Submitted:
30 June 2023
Posted:
04 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
3.1. Genomic Databases
3.1.1. CancerResource
3.1.2. Cancer Specific Databases
3.2. Proteomic Databases
3.3. Lipidomics
3.4. Glyco Databases
3.5. Clinical Trial Databases
3.6. Other Cancer Databases
3.7. Web-based Servers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgements
Conflicts of Interest
References
- G. B. Faguet, “A brief history of cancer: Age-old milestones underlying our current knowledge database,” Int J Cancer, vol. 136, no. 9, pp. 2022–2036, May 2015. [CrossRef]
- B. Weinstein and K. Case, “The History of Cancer Research: Introducing an AACR Centennial Series,” Cancer Res, vol. 68, no. 17, pp. 6861–6862, Sep. 2008. [CrossRef]
- “SEER Training Modules, Cancer Facts and the War on Cancer,” National Cancer Institutes.
- “SEER Training Modules, Brief History of Cancer Registration,” National Cancer Institute.
- G. Ursin, “Cancer registration in the era of modern oncology and GDPR,” https://doi.org/10.1080/0284186X.2019.1657586, vol. 58, no. 11, pp. 1547–1548, Nov. 2019. [CrossRef]
- K. Tomczak, P. Czerwińska, and M. Wiznerowicz, “The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge,” Wspolczesna Onkologia, vol. 1A. Termedia Publishing House Ltd., pp. A68–A77, 2015. [CrossRef]
- L. Sarver, A. E. Sarver, C. Yuan, and S. Subramanian, “OMCD: OncomiR Cancer Database,” BMC Cancer, vol. 18, no. 1, Dec. 2018. [CrossRef]
- S. Mei et al., “Cistrome cancer: A web resource for integrative gene regulation modeling in cancer,” Cancer Res, vol. 77, no. 21, pp. e19–e22, Nov. 2017. [CrossRef]
- E. Cerami et al., “The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data,” Cancer Discov, vol. 2, no. 5, pp. 401–404, May 2012. [CrossRef]
- J. Zhang et al., “The International Cancer Genome Consortium Data Portal,” Nature Biotechnology 2019 37:4, vol. 37, no. 4, pp. 367–369, Mar. 2019. [CrossRef]
- W. J. Kent et al., “The Human Genome Browser at UCSC,” Genome Res, vol. 12, no. 6, pp. 996–1006, Jun. 2002. [CrossRef]
- “The Human Genome Browser at UCSC.” https://genome.cshlp.org/content/12/6/996.short (accessed Feb. 06, 2023).
- E. Clough and T. Barrett, “The Gene Expression Omnibus database,” Methods in Molecular Biology, vol. 1418, pp. 93–110, 2016. [CrossRef]
- P. Flicek et al., “Ensembl 2014,” Nucleic Acids Res, vol. 42, no. D1, Jan. 2014. [CrossRef]
- D. R. Zerbino et al., “Ensembl 2018,” Nucleic Acids Res, vol. 46, no. D1, pp. D754–D761, Jan. 2018. [CrossRef]
- F. J. Martin et al., “Ensembl 2023,” Nucleic Acids Res, vol. 51, no. D1, pp. D933–D941, Jan. 2023. [CrossRef]
- J. Küntzer, D. Maisel, H. P. Lenhof, S. Klostermann, and H. Burtscher, “The Roche Cancer Genome Database 2.0,” BMC Med Genomics, vol. 4, p. 43, 2011. [CrossRef]
- M. A. Jensen, V. Ferretti, R. L. Grossman, and L. M. Staudt, “The NCI Genomic Data Commons as an engine for precision medicine,” Blood, vol. 130, no. 4, pp. 453–459, Jul. 2017. [CrossRef]
- “GDC.” https://portal.gdc.cancer.gov/ (accessed Feb. 15, 2023).
- E. Cappelli et al., “OpenGDC: Unifying, Modeling, Integrating Cancer Genomic Data and Clinical Metadata,” Applied Sciences 2020, Vol. 10, Page 6367, vol. 10, no. 18, p. 6367, Sep. 2020. [CrossRef]
- P. A. Futreal et al., “A census of human cancer genes,” Nature Reviews Cancer 2004 4:3, vol. 4, no. 3, pp. 177–183, 2004. [CrossRef]
- D. Repana et al., “The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens 06 Biological Sciences 0604 Genetics 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis 06 Biological Sciences 0601 Biochemistry and Cell Biology,” Genome Biol, vol. 20, no. 1, pp. 1–12, Jan. 2019. [CrossRef]
- M. E. Higgins, M. Claremont, J. E. Major, C. Sander, and A. E. Lash, “CancerGenes: a gene selection resource for cancer genome projects,” Nucleic Acids Res, vol. 35, no. suppl_1, pp. D721–D726, Jan. 2007. [CrossRef]
- D. Zhang et al., “CHG: A Systematically Integrated Database of Cancer Hallmark Genes,” Front Genet, vol. 11, p. 29, Feb. 2020. [CrossRef]
- S. Bamford et al., “The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website,” Br J Cancer, vol. 91, no. 2, pp. 355–358, Jul. 2004. [CrossRef]
- J. G. Tate et al., “COSMIC: the Catalogue Of Somatic Mutations In Cancer,” Nucleic Acids Res, vol. 47, no. D1, pp. D941–D947, Jan. 2019. [CrossRef]
- L. Brown, M. Li, A. Goncearenco, and A. R. Panchenko, “Finding driver mutations in cancer: Elucidating the role of background mutational processes,” PLoS Comput Biol, vol. 15, no. 4, 2019. [CrossRef]
- Q. Huang, P. Carrio-Cordo, B. Gao, R. Paloots, and M. Baudis, “The Progenetix oncogenomic resource in 2021,” Database, vol. 2021, no. 0, pp. 1–9, Sep. 2021. [CrossRef]
- “Progenetix.” https://progenetix.org/ (accessed Feb. 15, 2023).
