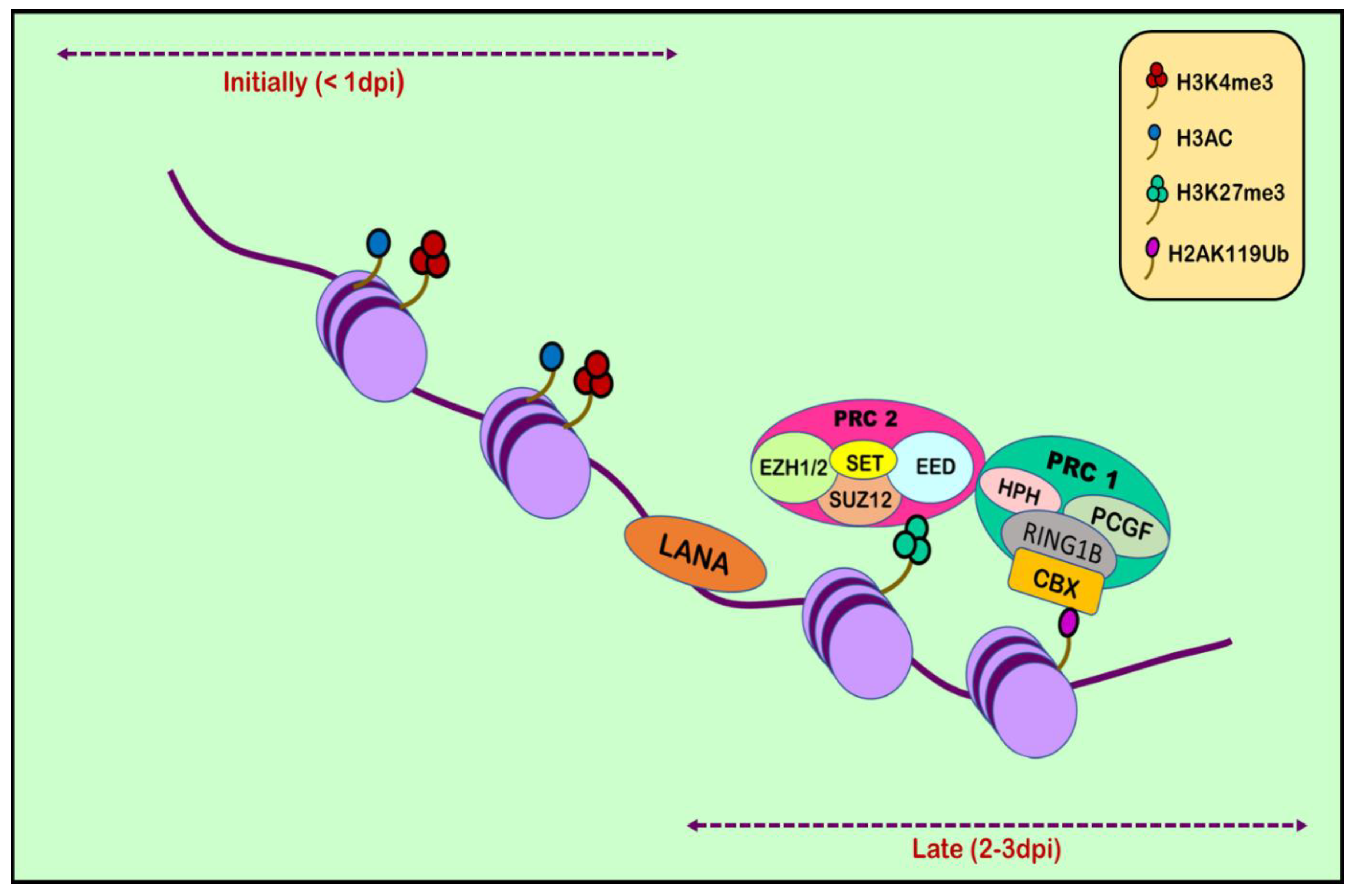
6. Histone Modification in KSHV infection:
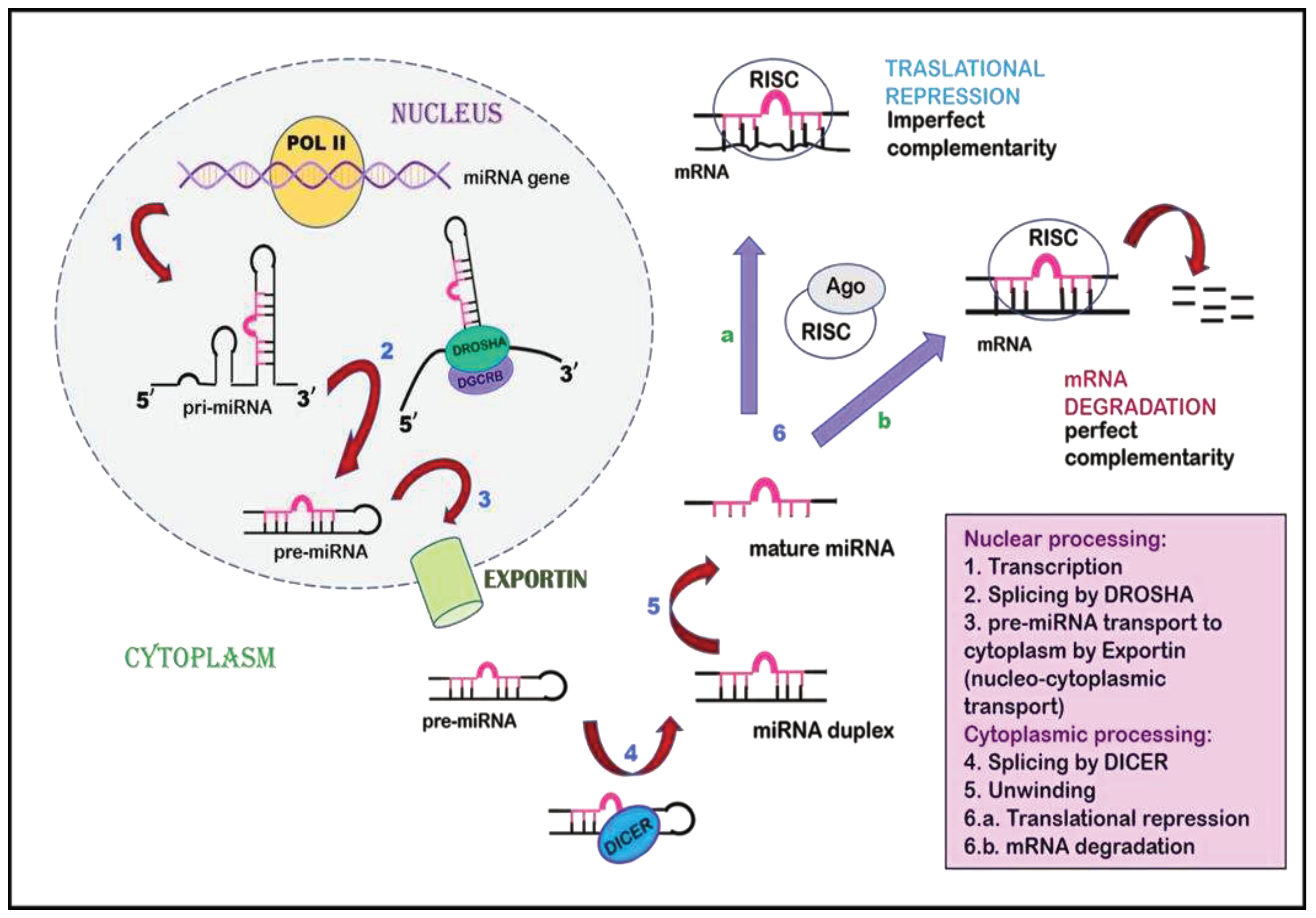
7. Non-coding RNAs
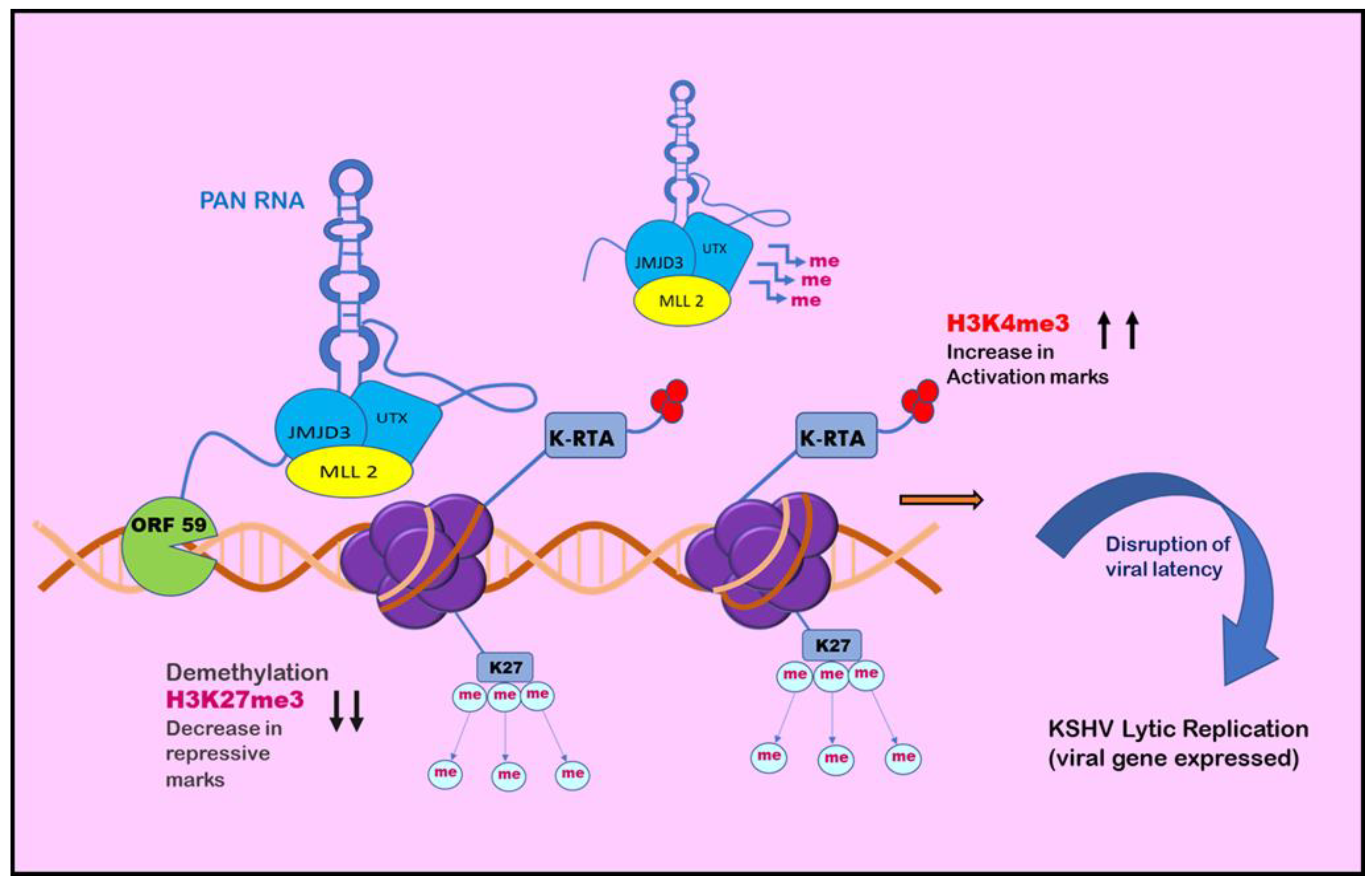
8. Role of Non-Coding RNAs in KSHV biology
Funding
Conflicts of Interest
References
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M.; Whitby, D. Kaposi sarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Cesarman, E.; Boshoff, C. Kaposi's sarcoma and its associated herpesvirus. Nat. Rev. Cancer 2010, 10, 707–719. [Google Scholar] [CrossRef]
- Polizzotto, M.N.; Uldrick, T.S.; Wyvill, K.M.; Aleman, K.; Marshall, V.; Wang, V.; Whitby, D.; Pittaluga, S.; Jaffe, E.S.; Millo, C.; et al. Clinical Features and Outcomes of Patients With Symptomatic Kaposi Sarcoma Herpesvirus (KSHV)-associated Inflammation: Prospective Characterization of KSHV Inflammatory Cytokine Syndrome (KICS). Clin. Infect. Dis. 2015, 62, 730–738. [Google Scholar] [CrossRef]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features. PLOS Pathog. 2014, 10, e1003847. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.d.O.; Marinho, P.D.N.; Medeiros, L.D.d.S.; de Paula, V.S. Human Gammaherpesvirus 8 Oncogenes Associated with Kaposi’s Sarcoma. Int. J. Mol. Sci. 2022, 23, 7203. [Google Scholar] [CrossRef] [PubMed]
- Lagunoff, M.; Ganem, D. The Structure and Coding Organization of the Genomic Termini of Kaposi's Sarcoma-Associated Herpesvirus (Human Herpesvirus 8). Virology 1997, 236, 147–154. [Google Scholar] [CrossRef]
- Wen, K.W.; Damania, B. Kaposi sarcoma-associated herpesvirus (KSHV): Molecular biology and oncogenesis. Cancer Lett. 2010, 289, 140–150. [Google Scholar] [CrossRef]
- Juillard, F.; Tan, M.; Li, S.; Kaye, K.M. Kaposi’s Sarcoma Herpesvirus Genome Persistence. Front. Microbiol. 2016, 7, 1149. [Google Scholar] [CrossRef]
- Kumar, A.; Lyu, Y.; Yanagihashi, Y.; Chantarasrivong, C.; Majerciak, V.; Salemi, M.; Wang, K.-H.; Inagaki, T.; Chuang, F.; Davis, R.R.; et al. KSHV episome tethering sites on host chromosomes and regulation of latency-lytic switch by CHD4. Cell Rep. 2022, 39, 110788–110788. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Yuan, Y. Reactivation and Lytic Replication of Kaposi’s Sarcoma-Associated Herpesvirus: An Update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef]
- Broussard, G.; Damania, B. Regulation of KSHV Latency and Lytic Reactivation. Viruses 2020, 12, 1034. [Google Scholar] [CrossRef]
- Lucas, M.; Karrer, U.; Lucas, A.; Klenerman, P. Viral escape mechanisms - escapology taught by viruses. Int. J. Exp. Pathol. 2008, 82, 269–286. [Google Scholar] [CrossRef]
