Submitted:
27 June 2023
Posted:
29 June 2023
You are already at the latest version
Abstract
Keywords:
Introduction
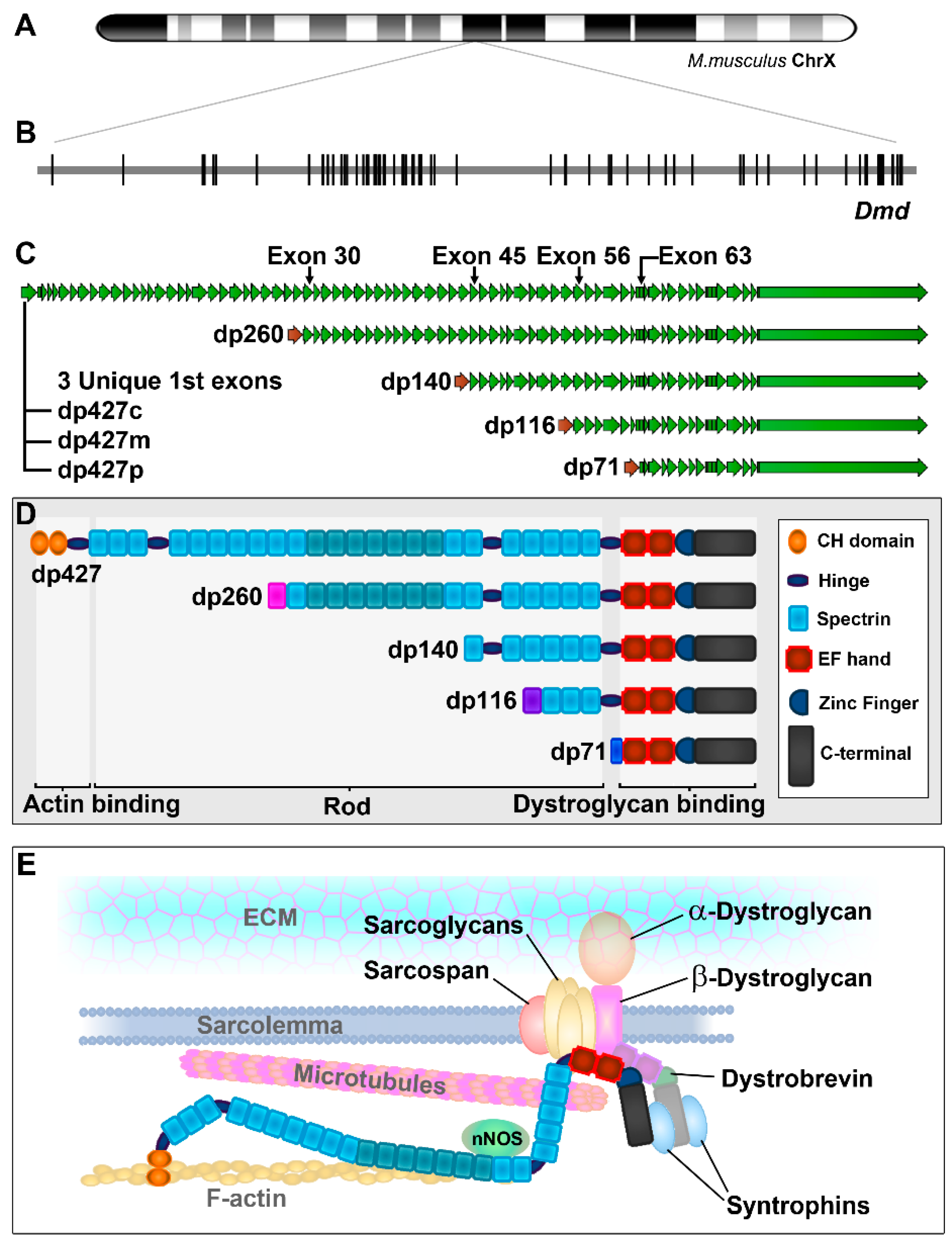
The dystrophin gene
Dystrophin and muscular dystrophy
Quantifying dystrophin protein
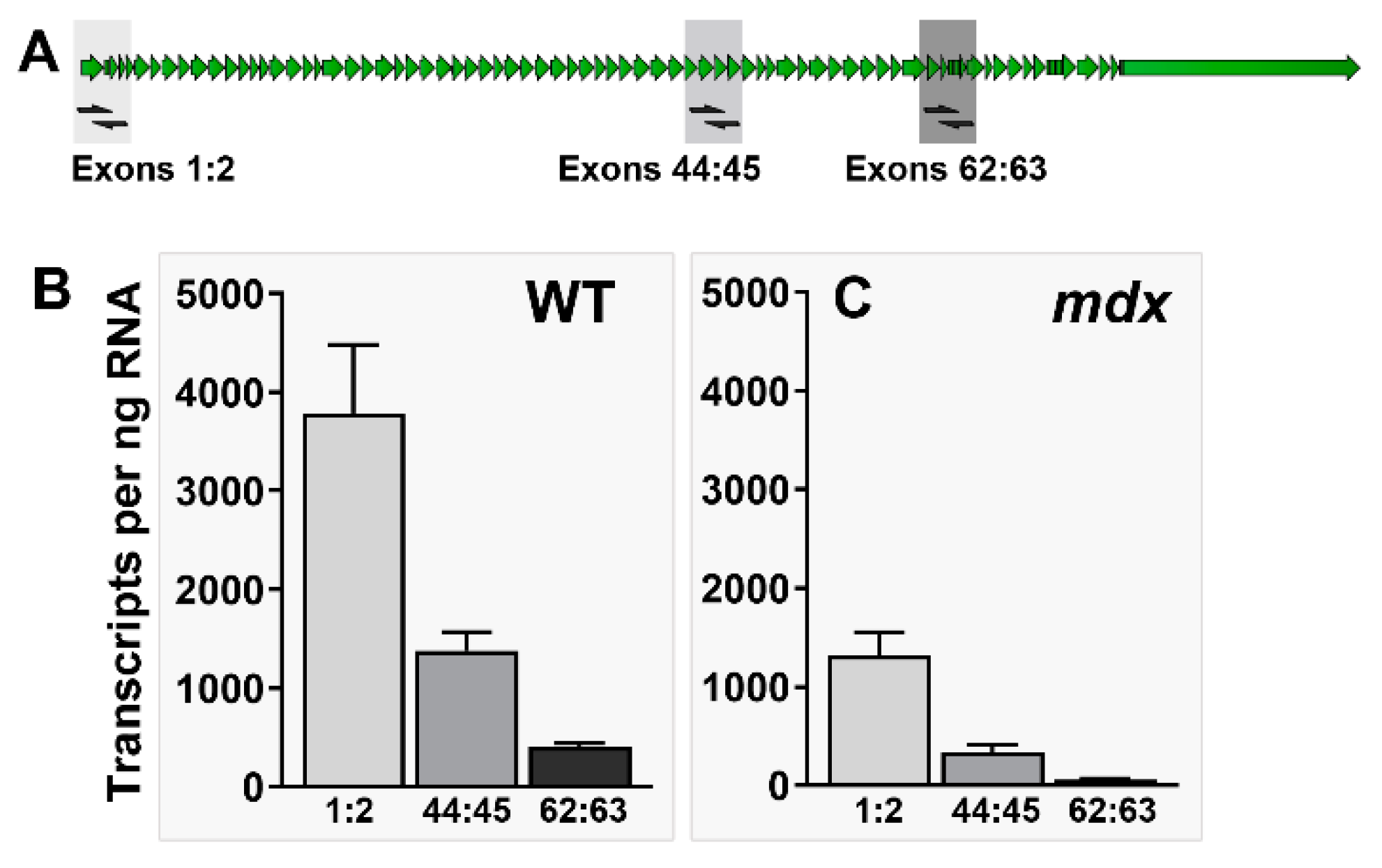
Quantifying dystrophin mRNA and transcript imbalance
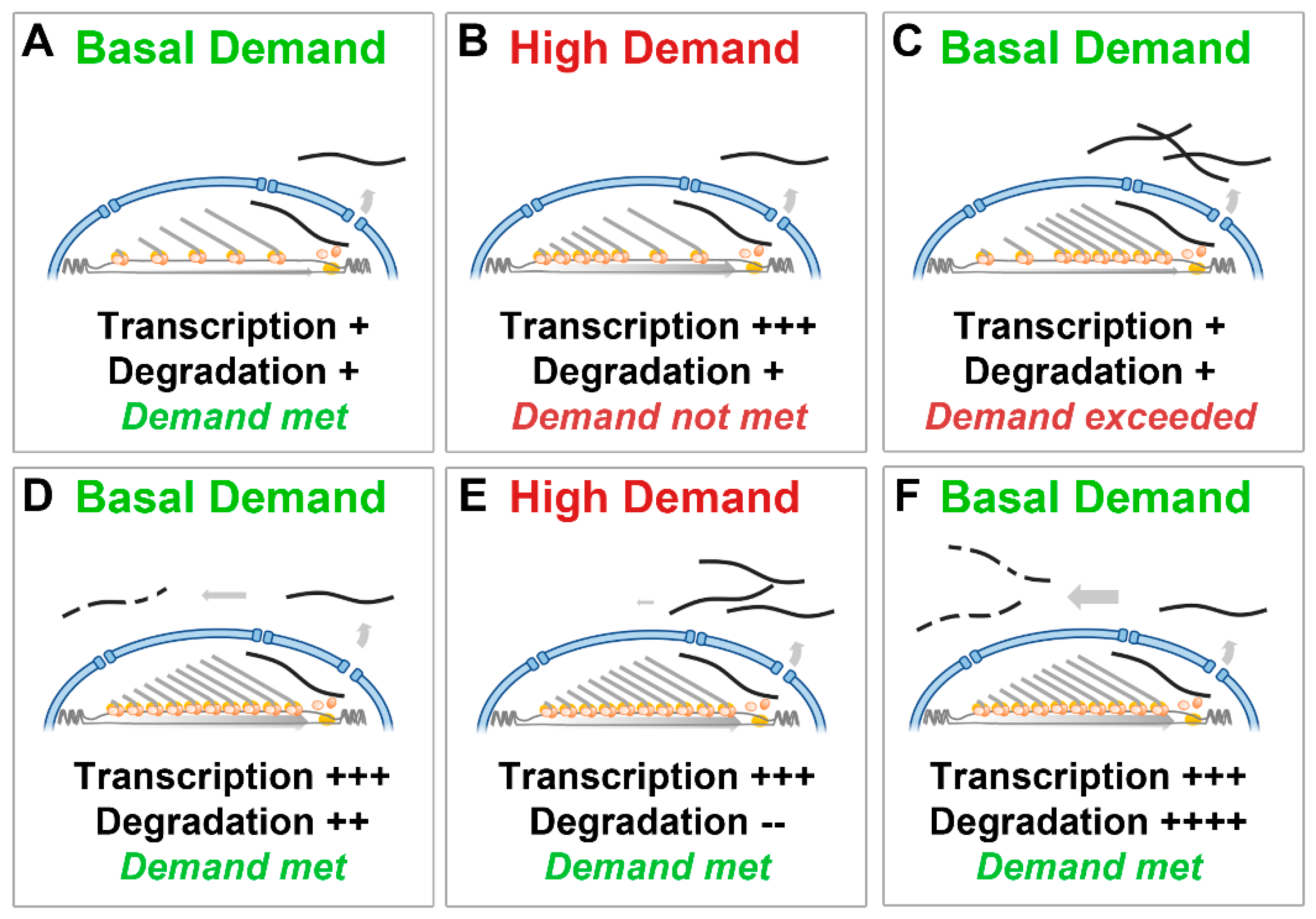
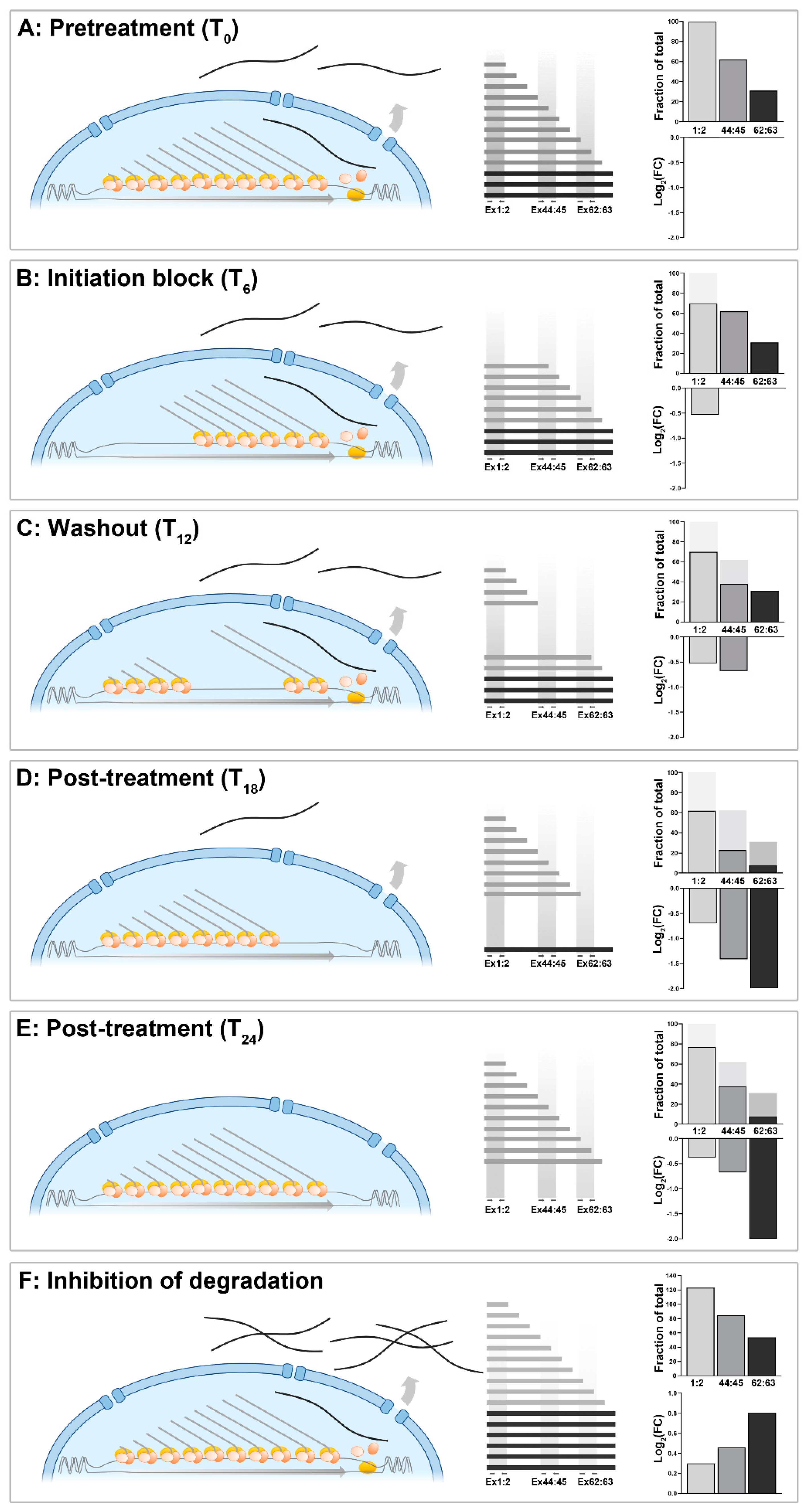
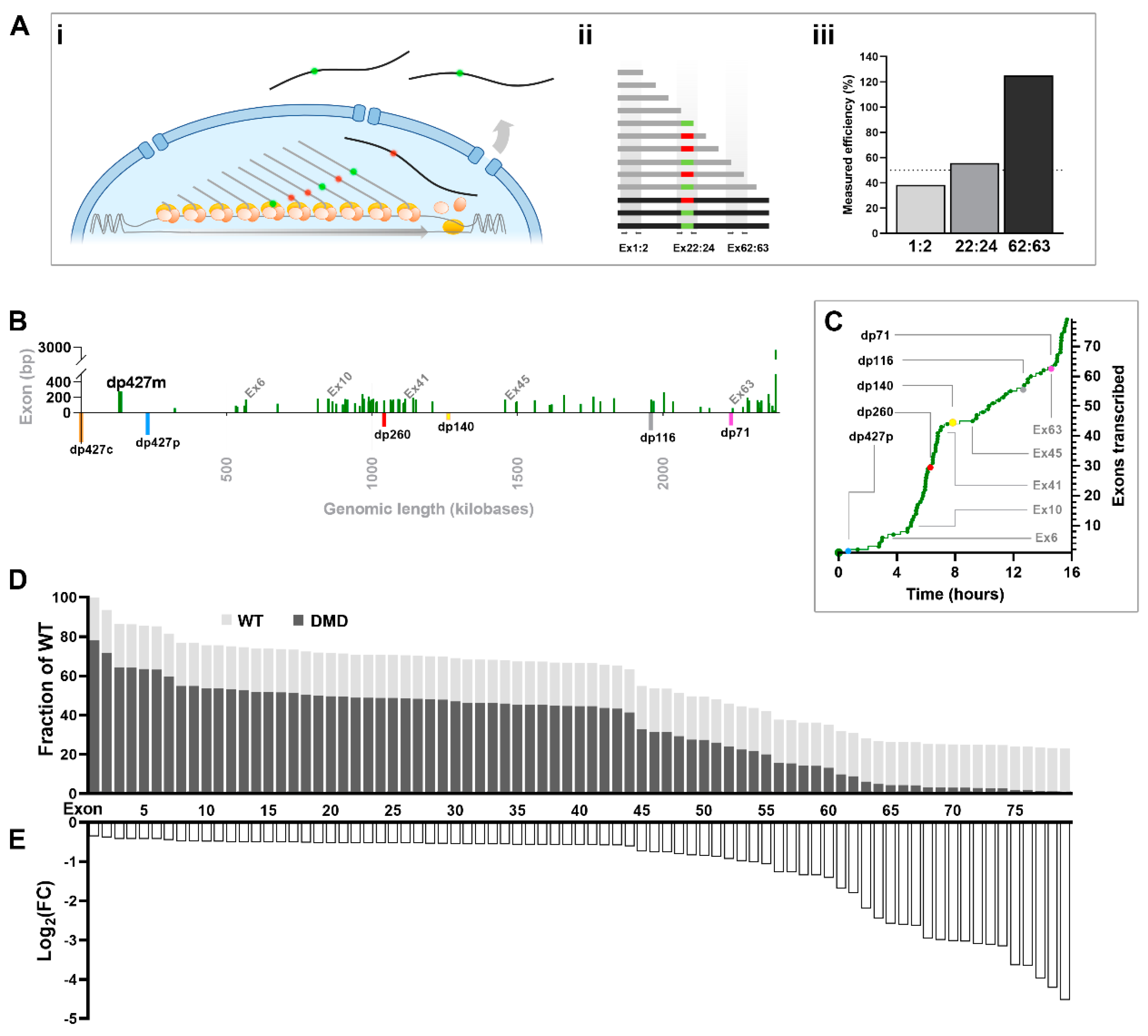
The dystrophin transcriptional model
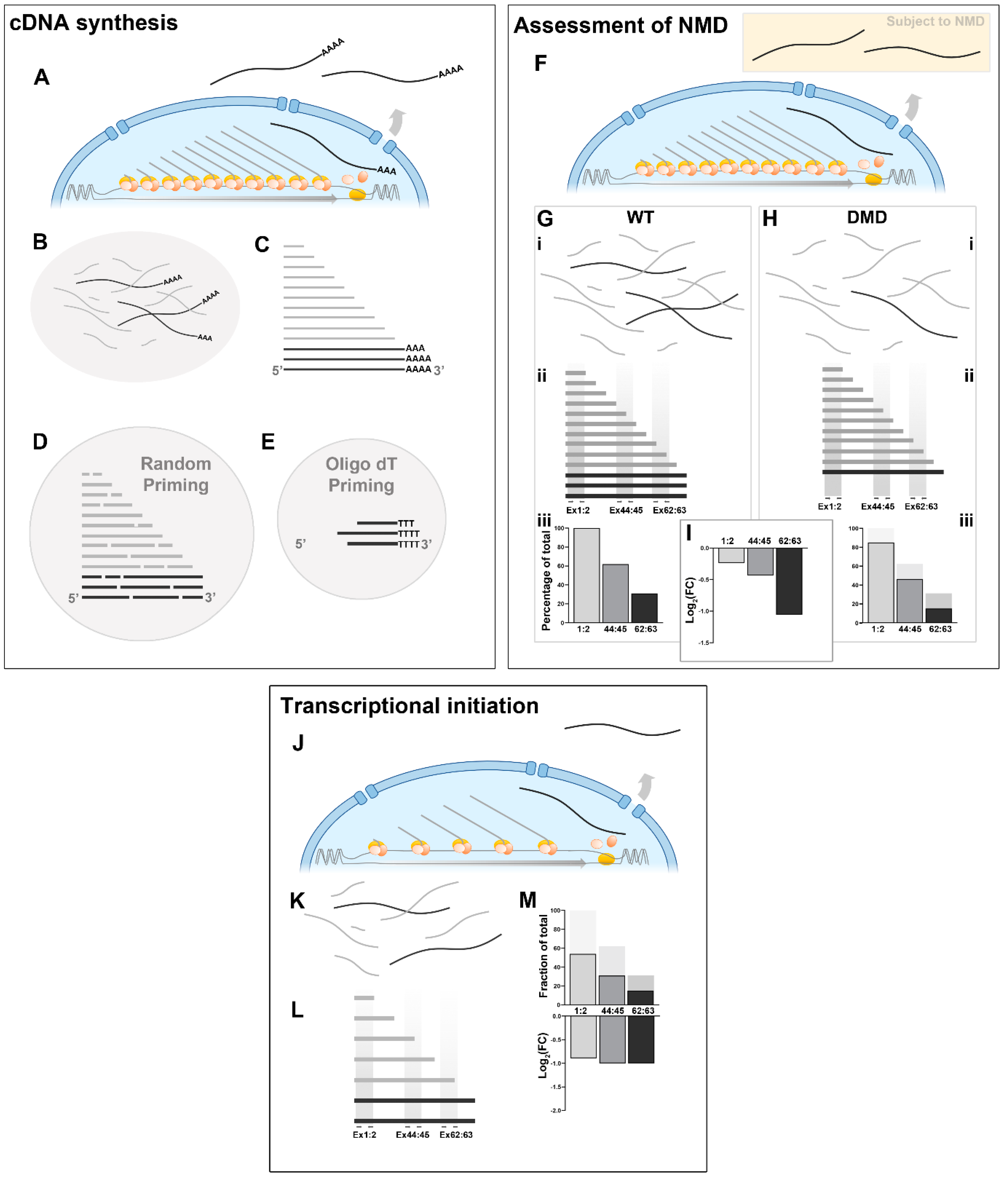
Quantitative measurement of unconventional dystrophin expression