- J. Ping et al., “MutEx: a multifaceted gateway for exploring integrative pan-cancer genomic data,” Brief Bioinform, vol. 21, no. 4, pp. 1479–1486, Jul. 2020. [CrossRef]
- D. R. Rhodes et al., “ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform 1,” 2004. [Online]. Available: www.oncomine.
- D. R. Rhodes et al., “Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles,” Neoplasia, vol. 9, no. 2, pp. 166–180, Feb. 2007. [CrossRef]
- L. K. Vestergaard, D. N. P. Oliveira, T. S. Poulsen, C. K. Høgdall, and E. V. Høgdall, “OncomineTM comprehensive assay v3 vs. OncomineTM comprehensive assay plus,” Cancers (Basel), vol. 13, no. 20, p. 5230, Oct. 2021. [CrossRef]
- J. Ahmed et al., “CancerResource: a comprehensive database of cancer-relevant proteins and compound interactions supported by experimental knowledge,” Nucleic Acids Res, vol. 39, no. Database issue, p. D960, Jan. 2011. [CrossRef]
- P. Davis et al., “Comparative Toxicogenomics Database (CTD): update 2021,” Nucleic Acids Res, vol. 49, no. D1, pp. D1138–D1143, Jan. 2021. [CrossRef]
- X. Chen, Z. L. Ji, and Y. Z. Chen, “TTD: Therapeutic Target Database,” Nucleic Acids Res, vol. 30, no. 1, pp. 412–415, Jan. 2002. [CrossRef]
- Y. Wang et al., “Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics,” Nucleic Acids Res, vol. 48, no. D1, pp. D1031–D1041, Jan. 2020. [CrossRef]
- C. F. Thorn, T. E. Klein, and R. B. Altman, “PharmGKB: The pharmacogenomics knowledge base,” Methods in Molecular Biology, vol. 1015, pp. 311–320, 2013. [CrossRef]
- L. Gong, M. Whirl-Carrillo, and T. E. Klein, “PharmGKB, an Integrated Resource of Pharmacogenomic Knowledge,” Curr Protoc, vol. 1, no. 8, p. e226, Aug. 2021. [CrossRef]
- D. S. Wishart et al., “DrugBank 5.0: a major update to the DrugBank database for 2018,” Nucleic Acids Res, vol. 46, no. D1, pp. D1074–D1082, Jan. 2018. [CrossRef]
- B. O. Gohlke, J. Nickel, R. Otto, M. Dunkel, and R. Preissner, “CancerResource—updated database of cancer-relevant proteins, mutations and interacting drugs,” Nucleic Acids Res, vol. 44, no. D1, pp. D932–D937, Jan. 2016. [CrossRef]
- L. Cai et al., “LCE: an open web portal to explore gene expression and clinical associations in lung cancer,” Oncogene 2018 38:14, vol. 38, no. 14, pp. 2551–2564, Dec. 2018. [CrossRef]
- V. S. Koshkin et al., “PROMISE: a real-world clinical-genomic database to address knowledge gaps in prostate cancer,” Prostate Cancer and Prostatic Diseases 2021 25:3, vol. 25, no. 3, pp. 388–396, Aug. 2021. [CrossRef]
- Q. Lian et al., “HCCDB: A Database of Hepatocellular Carcinoma Expression Atlas,” Genomics Proteomics Bioinformatics, vol. 16, no. 4, pp. 269–275, Aug. 2018. [CrossRef]
- “The oncoReveal Dx Lung and”.
- N. J. Edwards et al., “The CPTAC data portal: A resource for cancer proteomics research,” J Proteome Res, vol. 14, no. 6, pp. 2707–2713, Jun. 2015. [CrossRef]
- “Clinical Proteomic Tumor Analysis Consortium (CPTAC) | NCI Genomic Data Commons.” https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/clinical-proteomic-tumor-analysis-consortium-cptac (accessed Feb. 06, 2023).
- C. M. Lindgren et al., “Simplified and Unified Access to Cancer Proteogenomic Data,” J Proteome Res, vol. 20, no. 4, pp. 1902–1910, Apr. 2021. https://doi.org/10.1021/ACS.JPROTEOME.0C00919/ASSET/IMAGES/LARGE/PR0C00919_0003.JPEG. [CrossRef]
- D. Szklarczyk et al., “Correction to ‘The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets,’” Nucleic Acids Res, vol. 49, no. 18, pp. 10800–10800, Oct. 2021. [CrossRef]
- D. S. Chandrashekar et al., “UALCAN: An update to the integrated cancer data analysis platform,” Neoplasia, vol. 25, pp. 18–27, Mar. 2022. [CrossRef]
- M. Zhang et al., “CanProVar 2.0: An Updated Database of Human Cancer Proteome Variation,” J Proteome Res, vol. 16, no. 2, pp. 421–432, Feb. 2017. [CrossRef]
- P. W. Rose et al., “The RCSB Protein Data Bank: views of structural biology for basic and applied research and education,” Nucleic Acids Res, vol. 43, no. D1, pp. D345–D356, Jan. 2015. [CrossRef]
- T. U. Consortium, “Activities at the Universal Protein Resource (UniProt),” Nucleic Acids Res, vol. 42, no. 11, pp. 7486–7486, Jun. 2014. [CrossRef]
- Bateman, “UniProt: a worldwide hub of protein knowledge,” Nucleic Acids Res, vol. 47, no. D1, pp. D506–D515, Jan. 2019. [CrossRef]
- C. Orsburn, “Proteome Discoverer—A Community Enhanced Data Processing Suite for Protein Informatics,” Proteomes 2021, Vol. 9, Page 15, vol. 9, no. 1, p. 15, Mar. 2021. [CrossRef]