- Lee, H.-R.; Lee, S.; Chaudhary, P.M.; Gill, P.; Jung, J.U.; Hewitt, P.; Walijew, A.; Sakakibara, S.; Tosato, G.; E Ballestas, M.; et al. Immune evasion by Kaposi’s sarcoma-associated herpesvirus. Futur. Microbiol. 2010, 5, 1349–1365. [Google Scholar] [CrossRef]
- Campbell, M.; Yang, W.-S.; Yeh, W.W.; Kao, C.-H.; Chang, P.-C. Epigenetic Regulation of Kaposi’s Sarcoma-Associated Herpesvirus Latency. Front. Microbiol. 2020, 11, 850. [Google Scholar] [CrossRef]
- Sakakibara, S.; Ueda, K.; Nishimura, K.; Do, E.; Ohsaki, E.; Okuno, T.; Yamanishi, K. Accumulation of Heterochromatin Components on the Terminal Repeat Sequence of Kaposi's Sarcoma-Associated Herpesvirus Mediated by the Latency-Associated Nuclear Antigen. J. Virol. 2004, 78, 7299–7310. [Google Scholar] [CrossRef]
- Toth, Z.; Brulois, K.; Lee, H.-R.; Izumiya, Y.; Tepper, C.; Kung, H.-J.; Jung, J.U. Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following De Novo Infection. PLOS Pathog. 2013, 9, e1003813. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Uppal, T.; Verma, S.C. Molecular Biology of KSHV Lytic Reactivation. Viruses 2015, 7, 116–153. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Bose, D.; Robertson, E.S. Epigenetic Reprogramming of Kaposi’s Sarcoma-Associated Herpesvirus during Hypoxic Reactivation. Cancers 2022, 14, 5396. [Google Scholar] [CrossRef]
- Traylen, C.M.; Patel, H.R.; Fondaw, W.; Mahatme, S.; Williams, J.F.; Walker, L.R.; Dyson, O.F.; Arce, S.; Akula, S.M. Virus reactivation: a panoramic view in human infections. Futur. Virol. 2011, 6, 451–463. [Google Scholar] [CrossRef]
- Watanabe, T.; Sugaya, M.; Atkins, A.M.; Aquilino, E.A.; Yang, A.; Borris, D.L.; Brady, J.; Blauvelt, A. Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Prolongs the Life Span of Primary Human Umbilical Vein Endothelial Cells. J. Virol. 2003, 77, 6188–6196. [Google Scholar] [CrossRef] [PubMed]
- Uppal, T.; Banerjee, S.; Sun, Z.; Verma, S.C.; Robertson, E.S. KSHV LANA—The Master Regulator of KSHV Latency. Viruses 2014, 6, 4961–4998. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.C.; Lan, K.; Robertson, E. Structure and Function of Latency-Associated Nuclear Antigen. Poxviruses 2007, 312, 101–136. [Google Scholar] [CrossRef]
- Jones, T.; da Silva, S.R.; Bedolla, R.; Ye, F.; Zhou, F.; Gao, S. Viral Cyclin promotes KSHV-induced cellular transformation and tumorigenesis by overriding contact inhibition. Cell Cycle 2014, 13, 845–858. [Google Scholar] [CrossRef]
- Ballon, G.; Chen, K.; Perez, R.; Tam, W.; Cesarman, E. Kaposi sarcoma herpesvirus (KSHV) vFLIP oncoprotein induces B cell transdifferentiation and tumorigenesis in mice. J. Clin. Investig. 2011, 121, 1141–1153. [Google Scholar] [CrossRef]
- Singh, R.K.; Pei, Y.; Bose, D.; Lamplugh, Z.L.; Sun, K.; Yuan, Y.; Lieberman, P.; You, J.; Robertson, E.S. KSHV-encoded vCyclin can modulate HIF1α levels to promote DNA replication in hypoxia. eLife 2021, 10. [Google Scholar] [CrossRef]
