Introduction
The feet must bear the weight of the entire body, and the talus, as a connector between the foot and leg that makes up the lower part of the ankle joint, helps to transfer pressure forces from the upper body [
1]. Due to regularly arising disturbances of normal muscle and talus collapse, structural and postural abnormalities in the feet regularly strain the lumbar spine, resulting in chronic low back pain with increasing age [
2].
The collapse of the talar dome leads to degenerative changes and disability of the ankle, and eventually avascular necrosis. Surgical treatment usually applies ankle fusion to delay the development of symptoms [
3]. However, the bones may not join together properly due to the anatomical position and weak blood supply of the talus [
3,
4]. Therefore, talus implant treatment is a suitable way to relieve pain while preserving functional range of motion so patients will have less pain and better function during activity [
5,
6]. Custom total talus replacement, which involves designing a prosthesis component with tailored dimensions, is a viable alternative [
7,
8,
9]. Nevertheless, quantifying variations of the talus is fundamental before talus implant design. When the surgeon is satisfied with the talar implant, a 3D model is used to recreate the talar surface and design the implant accurately.
Rendered 3D images provide detailed information for quantifying the talus [
10]. Thin-slice computed tomography (CT) is a favorable candidate for 3D rendering, especially for bony structures. The most crucial step towards designing a custom-made talar body prosthesis is to achieve automatic segmentation of the talus on CT slice by slice. However, manual segmentation is time-consuming and subjective due to the enormous amount of image data and differences in expert experience. Recently, deep learning has emerged as a promising technique to achieve semantic segmentation, help doctors diagnose the type of disease, and evaluate the effects of treatment. Compared with traditional machine learning, it has advantages in accuracy and efficiency of segmentation [
11,
12,
13].
The increase in segmentation has led to increased demand for automatic segmentation techniques. Waseem et al. [
14] proposed a segmentation algorithm that uses a probability-based variation of the watershed transform to perform segmentation based on the intensity and position information of the image pixels in a fractured bone. Ida et al. [
15] presented a structure-based algorithm for separating cortical bone from the trabecular bone in binarized images. Jingting et al. [
16] employed a statistical shape model based on the Bayesian model to improve deep neural network segmentation in pancreas CT images. The experimental results demonstrated the superior performance of their method in localizing the blurry boundaries. In the field of bone segmentation, Wahyu et al. [
17] proposed a method for the automated segmentation and detection of calcaneal fractures for real-time application, highlighting bone fractures in processed images. Many researchers have focused on deep learning approaches that promote dataset statistics and domain-specific segmentation. Arnuad et al. [
18] optimized a single segmentation network over the union of pediatric imaging datasets acquired on separate anatomical regions in MR images. Courtis et al. [
19] evaluated the accuracy of the X-ray image segmentation system for a patient-specific instrument that requires 3D modeling of the knee. Their results demonstrated that X-ray-based 2D/3D segmentation is a viable tool in orthopedic applications requiring accurate 3D segmentation of knee bones. Fabian et al. [
20] proposed a framework to segment brain tumors, and it achieved DSC of 88.65, 85.06, and 82.03 for tumor segmentation.
To attain better talus segmentation than the methods presented above, it is necessary to more comprehensively characterize the shape of the talus by accurately measuring the anatomical parameters of the talus and utilizing a deep fusion model. In this work, we investigated nnU-Net’s suitability for talus segmentation and exploited an efficiency framework to achieve talus segmentation as a technology supporting the treatment of scoliosis deformity in clinical trials.
Materials and Methods
Manual Annotations
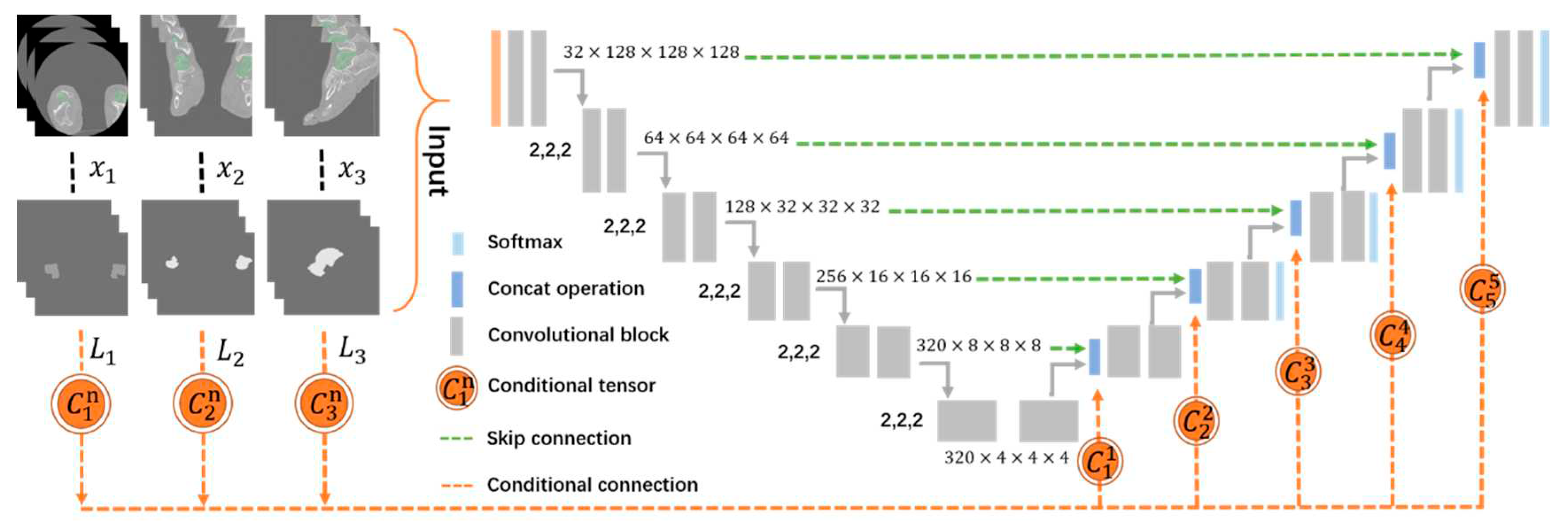
Initially, 80 cases were identified, but 6 cases were excluded because they had poor image quality. Finally, 74 CT scans of healthy subjects aged 18–84 years were collected from Southern University of Science and Technology Hospital. A set of 512 × 512 images was included, and slice thickness ranged from 0.450–1.250 mm. The radiologist screened all talus CT scans to ensure the subjects were normal. The talus was first segmented from the hindfoot by setting the threshold in Mimics 19.0 to 266 HU. The segmented regions were reconstructed to acquire the 3D morphologies of the talus separated from the surrounding tissue. To obtain a geometric model with high-quality surfaces, 148 reconstructed talus models were smoothed and adjusted to a smoothing degree at 0.8 mm. The 3D objects were saved in STL format on Mimics and translated to NIFTI images (nii format) on Slicer for training. The talus CT datasets were first converted into NIFTI images using Python. Additionally, orthopedists manually segmented the datasets slice by slice in Slicer 5.0.3 and evaluated the segmentation results in follow-up, as shown in
Figure 1.