- O’Donovan, M. J. Martin, A. Gattiker, E. Gasteiger, A. Bairoch, and R. Apweiler, “High-quality protein knowledge resource: SWISS-PROT and TrEMBL,” Brief Bioinform, vol. 3, no. 3, pp. 275–284, Sep. 2002. [CrossRef]
- Y. Moriya et al., “The jPOST environment: an integrated proteomics data repository and database,” Nucleic Acids Res, vol. 47, no. D1, pp. D1218–D1224, Jan. 2019. [CrossRef]
- X. Shao, I. N. Taha, K. R. Clauser, Y. (Tom) Gao, and A. Naba, “MatrisomeDB: the ECM-protein knowledge database,” Nucleic Acids Res, vol. 48, no. D1, pp. D1136–D1144, Jan. 2020. [CrossRef]
- F. Yan, H. Zhao, and Y. Zeng, “Lipidomics: a promising cancer biomarker,” Clin Transl Med, vol. 7, no. 1, p. e21, Dec. 2018. [CrossRef]
- M. Buszewska-forajta et al., “Lipidomics as a diagnostic tool for prostate cancer,” Cancers (Basel), vol. 13, no. 9, p. 2000, May 2021. [CrossRef]
- Q. Wu et al., “DBLiPro: A Database for Lipids and Proteins in Human Lipid Metabolism,” Phenomics 2023, pp. 1–10, May 2023. [CrossRef]
- D. Cotter, A. Maer, C. Guda, B. Saunders, and S. Subramaniam, “LMPD: LIPID MAPS proteome database,” Nucleic Acids Res, vol. 34, no. suppl_1, pp. D507–D510, Jan. 2006. [CrossRef]
- M. Sud et al., “LMSD: LIPID MAPS structure database,” Nucleic Acids Res, vol. 35, no. suppl_1, pp. D527–D532, Jan. 2007. [CrossRef]
- G. Liebisch et al., “Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures,” J Lipid Res, vol. 61, no. 12, pp. 1539–1555, Dec. 2020. [CrossRef]
- B. B. Blair et al., “Increased circulating levels of galectin proteins in patients with breast, colon, and lung cancer,” Cancers (Basel), vol. 13, no. 19, Oct. 2021. [CrossRef]
- S. S. Pinho and C. A. Reis, “Glycosylation in cancer: mechanisms and clinical implications,” Nature Reviews Cancer 2015 15:9, vol. 15, no. 9, pp. 540–555, Aug. 2015. [CrossRef]
- F. T. Liu and S. R. Stowell, “The role of galectins in immunity and infection,” Nature Reviews Immunology 2023, pp. 1–16, Jan. 2023. [CrossRef]
- T. Funkhouser et al., “KIT Mutations Correlate with Higher Galectin Levels and Brain Metastasis in Breast and Non-Small Cell Lung Cancer,” Cancers (Basel), vol. 14, no. 11, Jun. 2022. [CrossRef]
- D. B. Hizal et al., “Glycoproteomic and glycomic databases,” Clin Proteomics, vol. 11, no. 1, pp. 1–10, Apr. 2014. [CrossRef]
- Y. Tian and H. Zhang, “Glycoproteomics and clinical applications,” Proteomics Clin Appl, vol. 4, no. 2, pp. 124–132, Feb. 2010. [CrossRef]
- E. H. Kim, & D. E. Misek, “Glycoproteomics-based identification of cancer biomarkers,” Int J Proteomics, 2011.
- S. Pan, R. Chen, R. Aebersold, and T. A. Brentnall, “Mass Spectrometry Based Glycoproteomics—From a Proteomics Perspective *,” Molecular & Cellular Proteomics, vol. 10, no. 1, p. R110.003251, Jan. 2011. [CrossRef]
- J. A. Ferreira, M. Relvas-Santos, A. Peixoto, A. M.N. Silva, and L. Lara Santos, “Glycoproteogenomics: Setting the Course for Next-generation Cancer Neoantigen Discovery for Cancer Vaccines,” Genomics, Proteomics and Bioinformatics, vol. 19, no. 1. Beijing Genomics Institute, pp. 25–43, Feb. 01, 2021. [CrossRef]
- C. A. Cooper, M. J. Harrison, M. R. Wilkins, and N. H. Packer, “GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources,” Nucleic Acids Res, vol. 29, no. 1, pp. 332–335, Jan. 2001. [CrossRef]
- C. A. Hayes et al., “UniCarb-DB: a database resource for glycomic discovery,” Bioinformatics, vol. 27, no. 9, pp. 1343–1344, May 2011. 20 May. [CrossRef]
- C. W. Von Der Lieth et al., “EUROCarbDB: An open-access platform for glycoinformatics,” Glycobiology, vol. 21, no. 4, pp. 493–502, Apr. 2011. [CrossRef]
- H. Zhang et al., “UniPep - A database for human N-linked glycosites: A resource for biomarker discovery,” Genome Biol, vol. 7, no. 8, pp. 1–12, Aug. 2006. [CrossRef]
- Togayachi, K.-Y. Dae, T. Shikanai, and H. Narimatsu, “A Database System for Glycogenes (GGDB),” Experimental Glycoscience, pp. 423–425, Mar. 2008. [CrossRef]
- R. Ranzinger, M. Frank, C. W. Von der lieth, and S. Herget, “Glycome-DB.org: A portal for querying across the digital world of carbohydrate sequences,” Glycobiology, vol. 19, no. 12, pp. 1563–1567, Dec. 2009. [CrossRef]
- M. P. Campbell, L. M. P. Campbell, L. Royle, C. M. Radcliffe, R. A. Dwek, and P. M. Rudd, “GlycoBase and autoGU: tools for HPLC-based glycan analysis,” Bioinformatics, vol. 24, no. 9, pp. 1214–1216, May 2008. [CrossRef]
- S. Zhao et al., “GlycoStore: a database of retention properties for glycan analysis,” Bioinformatics, vol. 34, no. 18, pp. 3231–3232, Sep. 2018. [CrossRef]
- R. Ranzinger et al., “GlycoRDF: an ontology to standardize glycomics data in RDF,” Bioinformatics, vol. 31, no. 6, pp. 919–925, Mar. 2015. [CrossRef]