- Sun, S.C.; Cesarman, E. NF-kappaB as a target for oncogenic viruses. Curr Top Microbiol Immunol. 2011, 349, 197–244. [Google Scholar] [CrossRef]
- Ganem, D. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J. Clin. Investig. 2010, 120, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.V.; D´agostino, A.; Ma, Q.; Eroles, P.; Cavallin, L.; Chiozzini, C.; Sapochnik, D.; Cymeryng, C.; Hyjek, E.; Cesarman, E.; et al. KSHV G-protein coupled receptor vGPCR oncogenic signaling upregulation of Cyclooxygenase-2 expression mediates angiogenesis and tumorigenesis in Kaposi’s sarcoma. PLOS Pathog. 2020, 16, e1009006. [Google Scholar] [CrossRef] [PubMed]
- Sodhi A, Montaner S, Patel V, Zohar M, Bais C, Mesri EA, et al. The Kaposi's sarcoma-associated herpes virus G protein-coupled receptor up-regulates vascular endothelial growth factor expression and secretion through mitogen-activated protein kinase and p38 pathways acting on hypoxia-inducible factor 1alpha. Cancer Res. 2000;60(17):4873-80. [PubMed]
- Qin, J.; Li, W.; Gao, S.-J.; Lu, C. KSHV microRNAs: Tricks of the Devil. Trends Microbiol. 2017, 25, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Grundhoff, A.; Sullivan, C.S. Virus-encoded microRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef]
- Uppal, T.; Jha, H.C.; Verma, S.C.; Robertson, E.S. Chromatinization of the KSHV Genome During the KSHV Life Cycle. Cancers 2015, 7, 112–142. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Brulois, K.; Jung, J.U. The Chromatin Landscape of Kaposi’s Sarcoma-Associated Herpesvirus. Viruses 2013, 5, 1346–1373. [Google Scholar] [CrossRef] [PubMed]
- Guito, J.; Lukac, D.M. KSHV Reactivation and Novel Implications of Protein Isomerization on Lytic Switch Control. Viruses 2015, 7, 72–109. [Google Scholar] [CrossRef]
- Li, X.; Feng, J.; Sun, R. Oxidative Stress Induces Reactivation of Kaposi's Sarcoma-Associated Herpesvirus and Death of Primary Effusion Lymphoma Cells. J. Virol. 2011, 85, 715–724. [Google Scholar] [CrossRef]
- Davis, D.A.; Rinderknecht, A.S.; Zoeteweij, J.P.; Aoki, Y.; Read-Connole, E.L.; Tosato, G.; Blauvelt, A.; Yarchoan, R. Hypoxia induces lytic replication of Kaposi sarcoma–associated herpesvirus. Blood 2001, 97, 3244–3250. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.; Montani, M.S.G.; Angiolillo, C.; D’orazi, G.; Faggioni, A.; Cirone, M. Cytotoxic Drugs Activate KSHV Lytic Cycle in Latently Infected PEL Cells by Inducing a Moderate ROS Increase Controlled by HSF1, NRF2 and p62/SQSTM1. Viruses 2018, 11, 8. [Google Scholar] [CrossRef]