cDNA synthesis
Comparing healthy and dystrophic transcripts
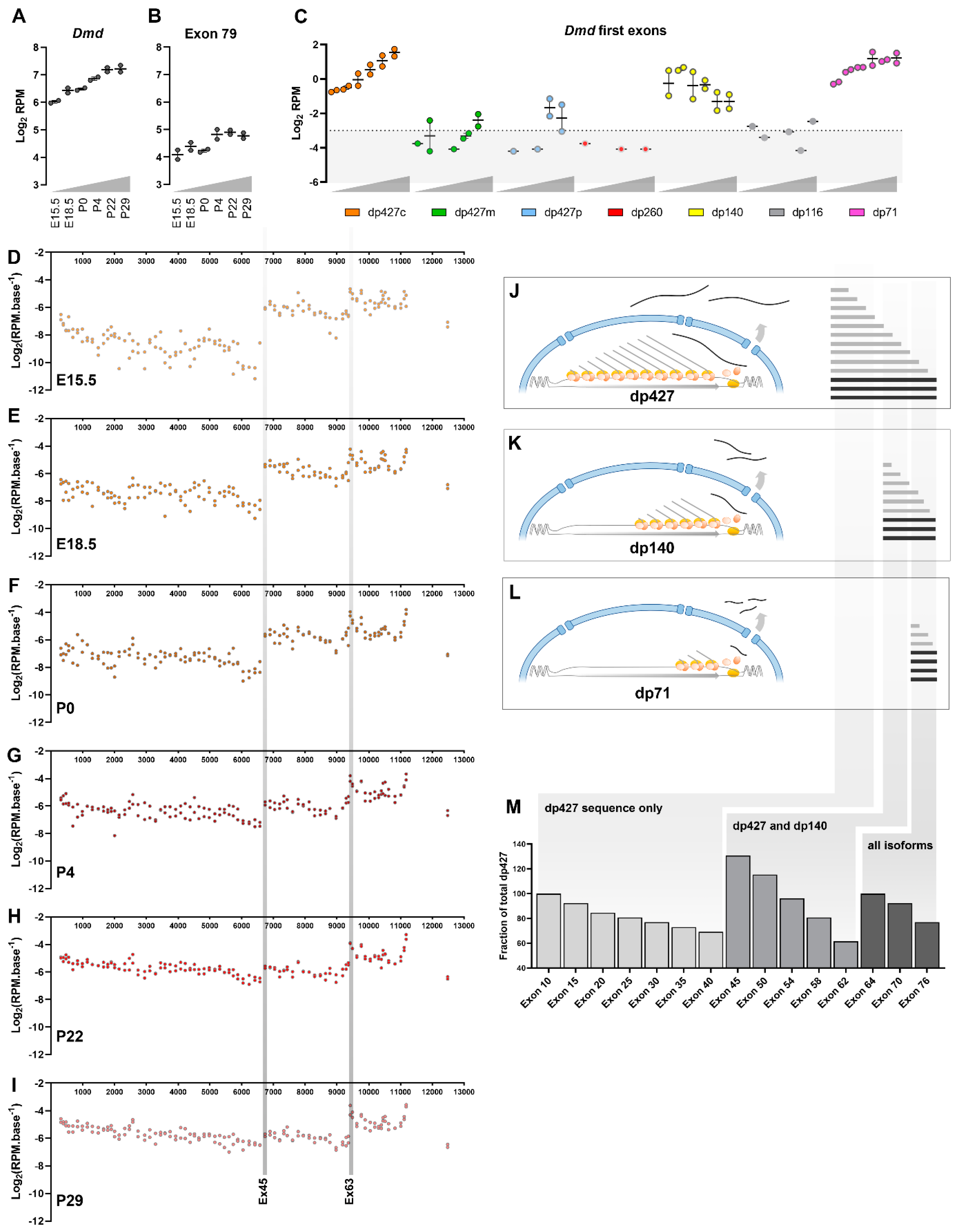
Measuring transcriptional changes over time
Quantifying exon skipping
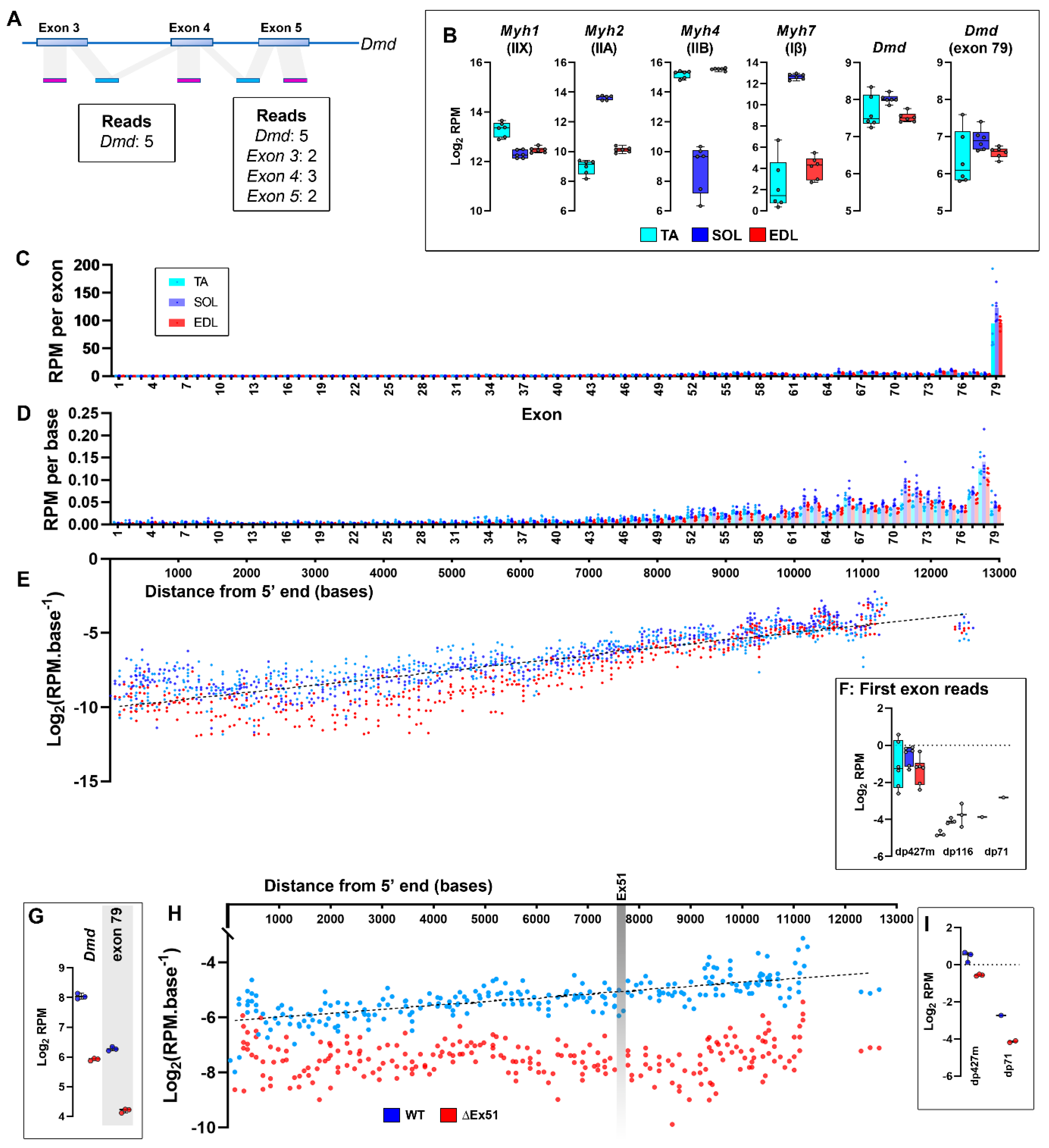
Dystrophin in the transcriptomic era
Discussion
The dystrophin transcriptional model
Dystrophin expression and RNAseq
Dystrophin transcription: caveats and alternative hypotheses
Conclusions
Methods
Sample collection and preparation
Multiplex FISH: sample preparation
RNAscope multiplex assay
Imaging
RNAseq analysis
References
- Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nature genetics. 1995;9(2):184-90. Epub 1995/02/01. doi: 10.1038/ng0295-184. PubMed PMID: 7719347. [CrossRef]
- Gazzoli I, Pulyakhina I, Verwey NE, Ariyurek Y, Laros JF, t Hoen PA, et al. Non-sequential and multi-step splicing of the dystrophin transcript. RNA Biol. 2016;13(3):290-305. Epub 2015/12/17. doi: 10.1080/15476286.2015.1125074. PubMed PMID: 26670121; PubMed Central PMCID: PMCPMC4829307. [CrossRef]
- Warner LE, DelloRusso C, Crawford RW, Rybakova IN, Patel JR, Ervasti JM, et al. Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy. Human molecular genetics. 2002;11(9):1095-105. Epub 2002/04/30. doi: 10.1093/hmg/11.9.1095. PubMed PMID: 11978768. [CrossRef]
- Molza AE, Mangat K, Le Rumeur E, Hubert JF, Menhart N, Delalande O. Structural Basis of Neuronal Nitric-oxide Synthase Interaction with Dystrophin Repeats 16 and 17. The Journal of biological chemistry. 2015;290(49):29531-41. Epub 2015/09/18. doi: 10.1074/jbc.M115.680660. PubMed PMID: 26378238; PubMed Central PMCID: PMCPMC4705953. [CrossRef]
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, Long C, et al. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. The Journal of clinical investigation. 2009;119(3):624-35. Epub 2009/02/21. doi: 10.1172/jci36612. PubMed PMID: 19229108; PubMed Central PMCID: PMCPmc2648692. [CrossRef]
- Belanto JJ, Mader TL, Eckhoff MD, Strandjord DM, Banks GB, Gardner MK, et al. Microtubule binding distinguishes dystrophin from utrophin. Proceedings of the National Academy of Sciences. 2014;111(15):5723. doi: 10.1073/pnas.1323842111. [CrossRef]
- Prins KW, Humston JL, Mehta A, Tate V, Ralston E, Ervasti JM. Dystrophin is a microtubule-associated protein. The Journal of cell biology. 2009;186(3):363-9. PubMed PMID: 19651889.