- D. B. Weatherly, F. S. Arpinar, M. Porterfield, M. Tiemeyer, W. S. York, and R. Ranzinger, “GRITS Toolbox—a freely available software for processing, annotating and archiving glycomics mass spectrometry data,” Glycobiology, vol. 29, no. 6, pp. 452–460, Jun. 2019. [CrossRef]
- M. Tiemeyer et al., “GlyTouCan: an accessible glycan structure repository,” Glycobiology, vol. 27, no. 10, pp. 915–919, Oct. 2017. [CrossRef]
- J. Hirabayashi, H. Tateno, T. Shikanai, K. F. Aoki-Kinoshita, and H. Narimatsu, “The Lectin Frontier Database (LfDB), and Data Generation Based on Frontal Affinity Chromatography,” Molecules 2015, Vol. 20, Pages 951-973, vol. 20, no. 1, pp. 951–973, Jan. 2015. [CrossRef]
- P. V. Toukach and A. I. Shirkovskaya, “Carbohydrate Structure Database and Other Glycan Databases as an Important Element of Glycoinformatics,” Russ J Bioorg Chem, vol. 48, no. 3, pp. 457–466, Jun. 2022. [CrossRef]
- B. D. Solomon, A. D. Nguyen, K. A. Bear, and T. G. Wolfsberg, “Clinical genomic database,” Proc Natl Acad Sci U S A, vol. 110, no. 24, pp. 9851–9855, Jun. 2013. [CrossRef]
- R. J. Hartmaier et al., “High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis,” Cancer Res, vol. 77, no. 9, pp. 2464–2475, May 2017. doi.org/10.1158/0008-5472.CAN-16-2479/657735/AM/HIGH-THROUGHPUT-GENOMIC-PROFILING-OF-ADULT-SOLID. 20 May. [CrossRef]
- D. P. Mudaranthakam et al., “A Curated Cancer Clinical Outcomes Database (C3OD) for accelerating patient recruitment in cancer clinical trials,” JAMIA Open, vol. 1, no. 2, pp. 166–171, Oct. 2018. [CrossRef]
- J. Overgaard, A. Jovanovic, C. Godballe, and J. Grau Eriksen, “The Danish Head and Neck Cancer database,” Clin Epidemiol, vol. 8, pp. 491–496, Oct. 2016. [CrossRef]
- R. M. McCabe, “National Cancer Database: The Past, Present, and Future of the Cancer Registry and Its Efforts to Improve the Quality of Cancer Care,” Semin Radiat Oncol, vol. 29, no. 4, pp. 323–325, Oct. 2019. [CrossRef]
- M. C. Daly and I. M. Paquette, “Surveillance, Epidemiology, and End Results (SEER) and SEER-Medicare Databases: Use in Clinical Research for Improving Colorectal Cancer Outcomes,” Clin Colon Rectal Surg, vol. 32, no. 01, pp. 061–068, 2019.
- M. J. Landrum and B. L. Kattman, “ClinVar at five years: Delivering on the promise,” Hum Mutat, vol. 39, no. 11, pp. 1623–1630, Nov. 2018. [CrossRef]
- J. S. Nanda, R. Kumar, and G. P. S. Raghava, “dbEM: A database of epigenetic modifiers curated from cancerous and normal genomes,” Sci Rep, vol. 6, Jan. 2016. [CrossRef]
- S. Ullah et al., “The Cancer Research Database (CRDB): Integrated Platform to Gain Statistical Insight into the Correlation between Cancer and COVID-19,” JMIR Cancer, vol. 8, no. 2, Apr. 2022. [CrossRef]
- H. Zheng et al., “Comprehensive Review of Web Servers and Bioinformatics Tools for Cancer Prognosis Analysis,” Front Oncol, vol. 10, p. 68, Feb. 2020. [CrossRef]
- C. P. Goswami and H. Nakshatri, “PROGgeneV2: Enhancements on the existing database,” BMC Cancer, vol. 14, no. 1, pp. 1–6, Dec. 2014. [CrossRef]
- R. Kumar et al., “CancerDR: Cancer drug resistance database,” Sci Rep, vol. 3, 2013. [CrossRef]
- S. H. Liu et al., “DriverDBv3: a multi-omics database for cancer driver gene research,” Nucleic Acids Res, vol. 48, no. D1, pp. D863–D870, Jan. 2020. [CrossRef]
- L. Cheng et al., “LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse,” Nucleic Acids Res, vol. 47, no. D1, pp. D140–D144, Jan. 2019. [CrossRef]
- Y. Gao et al., “Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data,” Nucleic Acids Res, vol. 49, no. D1, pp. D1251–D1258, Jan. 2021. [CrossRef]
- L. J. Carithers and H. M. Moore, “The Genotype-Tissue Expression (GTEx) Project,” https://home.liebertpub.com/bio, vol. 13, no. 5, pp. 307–308, Oct. 2015. [CrossRef]
- C. J. Liu, F. F. Hu, M. X. Xia, L. Han, Q. Zhang, and A. Y. Guo, “GSCALite: a web server for gene set cancer analysis,” Bioinformatics, vol. 34, no. 21, pp. 3771–3772, Nov. 2018. [CrossRef]
- Hamosh, J. S. Amberger, C. Bocchini, A. F. Scott, and S. A. Rasmussen, “Online Mendelian Inheritance in Man (OMIM®): Victor McKusick’s magnum opus,” Am J Med Genet A, vol. 185, no. 11, pp. 3259–3265, Nov. 2021. [CrossRef]
- Z. Tang, C. Li, B. Kang, G. Gao, C. Li, and Z. Zhang, “GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses,” Nucleic Acids Res, vol. 45, no. W1, pp. W98–W102, Jul. 2017. [CrossRef]
- B. Wen, X. Wang, and B. Zhang, “PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations,” Genome Res, vol. 29, pp. 485–493, Jan. 2019.