- Diederichs, S.; Bartsch, L.; Berkmann, J.C.; Fröse, K.; Heitmann, J.; Hoppe, C.; Iggena, D.; Jazmati, D.; Karschnia, P.; Linsenmeier, M.; et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol. Med. 2016, 8, 442–457. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2011, 81, 303–311. [Google Scholar] [CrossRef]
- Chang, Y.; Moore, P.S.; Weiss, R.A.; Yuan, C.; S. , M.P.; A., W.R.; C, M.; S, A. Human oncogenic viruses: nature and discovery. Philos. Trans. R. Soc. B: Biol. Sci. 2017, 372, 20160264. [Google Scholar] [CrossRef] [PubMed]
- Mui, U.N.; Haley, C.T.; Tyring, S.K. Viral Oncology: Molecular Biology and Pathogenesis. J. Clin. Med. 2017, 6, 111. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.S.K.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Munger, K. Viruses associated with human cancer. Biochim. et Biophys. Acta (BBA) - Mol. Basis Dis. 2008, 1782, 127–150. [Google Scholar] [CrossRef]
- Tempera, I.; Lieberman, P.M. Oncogenic Viruses as Entropic Drivers of Cancer Evolution. Front. A J. Women Stud. 2021, 1, 753366. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. Host defense, viruses and apoptosis. Cell Death Differ. 2001, 8, 113–126. [Google Scholar] [CrossRef]
- Flanagan, J.M. Host epigenetic modifications by oncogenic viruses. Br. J. Cancer 2017, 96, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Lang, F.; Pei, Y.; Jha, H.C.; Robertson, E.S. Metabolic reprogramming of Kaposi’s sarcoma associated herpes virus infected B-cells in hypoxia. PLOS Pathog. 2018, 14, e1007062. [Google Scholar] [CrossRef]
- Kim, J.K.; Samaranayake, M.; Pradhan, S. Epigenetic mechanisms in mammals. Cell. Mol. Life Sci. 2008, 66, 596–612. [Google Scholar] [CrossRef]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone Modifications and Non-Coding RNAs: Mutual Epigenetic Regulation and Role in Pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Mugal, C.F.; Arndt, P.F.; Holm, L.; Ellegren, H. Evolutionary Consequences of DNA Methylation on the GC Content in Vertebrate Genomes. G3 Genes|Genomes|Genetics 2015, 5, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gomez, P.; Bello, M.J.; E Alonso, M.; Arjona, D.; Lomas, J.; de Campos, J.M.; Isla, A.; A Rey, J. CpG island methylation status and mutation analysis of the RB1 gene essential promoter region and protein-binding pocket domain in nervous system tumours. Br. J. Cancer 2003, 88, 109–114. [Google Scholar] [CrossRef]
- Herman, J.G.; Latif, F.; Weng, Y.; I Lerman, M.; Zbar, B.; Liu, S.; Samid, D.; Duan, D.S.; Gnarra, J.R.; Linehan, W.M. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. 1994, 91, 9700–9704. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002, 21, 5427–5440. [Google Scholar] [CrossRef]