- Gao QQ, McNally EM. The Dystrophin Complex: Structure, Function, and Implications for Therapy. Comprehensive Physiology. 2015;5(3):1223-39. Epub 2015/07/04. doi: 10.1002/cphy.c140048. PubMed PMID: 26140716; PubMed Central PMCID: PMCPMC4767260. [CrossRef]
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2(12):731-40. doi: Doi 10.1016/S1474-4422(03)00585-4. PubMed PMID: WOS:000186665800016. [CrossRef]
- Boyce FM, Beggs AH, Feener C, Kunkel LM. Dystrophin is transcribed in brain from a distant upstream promoter. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(4):1276-80. Epub 1991/02/15. doi: 10.1073/pnas.88.4.1276. PubMed PMID: 1996328; PubMed Central PMCID: PMCPMC51000. [CrossRef]
- Gorecki DC, Monaco AP, Derry JM, Walker AP, Barnard EA, Barnard PJ. Expression of four alternative dystrophin transcripts in brain regions regulated by different promoters. Human molecular genetics. 1992;1(7):505-10. Epub 1992/10/01. doi: 10.1093/hmg/1.7.505. PubMed PMID: 1307251. [CrossRef]
- Klamut HJ, Gangopadhyay SB, Worton RG, Ray PN. Molecular and functional analysis of the muscle-specific promoter region of the Duchenne muscular dystrophy gene. Molecular and cellular biology. 1990;10(1):193-205. Epub 1990/01/01. doi: 10.1128/mcb.10.1.193. PubMed PMID: 2403634; PubMed Central PMCID: PMCPMC360727. [CrossRef]
- Nudel U, Zuk D, Einat P, Zeelon E, Levy Z, Neuman S, et al. Duchenne muscular dystrophy gene product is not identical in muscle and brain. Nature. 1989;337(6202):76-8. Epub 1989/01/05. doi: 10.1038/337076a0. PubMed PMID: 2909892. [CrossRef]
- Lidov HG, Selig S, Kunkel LM. Dp140: a novel 140 kDa CNS transcript from the dystrophin locus. Human molecular genetics. 1995;4(3):329-35. Epub 1995/03/01. doi: 10.1093/hmg/4.3.329. PubMed PMID: 7795584. [CrossRef]
- Crawford AH, Hildyard JCW, Rushing SAM, Wells DJ, Diez-Leon M, Piercy RJ. Validation of DE50-MD dogs as a model for the brain phenotype of Duchenne muscular dystrophy. Disease models & mechanisms. 2022;15(3). Epub 2022/01/13. doi: 10.1242/dmm.049291. PubMed PMID: 35019137; PubMed Central PMCID: PMCPMC8906169. [CrossRef]
- D’Souza VN, Nguyen TM, Morris GE, Karges W, Pillers DA, Ray PN. A novel dystrophin isoform is required for normal retinal electrophysiology. Human molecular genetics. 1995;4(5):837-42. Epub 1995/05/01. doi: 10.1093/hmg/4.5.837. PubMed PMID: 7633443. [CrossRef]
- Byers TJ, Lidov HG, Kunkel LM. An alternative dystrophin transcript specific to peripheral nerve. Nature genetics. 1993;4(1):77-81. Epub 1993/05/01. doi: 10.1038/ng0593-77. PubMed PMID: 8513330. [CrossRef]
- Bar S, Barnea E, Levy Z, Neuman S, Yaffe D, Nudel U. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem J. 1990;272(2):557-60. Epub 1990/12/01. doi: 10.1042/bj2720557. PubMed PMID: 2176467; PubMed Central PMCID: PMCPMC1149740. [CrossRef]
- Durbeej M, Jung D, Hjalt T, Campbell KP, Ekblom P. Transient expression of Dp140, a product of the Duchenne muscular dystrophy locus, during kidney tubulogenesis. Developmental biology. 1997;181(2):156-67. Epub 1997/01/15. doi: 10.1006/dbio.1996.8430. PubMed PMID: 9013927. [CrossRef]
- Doorenweerd N, Mahfouz A, van Putten M, Kaliyaperumal R, T’ Hoen PAC, Hendriksen JGM, et al. Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Scientific reports. 2017;7(1):12575-. doi: 10.1038/s41598-017-12981-5. PubMed PMID: 28974727. [CrossRef]
- Hildyard JCW, Crawford AH, Rawson F, Riddell DO, Harron RCM, Piercy RJ. Single-transcript multiplex in situ hybridisation reveals unique patterns of dystrophin isoform expression in the developing mammalian embryo. Wellcome Open Research. 2020;5(76). doi: 10.12688/wellcomeopenres.15762.2. [CrossRef]
- Jin H, Tan S, Hermanowski J, Böhm S, Pacheco S, McCauley JM, et al. The dystrotelin, dystrophin and dystrobrevin superfamily: new paralogues and old isoforms. BMC genomics. 2007;8:19-. doi: 10.1186/1471-2164-8-19. PubMed PMID: 17233888. [CrossRef]
- Mendell JR, Shilling C, Leslie ND, Flanigan KM, al-Dahhak R, Gastier-Foster J, et al. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Annals of neurology. 2012;71(3):304-13. Epub 2012/03/28. doi: 10.1002/ana.23528. PubMed PMID: 22451200. [CrossRef]
- Segurel L, Wyman MJ, Przeworski M. Determinants of mutation rate variation in the human germline. Annu Rev Genomics Hum Genet. 2014;15:47-70. Epub 2014/07/09. doi: 10.1146/annurev-genom-031714-125740. PubMed PMID: 25000986. [CrossRef]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471-5. Epub 2012/08/24. doi: 10.1038/nature11396. PubMed PMID: 22914163; PubMed Central PMCID: PMCPMC3548427. [CrossRef]
- White SJ, den Dunnen JT. Copy number variation in the genome; the human DMD gene as an example. Cytogenet Genome Res. 2006;115(3-4):240-6. Epub 2006/11/25. doi: 10.1159/000095920. PubMed PMID: 17124406. [CrossRef]
- Ankala A, Kohn JN, Hegde A, Meka A, Ephrem CL, Askree SH, et al. Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene. Genome Res. 2012;22(1):25-34. Epub 2011/11/18. doi: 10.1101/gr.123463.111. PubMed PMID: 22090376; PubMed Central PMCID: PMCPMC3246204. [CrossRef]
- Hoffman EP, Brown RH, Jr., Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919-28. Epub 1987/12/24. PubMed PMID: 3319190.
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science (New York, NY). 1989;244(4912):1578-80. Epub 1989/06/30. PubMed PMID: 2662404; PubMed Central PMCID: PMC2662404.
- Bushby KM, Gardner-Medwin D. The clinical, genetic and dystrophin characteristics of Becker muscular dystrophy. I. Natural history. J Neurol. 1993;240(2):98-104. Epub 1993/02/01. doi: 10.1007/BF00858725. PubMed PMID: 8437027. [CrossRef]
- Comi GP, Prelle A, Bresolin N, Moggio M, Bardoni A, Gallanti A, et al. Clinical variability in Becker muscular dystrophy. Genetic, biochemical and immunohistochemical correlates. Brain. 1994;117 ( Pt 1):1-14. Epub 1994/02/01. doi: 10.1093/brain/117.1.1-a. PubMed PMID: 8149204. [CrossRef]
- Morandi L, Mora M, Bernasconi P, Mantegazza R, Gebbia M, Balestrini MR, et al. Very small dystrophin molecule in a family with a mild form of Becker dystrophy. Neuromuscular disorders : NMD. 1993;3(1):65-70. Epub 1993/01/01. doi: 10.1016/0960-8966(93)90043-j. PubMed PMID: 8329891. [CrossRef]
- Farrokhi V, Walsh J, Palandra J, Brodfuehrer J, Caiazzo T, Owens J, et al. Dystrophin and mini-dystrophin quantification by mass spectrometry in skeletal muscle for gene therapy development in Duchenne muscular dystrophy. Gene therapy. 2022;29(10-11):608-15. Epub 2021/11/06. doi: 10.1038/s41434-021-00300-7. PubMed PMID: 34737451; PubMed Central PMCID: PMCPMC9068826. [CrossRef]
- Aartsma-Rus A, Morgan J, Lonkar P, Neubert H, Owens J, Binks M, et al. Report of a TREAT-NMD/World Duchenne Organisation Meeting on Dystrophin Quantification Methodology. J Neuromuscul Dis. 2019;6(1):147-59. Epub 2019/01/08. doi: 10.3233/JND-180357. PubMed PMID: 30614809; PubMed Central PMCID: PMCPMC6398559. [CrossRef]
- Godfrey C, Muses S, McClorey G, Wells KE, Coursindel T, Terry RL, et al. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. (1460-2083 (Electronic)).