- M. D. Wilkinson et al., “The FAIR Guiding Principles for scientific data management and stewardship,” Scientific Data 2016 3:1, vol. 3, no. 1, pp. 1–9, Mar. 2016. [CrossRef]
- Pavlopoulou, D. A. Spandidos, and I. Michalopoulos, “Human cancer databases (Review),” Oncology Reports, vol. 33, no. 1. Spandidos Publications, pp. 3–18, Jan. 01, 2015. [CrossRef]
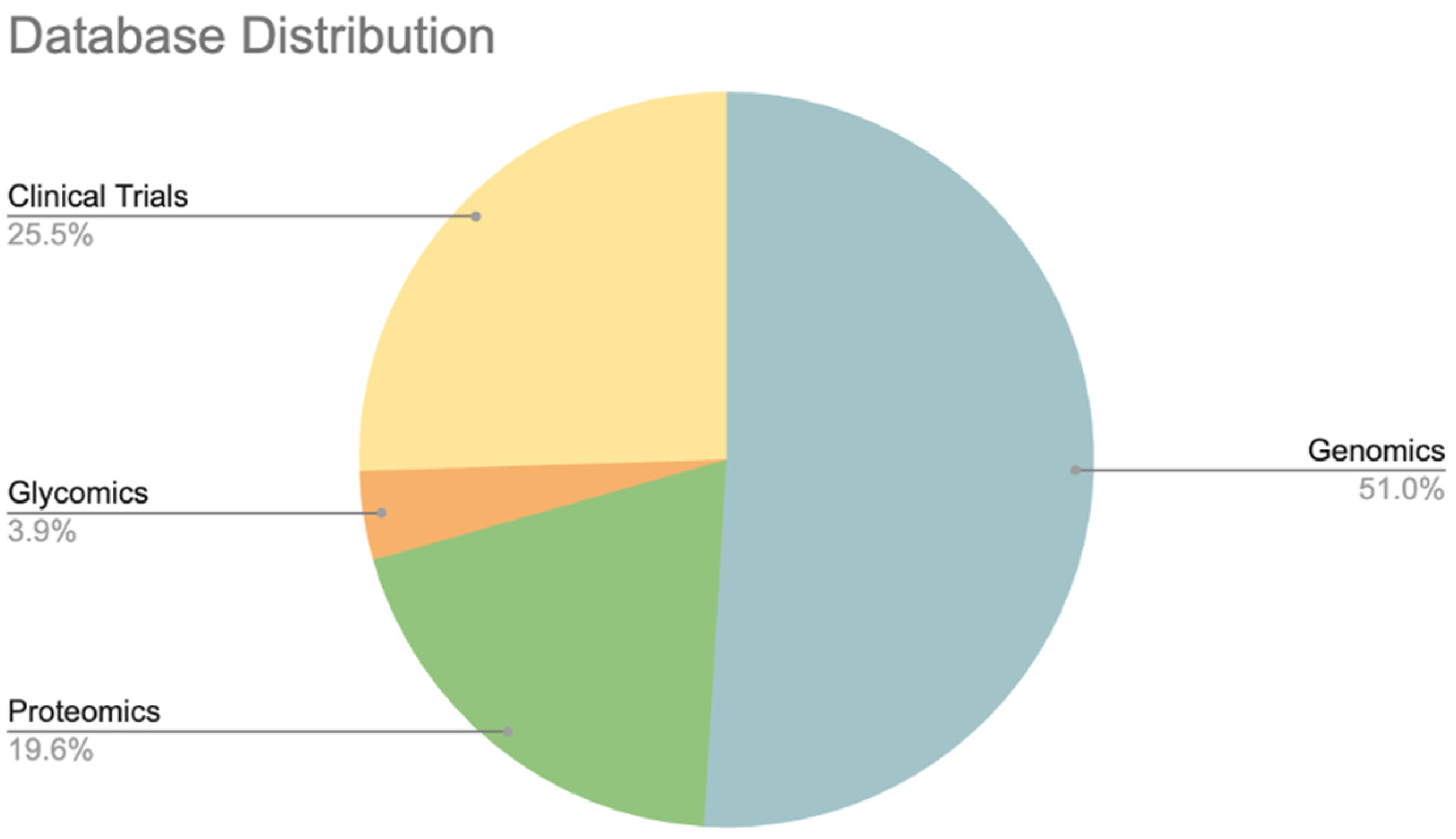
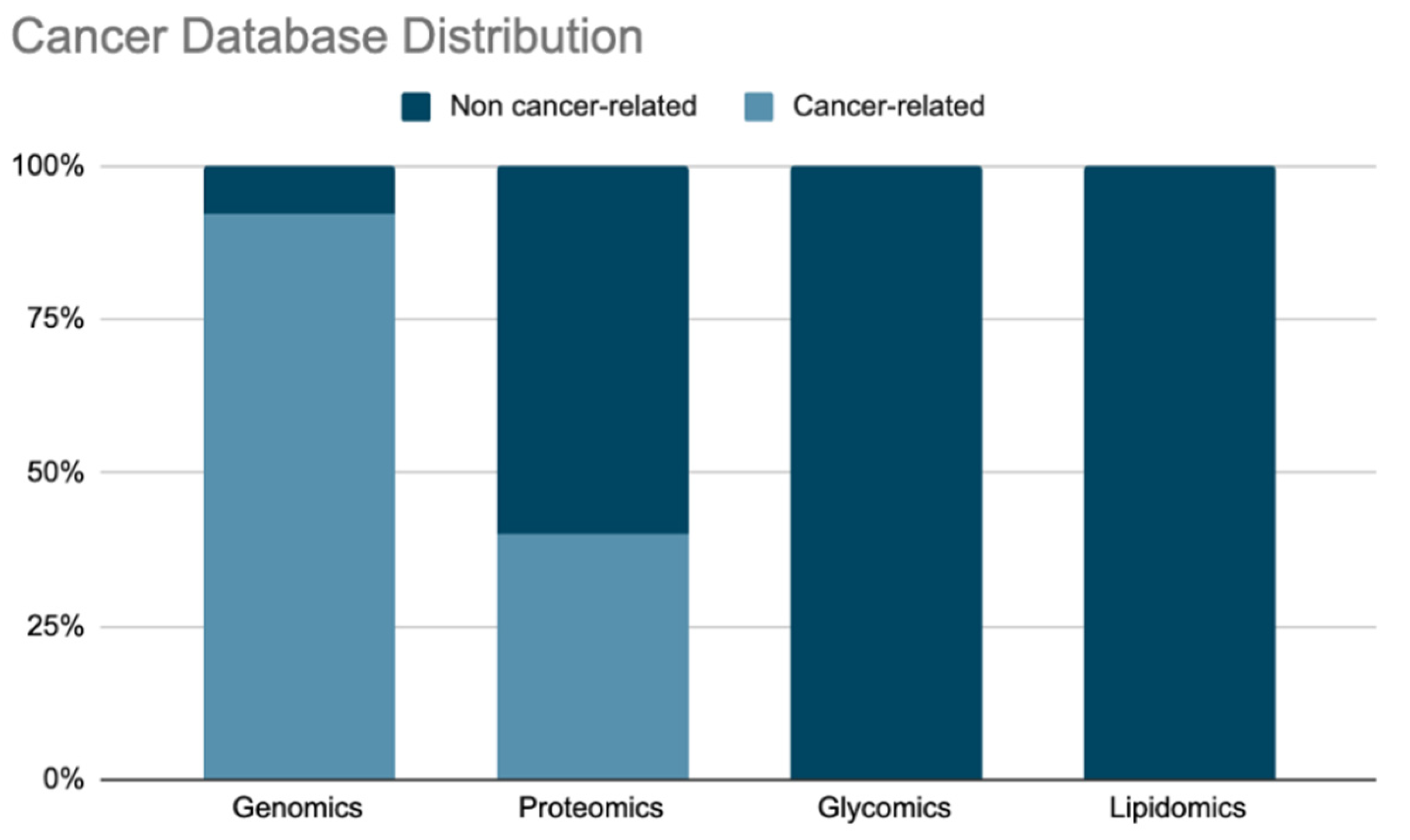
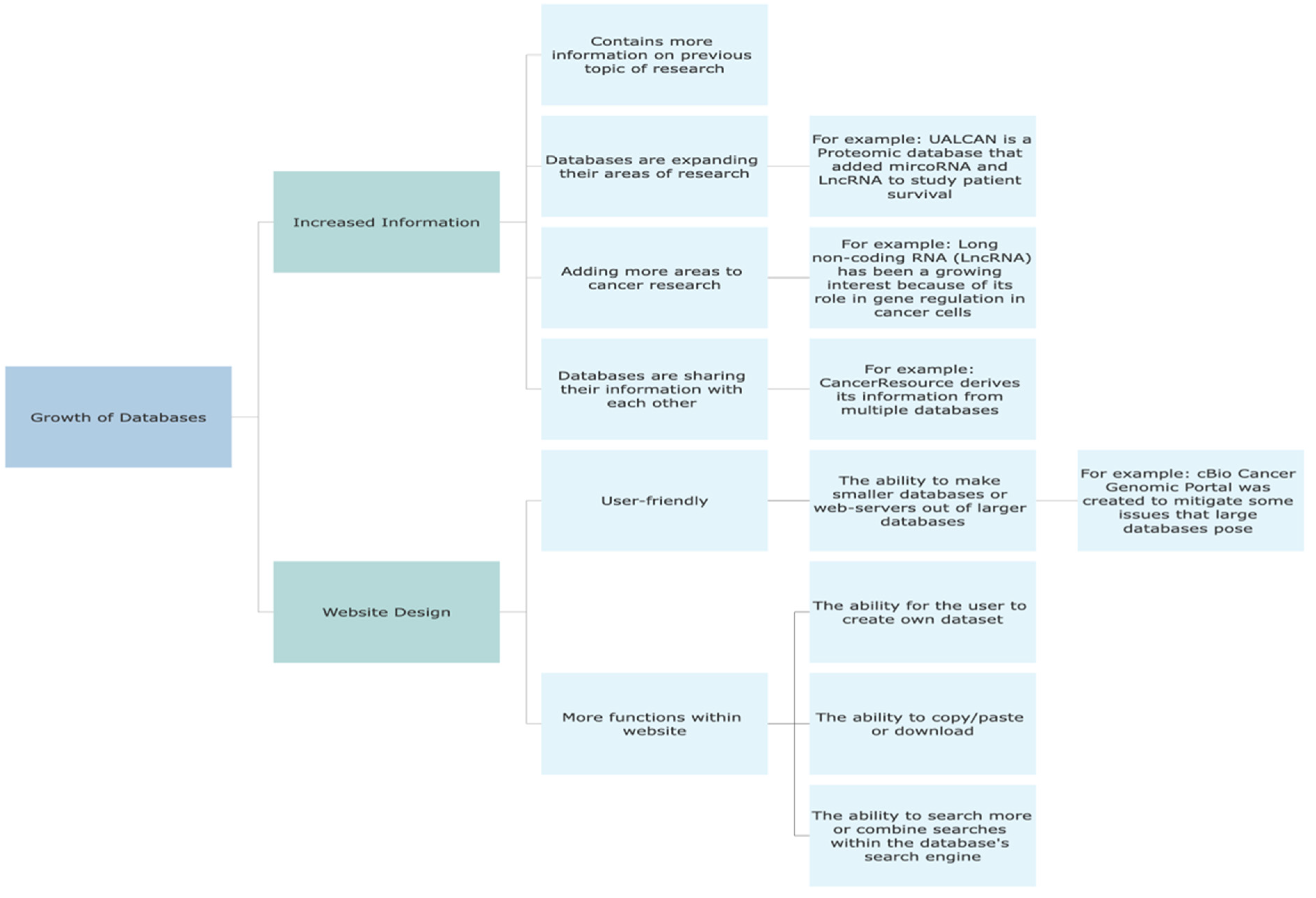
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| The Cancer Genome Atlas Cases= 11,315 |
Genome sequencing across 33 tumor types | Yes | Yes | Yes | F, A, I, R | https://www.cancer.gov/ccg/research/genome-sequencing/tcga |
| OncomiR Cancer Database OMCD Cases= 9500 |
Comparative genomic analysis of miRNA data sequencing | Yes | N/A | Yes | F, A, I | http://www.oncomir.org/cgi-bin/dbSearch.cgi |
| cBio Cancer Genomic Portal | Genomic analysis of cancer-related genes | Yes | Yes | Yes | F, A, I, R | https://www.cbioportal.org/ |
| International Cancer Genome Consortium (ICGC) Donors~ 24,500 |
Catalog of mutational abnormalities in the major tumor types | Yes | Yes | Yes | F, A, I, R | https://dcc.icgc.org/ |
| Human Genome Browser at USCS | Genomic data | Yes | Yes | F, A, R | https://genome.ucsc.edu/index.html | |
| Gene Expression Omnibus Database (GEO) | Gene expression data | Yes | Yes | F, A, R | https://www.ncbi.nlm.nih.gov/geo/ | |
| Ensembl | Genomic analysis | Yes | Yes | F, A, R | https://www.ensembl.org/index.html | |
| Roche Cancer Genome Database (RCGDB) | ||||||
| National Cancer Institute Genomic Commons (GDC) Cases= 22,000 |
Storage, analysis, and sharing of clinical data of patients | Yes | Yes | Yes | F, A, I, R | https://portal.gdc.cancer.gov/ |
| Network of Cancer Genes | Cancer genes, healthy drivers and their properties | Yes | Yes | Yes | F, A, I, R | http://ncg.kcl.ac.uk/index.php |
| CancerGenes | Could not find | |||||
| Catalogue of Somatic Mutation in Cancer (COSMIC) | Genetic mechanisms that promote cancer | Yes | Yes | Yes | F, A, I, R | https://cancer.sanger.ac.uk/cosmic |
| Mutagene | Mutational profiles in 37 cancer types | Yes | Yes | Yes | F, A, I, R | https://www.ncbi.nlm.nih.gov/research/mutagene/ |
| Progenetix Samples= 142,063 |
Cancer Copy Number Abnormalities (CNA) | Yes | Yes | Yes | F, A, I, R | https://progenetix.org/ |