- Gama-Sosa, M.A.; Slagel, V.A.; Trewyn, R.W.; Oxenhandler, R.; Kuo, K.C.; Gehrke, C.W.; Ehrlich, M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983, 11, 6883–6894. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef]
- Ehrlich, M.; Fang, C.; Zou, C.; Fu, Y.; Li, J.; Li, Y.; Ma, Y.; Zhao, S.; Li, C.; A Hore, T.; et al. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Hoffmann, M.J.; A Schulz, W. Causes and consequences of DNA hypomethylation in human cancer. Biochem. Cell Biol. 2005, 83, 296–321. [Google Scholar] [CrossRef]
- Kurkjian, C.; Kummar, S.; Murgo, A. DNA Methylation: Its Role in Cancer Development and Therapy. Curr. Probl. Cancer 2008, 32, 187–235. [Google Scholar] [CrossRef]
- Gómez-Díaz, E.; Jordà, M.; Peinado, M.A.; Rivero, A. Epigenetics of Host–Pathogen Interactions: The Road Ahead and the Road Behind. PLOS Pathog. 2012, 8, e1003007. [Google Scholar] [CrossRef]
- Ueda, K.; Ito, E.; Karayama, M.; Ohsaki, E.; Nakano, K.; Watanabe, S. KSHV-infected PEL cell lines exhibit a distinct gene expression profile. Biochem. Biophys. Res. Commun. 2010, 394, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Journo, G.; Tushinsky, C.; Shterngas, A.; Avital, N.; Eran, Y.; Karpuj, M.V.; Frenkel-Morgenstern, M.; Shamay, M. Modulation of Cellular CpG DNA Methylation by Kaposi's Sarcoma-Associated Herpesvirus. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Westrich, J.A.; Pyeon, D. DNA Tumor Virus Regulation of Host DNA Methylation and Its Implications for Immune Evasion and Oncogenesis. Viruses 2018, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- E Ballestas, M.; Kaye, K.M.; Hewitt, P.; Walijew, A.; Sakakibara, S.; Tosato, G.; A Phillips, K.; Veenstra, D.; Van Bebber, S.; Sakowski, J.; et al. The latency-associated nuclear antigen, a multifunctional protein central to Kaposi’s sarcoma-associated herpesvirus latency. Futur. Microbiol. 2011, 6, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Lei, X.; Gao, S.-J. Mechanisms of Kaposi's Sarcoma-Associated Herpesvirus Latency and Reactivation. Adv. Virol. 2011, 2011, 193860. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Grundhoff, A. The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes. PLOS Pathog. 2010, 6, e1000935. [Google Scholar] [CrossRef]
- Darst, R.P.; Haecker, I.; Pardo, C.E.; Renne, R.; Kladde, M.P. Epigenetic diversity of Kaposi’s sarcoma–associated herpesvirus. Nucleic Acids Res. 2013, 41, 2993–3009. [Google Scholar] [CrossRef] [PubMed]
- Journo, G.; Ahuja, A.; Dias-Polak, D.; Eran, Y.; Bergman, R.; Shamay, M. Global CpG DNA Methylation Footprint in Kaposi’s Sarcoma. Front. Cell. Infect. Microbiol. 2021, 11, 666143. [Google Scholar] [CrossRef]
- Luger, K.; Dechassa, M.L.; Tremethick, D.J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012, 13, 436–447. [Google Scholar] [CrossRef]