- Morin A, Stantzou A, Petrova ON, Hildyard J, Tensorer T, Matouk M, et al. Dystrophin myonuclear domain restoration governs treatment efficacy in dystrophic muscle. Proceedings of the National Academy of Sciences of the United States of America. 2023;120(2):e2206324120. Epub 2023/01/04. doi: 10.1073/pnas.2206324120. PubMed PMID: 36595689; PubMed Central PMCID: PMCPMC9926233. [CrossRef]
- Garcia-Rodriguez R, Hiller M, Jimenez-Gracia L, van der Pal Z, Balog J, Adamzek K, et al. Premature termination codons in the DMD gene cause reduced local mRNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(28):16456-64. Epub 2020/07/04. doi: 10.1073/pnas.1910456117. PubMed PMID: 32616572; PubMed Central PMCID: PMCPMC7368324. [CrossRef]
- Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142(3):368-74. Epub 2010/08/10. doi: 10.1016/j.cell.2010.07.022. PubMed PMID: 20691898; PubMed Central PMCID: PMCPMC2950652. [CrossRef]
- Hildyard JCW, Rawson F, Wells DJ, Piercy RJ. Multiplex in situ hybridization within a single transcript: RNAscope reveals dystrophin mRNA dynamics. PLoS One. 2020;15(9):e0239467. Epub 2020/09/25. doi: 10.1371/journal.pone.0239467. PubMed PMID: 32970731; PubMed Central PMCID: PMCPMC7514052. [CrossRef]
- Spitali P, van den Bergen JC, Verhaart IE, Wokke B, Janson AA, van den Eijnde R, et al. DMD transcript imbalance determines dystrophin levels. FASEB J. 2013;27(12):4909-16. Epub 2013/08/27. doi: 10.1096/fj.13-232025. PubMed PMID: 23975932. [CrossRef]
- Tennyson CN, Shi Q, Worton RG. Stability of the human dystrophin transcript in muscle. Nucleic acids research. 1996;24(15):3059-64. Epub 1996/08/01. doi: 10.1093/nar/24.15.3059. PubMed PMID: 8760894; PubMed Central PMCID: PMCPMC146056. [CrossRef]
- Waldrop MA, Moore SA, Mathews KD, Darbro BW, Medne L, Finkel R, et al. Intron mutations and early transcription termination in Duchenne and Becker muscular dystrophy. Human mutation. 2022;43(4):511-28. Epub 2022/02/16. doi: 10.1002/humu.24343. PubMed PMID: 35165973; PubMed Central PMCID: PMCPMC9901284. [CrossRef]
- Muniz L, Nicolas E, Trouche D. RNA polymerase II speed: a key player in controlling and adapting transcriptome composition. EMBO J. 2021;40(15):e105740. Epub 2021/07/14. doi: 10.15252/embj.2020105740. PubMed PMID: 34254686; PubMed Central PMCID: PMCPMC8327950. [CrossRef]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16(11):1128-33. Epub 2009/10/13. doi: 10.1038/nsmb.1666. PubMed PMID: 19820712; PubMed Central PMCID: PMCPMC2783620. [CrossRef]
- Hildyard JCW, Riddell DO, Harron RCM, Rawson F, Foster EMA, Massey C, et al. The skeletal muscle phenotype of the DE50-MD dog model of Duchenne muscular dystrophy. Wellcome Open Res. 2022;7:238. Epub 2023/03/04. doi: 10.12688/wellcomeopenres.18251.1. PubMed PMID: 36865375; PubMed Central PMCID: PMCPMC9971692. [CrossRef]
- Hildyard JCW, Wells DJ, Piercy RJ. Identification of qPCR reference genes suitable for normalising gene expression in the developing mouse embryo. Wellcome Open Res. 2021;6:197. Epub 2022/05/06. doi: 10.12688/wellcomeopenres.16972.1. PubMed PMID: 35509373; PubMed Central PMCID: PMCPMC9024131. [CrossRef]
- Donandt T, Todorow V, Hintze S, Graupner A, Schoser B, Walter MC, et al. Nuclear Small Dystrophin Isoforms during Muscle Differentiation. Life. 2023;13(6). doi: 10.3390/life13061367. [CrossRef]
- Terry EE, Zhang X, Hoffmann C, Hughes LD, Lewis SA, Li J, et al. Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Elife. 2018;7. Epub 2018/05/29. doi: 10.7554/eLife.34613. PubMed PMID: 29809149; PubMed Central PMCID: PMCPMC6008051. [CrossRef]
- Chemello F, Wang Z, Li H, McAnally JR, Liu N, Bassel-Duby R, et al. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(47):29691-701. Epub 2020/11/06. doi: 10.1073/pnas.2018391117. PubMed PMID: 33148801; PubMed Central PMCID: PMCPMC7703557. [CrossRef]
- Schmitt BM, Rudolph KL, Karagianni P, Fonseca NA, White RJ, Talianidis I, et al. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome Res. 2014;24(11):1797-807. Epub 2014/08/15. doi: 10.1101/gr.176784.114. PubMed PMID: 25122613; PubMed Central PMCID: PMCPMC4216921. [CrossRef]
- Barthelemy F, Defour A, Levy N, Krahn M, Bartoli M. Muscle Cells Fix Breaches by Orchestrating a Membrane Repair Ballet. J Neuromuscul Dis. 2018;5(1):21-8. Epub 2018/02/27. doi: 10.3233/JND-170251. PubMed PMID: 29480214; PubMed Central PMCID: PMCPMC5836414. [CrossRef]
- Carmeille R, Bouvet F, Tan S, Croissant C, Gounou C, Mamchaoui K, et al. Membrane repair of human skeletal muscle cells requires Annexin-A5. Biochimica et biophysica acta. 2016;1863(9):2267-79. Epub 2016/06/12. doi: 10.1016/j.bbamcr.2016.06.003. PubMed PMID: 27286750. [CrossRef]
- Hershey JWB, Sonenberg N, Mathews MB. Principles of Translational Control. Cold Spring Harb Perspect Biol. 2019;11(9). Epub 2018/07/01. doi: 10.1101/cshperspect.a032607. PubMed PMID: 29959195; PubMed Central PMCID: PMCPMC6719596. [CrossRef]
- Larsen CA, Howard MT. Conserved regions of the DMD 3’ UTR regulate translation and mRNA abundance in cultured myotubes. Neuromuscular disorders : NMD. 2014;24(8):693-706. Epub 2014/06/15. doi: 10.1016/j.nmd.2014.05.006. PubMed PMID: 24928536; PubMed Central PMCID: PMCPMC4114305. [CrossRef]
- Hogg JR, Goff SP. Upf1 senses 3’UTR length to potentiate mRNA decay. Cell. 2010;143(3):379-89. Epub 2010/10/30. doi: 10.1016/j.cell.2010.10.005. PubMed PMID: 21029861; PubMed Central PMCID: PMCPMC2981159. [CrossRef]
- Greener MJ, Sewry CA, Muntoni F, Roberts RG. The 3’-untranslated region of the dystrophin gene - conservation and consequences of loss. Eur J Hum Genet. 2002;10(7):413-20. Epub 2002/07/11. doi: 10.1038/sj.ejhg.5200822. PubMed PMID: 12107815. [CrossRef]
- Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29(12):691-9. Epub 2013/10/05. doi: 10.1016/j.tig.2013.09.002. PubMed PMID: 24091060; PubMed Central PMCID: PMCPMC3854950. [CrossRef]
- Verhaart IEC, van Vliet-van den Dool L, Sipkens JA, de Kimpe SJ, Kolfschoten IGM, van Deutekom JCT, et al. The Dynamics of Compound, Transcript, and Protein Effects After Treatment With 2OMePS Antisense Oligonucleotides in mdx Mice. Molecular therapy Nucleic acids. 2014;3(2):e148-e. doi: 10.1038/mtna.2014.1. PubMed PMID: 24549299. [CrossRef]
- Novak JS, Spathis R, Dang UJ, Fiorillo AA, Hindupur R, Tully CB, et al. Interrogation of Dystrophin and Dystroglycan Complex Protein Turnover After Exon Skipping Therapy. J Neuromuscul Dis. 2021;8(s2):S383-S402. Epub 2021/09/28. doi: 10.3233/JND-210696. PubMed PMID: 34569969; PubMed Central PMCID: PMCPMC8673539. [CrossRef]
- Wilton SD, Dye DE, Blechynden LM, Laing NG. Revertant fibres: a possible genetic therapy for Duchenne muscular dystrophy? Neuromuscular Disorders. 1997;7(5):329-35. doi: 10.1016/s0960-8966(97)00058-8. [CrossRef]
- Wilton SD, Dye DE, Laing NG. Dystrophin gene transcripts skipping the mdx mutation. Muscle & nerve. 1997;20(6):728-34. doi: 10.1002/(SICI)1097-4598(199706)20:6<728::AID-MUS10>3.0.CO;2-Q. [CrossRef]
- Bouge AL, Murauer E, Beyne E, Miro J, Varilh J, Taulan M, et al. Targeted RNA-Seq profiling of splicing pattern in the DMD gene: exons are mostly constitutively spliced in human skeletal muscle. Sci Rep. 2017;7:39094. Epub 2017/01/04. doi: 10.1038/srep39094. PubMed PMID: 28045018; PubMed Central PMCID: PMCPMC5206723. [CrossRef]
- Lederfein D, Levy Z, Augier N, Mornet D, Morris G, Fuchs O, et al. A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(12):5346-50. Epub 1992/06/15. doi: 10.1073/pnas.89.12.5346. PubMed PMID: 1319059; PubMed Central PMCID: PMCPMC49288. [CrossRef]
- Rau F, Laine J, Ramanoudjame L, Ferry A, Arandel L, Delalande O, et al. Abnormal splicing switch of DMD’s penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat Commun. 2015;6:7205. Epub 2015/05/29. doi: 10.1038/ncomms8205. PubMed PMID: 26018658; PubMed Central PMCID: PMCPMC4458869. [CrossRef]
- Naidoo M, Anthony K. Dystrophin Dp71 and the Neuropathophysiology of Duchenne Muscular Dystrophy. Mol Neurobiol. 2019. doi: 10.1007/s12035-019-01845-w. [CrossRef]
- Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic acids research. 2010;38(12):e131. Epub 2010/04/17. doi: 10.1093/nar/gkq224. PubMed PMID: 20395217; PubMed Central PMCID: PMCPMC2896536. [CrossRef]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17(6):831-40. Epub 2005/03/23. doi: 10.1016/j.molcel.2005.02.017. PubMed PMID: 15780939. [CrossRef]
- Carey LB. RNA polymerase errors cause splicing defects and can be regulated by differential expression of RNA polymerase subunits. Elife. 2015;4. Epub 2015/12/15. doi: 10.7554/eLife.09945. PubMed PMID: 26652005; PubMed Central PMCID: PMCPMC4868539. [CrossRef]
- Gout JF, Thomas WK, Smith Z, Okamoto K, Lynch M. Large-scale detection of in vivo transcription errors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18584-9. Epub 2013/10/30. doi: 10.1073/pnas.1309843110. PubMed PMID: 24167253; PubMed Central PMCID: PMCPMC3832031. [CrossRef]
- Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26(8):345-52. Epub 2010/07/03. doi: 10.1016/j.tig.2010.05.003. PubMed PMID: 20594608; PubMed Central PMCID: PMCPMC2910838. [CrossRef]
- Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nature Medicine. 2015;21(12):1455-63. doi: 10.1038/nm.3990. [CrossRef]
- Schultz E, Jaryszak DL, Valliere CR. Response of satellite cells to focal skeletal muscle injury. Muscle & nerve. 1985;8(3):217-22. Epub 1985/03/01. doi: 10.1002/mus.880080307. PubMed PMID: 4058466. [CrossRef]
- Kaczmarek A, Kaczmarek M, Cialowicz M, Clemente FM, Wolanski P, Badicu G, et al. The Role of Satellite Cells in Skeletal Muscle Regeneration-The Effect of Exercise and Age. Biology (Basel). 2021;10(10). Epub 2021/10/24. doi: 10.3390/biology10101056. PubMed PMID: 34681155; PubMed Central PMCID: PMCPMC8533525. [CrossRef]
- de Leon MB, Montanez C, Gomez P, Morales-Lazaro SL, Tapia-Ramirez V, Valadez-Graham V, et al. Dystrophin Dp71 expression is down-regulated during myogenesis: role of Sp1 and Sp3 on the Dp71 promoter activity. The Journal of biological chemistry. 2005;280(7):5290-9. Epub 2004/11/20. doi: 10.1074/jbc.M411571200. PubMed PMID: 15550398. [CrossRef]
- Farea M, Rani AQM, Maeta K, Nishio H, Matsuo M. Dystrophin Dp71ab is monoclonally expressed in human satellite cells and enhances proliferation of myoblast cells. Sci Rep. 2020;10(1):17123. Epub 2020/10/15. doi: 10.1038/s41598-020-74157-y. PubMed PMID: 33051488; PubMed Central PMCID: PMCPMC7553993. [CrossRef]
- Scherer S. A Short Guide to the Human Genome: Cold Spring Harbor Laboratory Press; 2009.
- Galaxy C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic acids research. 2022;50(W1):W345-W51. Epub 2022/04/22. doi: 10.1093/nar/gkac247. PubMed PMID: 35446428; PubMed Central PMCID: PMCPMC9252830. [CrossRef]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907-15. Epub 2019/08/04. doi: 10.1038/s41587-019-0201-4. PubMed PMID: 31375807; PubMed Central PMCID: PMCPMC7605509. [CrossRef]
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166-9. Epub 2014/09/28. doi: 10.1093/bioinformatics/btu638. PubMed PMID: 25260700; PubMed Central PMCID: PMCPMC4287950. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