| MutEx | Records the relationships between gene expression, somatic mutation, and survival data | Yes | Yes | Yes | F, A, I, R | http://www.innovebioinfo.com/Databases/Mutationdb_About.php |
| Oncomine | Precision oncology | Yes | Yes | Yes | F, A, I, R | https://www.oncomine.com/ |
| CancerResource | Server taken down | |||||
| Comparative Toxicogenomic Database (CTD) | Toxicological information | Yes | Yes | Yes | F, A, I, R | http://ctdbase.org/ |
| Therapeutic Target Database (TTD) | Pathway information and the drug/ligands directed at each target | Yes | Yes | Yes | F, A, I, R | https://db.idrblab.net/ttd/ |
| Pharmacogenomics Knowledge Base (PharmGKB) | Genotype, molecular, and clinical knowledge integrated into pathway representation | Yes | Yes | F, A, I, R | https://www.pharmgkb.org/ | |
| DrugBank | Molecular information about drugs, mechanisms, and interactions | Yes | Yes | F, A, I, R | https://go.drugbank.com/ | |
| Lung Cancer Explore (LCE) Entries= 356 |
Molecular information about drugs including interactions and targets | Yes | Yes | F, A, R | https://lce.biohpc.swmed.edu/lungcancer/imageset_tcga.php | |
| Prostate Cancer Precision Medicine Multi-Institutional Collaborative Effort PROMISE | DNA kit, analyzes genes and patient outcomes | Yes | Yes | F, A, I | https://www.prostatecancerpromise.org/research/ | |
| HCCDb | Contains information on hepatocellular carcinoma | Yes | Yes | F, A, R | http://lifeome.net/database/hccdb/home.html | |
| HCCDb | Contains information on hepatocellular carcinoma | Yes | Yes | F, A, R | http://lifeome.net/database/hccdb/home.html |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Clinical Proteomic Tumor Analysis Consortium (CPTAC) | Analyzes cancer biospecimens using mass spectrometry | Yes | Yes | Yes | F, A, I, R | https://proteomics.cancer.gov/programs/cptac |
| String Database | Protein interactions | Yes | Yes | Yes | F, A, I, R | https://string-db.org/ |
| Ualcan | Analyzes and delivers cancer transcriptome, proteomics, and patient survival | Yes | N/A | Yes | F, A, I | https://ualcan.path.uab.edu/ |
| CanProVar | Proteomic variations | Yes | Yes | F, A, R | http://119.3.70.71/CanProVar/index.html | |
| RCSB Protein Data Bank | Works with UniProt and looks at structures of proteins | Yes | Yes | Yes | F, A, I, R | https://www.rcsb.org/ |
| Universal Protein Resource (UniProt) | Contains protein structure and interaction | Yes | Yes | Yes | F, A, I, R | https://www.uniprot.org/ |