- Mariño-Ramírez, L.; Kann, M.G.; A Shoemaker, B.; Landsman, D. Histone structure and nucleosome stability. Expert Rev. Proteom. 2005, 2, 719–729. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Tan, S. Nucleosome Structure and Function. Chem. Rev. 2014, 115, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Ramazi, S.; Allahverdi, A.; Zahiri, J. Evaluation of post-translational modifications in histone proteins: A review on histone modification defects in developmental and neurological disorders. J. Biosci. 2020, 45, 1–29. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Zhu, Q.; Wani, A.A. Histone modifications: crucial elements for damage response and chromatin restoration. J. Cell. Physiol. 2010, 223, 283–288. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Clayton, A.L.; Hazzalin, C.A.; Mahadevan, L.C. Enhanced Histone Acetylation and Transcription: A Dynamic Perspective. Mol. Cell 2006, 23, 289–296. [Google Scholar] [CrossRef]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- Petty, E.; Pillus, L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013, 29, 621–629. [Google Scholar] [CrossRef]
- Gujral, P.; Mahajan, V.; Lissaman, A.C.; Ponnampalam, A.P. Histone acetylation and the role of histone deacetylases in normal cyclic endometrium. Reprod. Biol. Endocrinol. 2020, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Berger, S.L. Acetylation of Histones and Transcription-Related Factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef]
- Xia, C.; Tao, Y.; Li, M.; Che, T.; Qu, J. Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review). Exp. Ther. Med. 2020, 20, 2923–2940. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.C.; Denu, J.M. Chemical mechanisms of histone lysine and arginine modifications. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2009, 1789, 45–57. [Google Scholar] [CrossRef]
- Nottke, A.; ColaiácovoM. P.; Shi, Y. Developmental roles of the histone lysine demethylases. Development 2009, 136, 879–889. [Google Scholar] [CrossRef]
- Liang, Y.; Chang, J.; Lynch, S.J.; Lukac, D.M.; Ganem, D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 2002, 16, 1977–1989. [Google Scholar] [CrossRef] [PubMed]
- Papp, B.; Motlagh, N.; Smindak, R.J.; Jang, S.J.; Sharma, A.; Alonso, J.D.; Toth, Z. Genome-Wide Identification of Direct RTA Targets Reveals Key Host Factors for Kaposi’s Sarcoma-Associated Herpesvirus Lytic Reactivation. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Günther, T.; Theiss, J.M.; Fischer, N.; Grundhoff, A. Investigation of Viral and Host Chromatin by ChIP-PCR or ChIP-Seq Analysis. Curr. Protoc. Microbiol. 2016, 40, 1E–10. [Google Scholar] [CrossRef]
- Toth, Z.; Maglinte, D.T.; Lee, S.H.; Lee, H.-R.; Wong, L.-Y.; Brulois, K.F.; Lee, S.; Buckley, J.D.; Laird, P.W.; Marquez, V.E.; et al. Epigenetic Analysis of KSHV Latent and Lytic Genomes. PLOS Pathog. 2010, 6, e1001013. [Google Scholar] [CrossRef]
- Dochnal, S.A.; Francois, A.K.; Cliffe, A.R. De Novo Polycomb Recruitment: Lessons from Latent Herpesviruses. Viruses 2021, 13, 1470. [Google Scholar] [CrossRef]
- Toth, Z.; Papp, B.; Brulois, K.; Choi, Y.J.; Gao, S.-J.; Jung, J.U. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLOS Pathog. 2016, 12, e1005878. [Google Scholar] [CrossRef]
- Toth, Z.; Smindak, R.J.; Papp, B. Inhibition of the lytic cycle of Kaposi's sarcoma-associated herpesvirus by cohesin factors following de novo infection. Virology 2017, 512, 25–33. [Google Scholar] [CrossRef]
- Naik, N.G.; Nguyen, T.H.; Roberts, L.; Fischer, L.T.; Glickman, K.; Golas, G.; Papp, B.; Toth, Z. Epigenetic factor siRNA screen during primary KSHV infection identifies novel host restriction factors for the lytic cycle of KSHV. PLOS Pathog. 2020, 16, e1008268. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Gregory, T.R. The Case for Junk DNA. PLOS Genet. 2014, 10, e1004351. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: what is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-Coding RNAs: Multi-Tasking Molecules in the Cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs. Veter- Pathol. 2013, 51, 759–774. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2013, 9, 3–12. [Google Scholar] [CrossRef]
- Gaffar, F.Y.; Imani, J.; Karlovsky, P.; Koch, A.; Kogel, K.-H. Different Components of the RNA Interference Machinery Are Required for Conidiation, Ascosporogenesis, Virulence, Deoxynivalenol Production, and Fungal Inhibition by Exogenous Double-Stranded RNA in the Head Blight Pathogen Fusarium graminearum. Front. Microbiol. 2019, 10, 1662. [Google Scholar] [CrossRef]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA Interference: Biology, Mechanism, and Applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef] [PubMed]
- Ku, H.-Y.; Lin, H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl. Sci. Rev. 2014, 1, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.-Y.; Chang, D.C.; Lin, S.-L. The MicroRNA (miRNA): Overview of the RNA Genes that Modulate Gene Function. Mol. Biotechnol. 2007, 38, 257–268. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Reuter, J.A.; Spacek, D.V.; Snyder, M.P. High-Throughput Sequencing Technologies. Mol. Cell 2015, 58, 586–597. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lu, S.; Zhang, Z.; Gonzalez, C.M.; Damania, B.; Cullen, B.R. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. 2005, 102, 5570–5575. [Google Scholar] [CrossRef] [PubMed]