| Proteome Discover | Not free to access | |||||
| Swiss-Prot and TrEMBL | A part of the UniProt database | Yes | Yes | Yes | F, A, I, R | https://www.uniprot.org/uniprotkb?query=%2A |
| jPOST | Post-translational modifications on proteins | Yes | Yes | Yes | F, A, I, R | https://globe.jpostdb.org/ |
| MatrisomeDB | Proteomic data from studies on ECM | Yes | Yes | F, A, R | https://matrisomedb.org/ |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| DBLiPro | Yes | Yes | Yes | F, A, I, R | http://lipid.cloudna.cn/home | |
| Lipid Maps | Yes | Yes | Yes | F, A, I, R | https://www.lipidmaps.org/ |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| GlycoSuite | Was not able to find | |||||
| UniCarb-db | Carbohydrates characterized by LC-MS | Yes | Yes | Yes | F, A, I, R | https://unicarb-db.expasy.org/ |
| EuroCarbDB | Was not able to find | |||||
| UniPep | N-linked glycosites for proteomic analyses | Yes | Yes | F, A, R | https://unipep.systemsbiology.net/ | |
| GlycoGene (GGDB) | Contains all the information on glycogenes | Yes | Yes | F, A, R | https://www.glycogene.com/ | |
| Glycome-DB | A part of GlyTouCan database | http://www.glycome-db.org/ | ||||
| Glycobase | Was not able to find | |||||
| GlycoStore | Was not able to find | |||||
| GlycoRDF | Holds glycan publications and experimental data | Yes | Yes | F, A, R | https://github.com/glycoinfo/GlycoRDF/wiki | |
| GRITs Toolbox | Allows for archiving of research papers | Yes | Yes | Yes | F, A, I, R | http://www.grits-toolbox.org/ |
| GlyTouCan | Databases for publications and journals within glycan research | Yes | Yes | F, A, R | https://glytoucan.org/ | |
| The Lectin Frontier Database (LfDB) | Lectin-standard oligosaccharide interactions | Yes | F, A | https://acgg.asia/lfdb2/ | ||
| Carbohydrate Structure Database (CSDB) | Structural and biographical components of glycans | Yes | Yes, this is done by an external source | F, A | http://csdb.glycoscience.ru/database/index.html? help=credits |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Clinical Genomic Database (GCD) | Genetic information that pertains to patient care | Yes | Yes | F, A, R | https://research.nhgri.nih.gov/CGD/ | |
| Foundation Medicine Adult-Cancer-Clinical Dataset | Clinical relevance among rare alterations and diseases | Yes | Yes | Yes | F, A, I, R | https://gdc.cancer.gov/about-gdc/contributed-genomic-data-cancer-research/foundation-medicine/foundation-medicine |
| A Curated Cancer Clinical Outcome Database (C3OD) | Cannot access | |||||
| Danish Head and Neck Cancer Database | Contains patient data to be used in improved wait times | Yes | Yes | F, A, R | https://www.dahanca.dk/IndexPage | |
| National Cancer Database (NCDB) | Have to login for access | F | https://www.facs.org/quality-programs/cancer-programs/national-cancer-database/ | |||
| Surveillance Epidemiology and End Results (SEER) | Focus on colorectal cancer and improvements of patient care | Yes | F, A | https://seer.cancer.gov/ | ||
| ClinVar | Allows for the comparison of data among researchers | Yes | Yes | F, A, R | https://www.ncbi.nlm.nih.gov/clinvar/ |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Database of Epigenetics Modifiers (dbEM) | Contains genomic information on epigenetic modifiers/ proteins | Yes | F, A | https://webs.iiitd.edu.in/raghava/dbem/index.php | ||
| Cancer Research Database (CRDB) | Holds other databases in the fields of genomic, proteomic, mutations, etc. | Yes | F, A | https://www.habdsk.org/crdb | ||
| PROGgene | Prognosis | Yes | Yes | F, A, I | http://www.progtools.net/gene/index.php | |
| Cancer Drug Resistance (CancerDR) | Yes | Yes | F, A, R | https://webs.iiitd.edu.in/raghava/cancerdr/index.html | ||
| DriverDBv3 | Yes | Yes | Yes | F, A, I, R | http://driverdb.tms.cmu.edu.tw/ | |
| LncRNA2Target 2.0 | Was not able to access | |||||
| Lnc2Cancers 3.0 | Was not able to access | |||||
| Genotype Expression Project (GTEx) | Evaluate the relationships between genetic variations and gene expressions | Yes | Yes | F, A, R | https://www.gtexportal.org/home/ |
| Databases | Content | Web service | Downloadable | Analytics | Fairness | Website |
|---|---|---|---|---|---|---|
| Gene Set Cancer Analysis (GSCALite) | Analyzes gene and survival rates | Yes | F, A | http://bioinfo.life.hust.edu.cn/web/GSCALite/ | ||
| Online Mendelian Inheritance in Man (OMIM) | Includes multiple resources on genetic phenotype, DNA, proteins, etc. | Yes | Yes | F, A, R | https://www.omim.org/ | |
| Gene Expression Profiling Interactive Analysis (GEPIA) | Gene expression analysis, correlations analysis, and patient survival | Yes | Yes | Yes | F, A, I, R | http://gepia.cancer-pku.cn/ |
| PepQuery | Proteomic validations of genomic alterations | Yes | Yes | F, A, R | http://www.pepquery.org/ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