- Grundhoff, A.; Sullivan, C.S.; Ganem, D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 2006, 12, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Cullen, B.R. In-Depth Analysis of Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Expression Provides Insights into the Mammalian MicroRNA-Processing Machinery. J. Virol. 2010, 84, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral MicroRNA Targetome of KSHV-Infected Primary Effusion Lymphoma Cell Lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef]
- Schifano, J.M.; Corcoran, K.; Kelkar, H.; Dittmer, D.P. Expression of the Antisense-to-Latency Transcript Long Noncoding RNA in Kaposi's Sarcoma-Associated Herpesvirus. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Kung, H.-J.; Izumiya, Y. Long Non-Coding RNA and Epigenetic Gene Regulation of KSHV. Viruses 2014, 6, 4165–4177. [Google Scholar] [CrossRef]
- Campbell, M.; Izumiya, Y. PAN RNA: transcriptional exhaust from a viral engine. J. Biomed. Sci. 2020, 27, 1–10. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Creugny, A.; Fender, A.; Pfeffer, S. Regulation of primary microRNA processing. FEBS Lett. 2018, 592, 1980–1996. [Google Scholar] [CrossRef]
- Johanson, T.M.; Lew, A.M.; Chong, M.M.W. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013, 3, 130144. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 2017, 15, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- O'Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Iwakawa, H.-O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2021, 82, 30–43. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G.S. PAN’s Labyrinth: Molecular Biology of Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) PAN RNA, a Multifunctional Long Noncoding RNA. Viruses 2014, 6, 4212–4226. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA Associates with Demethylases UTX and JMJD3 to Activate Lytic Replication through a Physical Interaction with the Virus Genome. PLOS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef]
- Hiura, K.; Strahan, R.; Uppal, T.; Prince, B.; Rossetto, C.C.; Verma, S.C. KSHV ORF59 and PAN RNA Recruit Histone Demethylases to the Viral Chromatin during Lytic Reactivation. Viruses 2020, 12, 420. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of Viral and Cellular Gene Expression by Kaposi's Sarcoma-Associated Herpesvirus Polyadenylated Nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef]
- Withers, J.B.; Li, E.S.; Vallery, T.K.; Yario, T.A.; Steitz, J.A. Two herpesviral noncoding PAN RNAs are functionally homologous but do not associate with common chromatin loci. PLOS Pathog. 2018, 14, e1007389. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