Network Architecture
We chose nnU-Net as the segmentation framework, which is a deep learning-based segmentation network that can be used to perform tasks in the biomedical domain [
20]. The adopted network consists of an encoder and a decoder, which are interconnected by skip connections forming a 3D U-Net pattern. It only relies on plain convolutions for feature extraction and does not apply any of the recently proposed architectural variations. We merged the feature maps of the two parts through a cascade operation that could bring richer semantic information and lead to higher-accuracy talus segmentation. We implemented the nnU-Net models on the PyTorch deep learning platform on a PC with an Intel Core i7-10700K CPU. The input patch size was 128 × 128 × 128, the batch size was 2, and training ran for 1000 epochs to ensure the whole talus was in the middle of the patch. Then, the segmented parts were reassembled by zero-padding that considered the localization of the extracted features.
Results
The practicality of nnU-Net was verified by its accuracy in talus segmentation. To investigate the performance of nnU-Net, we performed five-fold cross-validation on the training dataset, which was composed of 74 CT scans of a healthy talus. Examination of the original CT images revealed that the talus could be more easily distinguished from other bones. We applied three configurations of U-Net to evaluate which one had the best network architecture, and the three U-Net configurations performed well, resulting in DSCs of 0.982, 0.975, and 0.983 and HD95 values of 0.562, 0.611, and 0.566, respectively. The evaluation metric statistics, presented in
Table 1, indicate that the proposed method performs well on the whole shape of the talus. The DSC and HD95 were observed between the estimated gold standard.
Table 2 shows the inference time in seconds. The 3D full resolution U-Net was nearly 14 times faster than manual segmentation, and the 3D U-Net cascade (3D low-resolution U-Net) was 10 times faster than the 3D full resolution U-Net. Additionally, in 3D U-Net cascade, 3D full resolution U-Net performed better in talus segmentation than 3D low-resolution U-Net, but 3D full resolution U-Net takes too much time. The 3D full-resolution U-Net demonstrated high accuracy in the detection of small bone spurs.
Conclusion and discussion
Conclusion
The segmentation of talus structure continues to be a critical component to help the physician understand variations in bone with increasing age, even early interventions to prevent scoliosis deformity. In this work, we developed a nnU-Net-based method for segmentation of the talus bone in CT images of Chinese people. The segmentation performance of the 3D full resolution U-Net was better than the other two configurations but inferior to 3D full resolution U-Net (3D U-Net cascade). The proposed three configurations of the U-Net network architecture achieved a DSC of 0.982±0.002 and HD95 of 0.562±0.059 on the experimental dataset. The results presented in
Table 1 demonstrate that the framework outperformed existing networks for labeled bone CT data learning. The effect of nnU-Net segmentation was almost the same as that of manual segmentation, so it was validated for the automatic segmentation of the talus bone, improving the workflow and efficiency of talus implant treatment planning for early intervention in scoliosis deformity.
Discussion
In this study, we proposed an automatic segmentation algorithm to generate a segmented mask that compares manually segmented masks and automatically generated masks in talus CT scans. Robust and accurate segmentation is essential to talus treatments, such as diagnostic intervention and computer-aided implant design. Our research indicated that the conditional nnU-Net could effectively utilize the prior conditional information to categorize the location of bone at the pixel level, thereby significantly improving the spatial information recovery and segmentation accuracy. Moreover, compared with Engström Messén research [
12], our research methods show a high accuracy in talus segmentation. We demonstrated that U-net configurations be successfully applied for the talus segmentation task, allowing CT images to learn local and global contexts for talus segmentation at multiple scales. The performance of nnU-Net was superior to manual segmentation in terms of inference time and DSC score. Although nnU-Net performs well in other types of image segmentation (such as brain tumor segmentation), it performs extremely well in bone segmentation. We tested the performance of three configurations in segmenting the talus based on DSC score and HD95. Furthermore, we investigated the inference time of the three different configurations. The results in
Table 1 show that 3D low-resolution U-Net converged faster than the other configurations and would be more suitable for talus segmentation. All three configurations had much higher DSC scores than manual segmentation did. Additionally, the generalizability of nnU-Net was demonstrated by its good segmentation in CT scans. The performance of the three configurations was close to the ground truth, which shows high accuracy in talus segmentation.
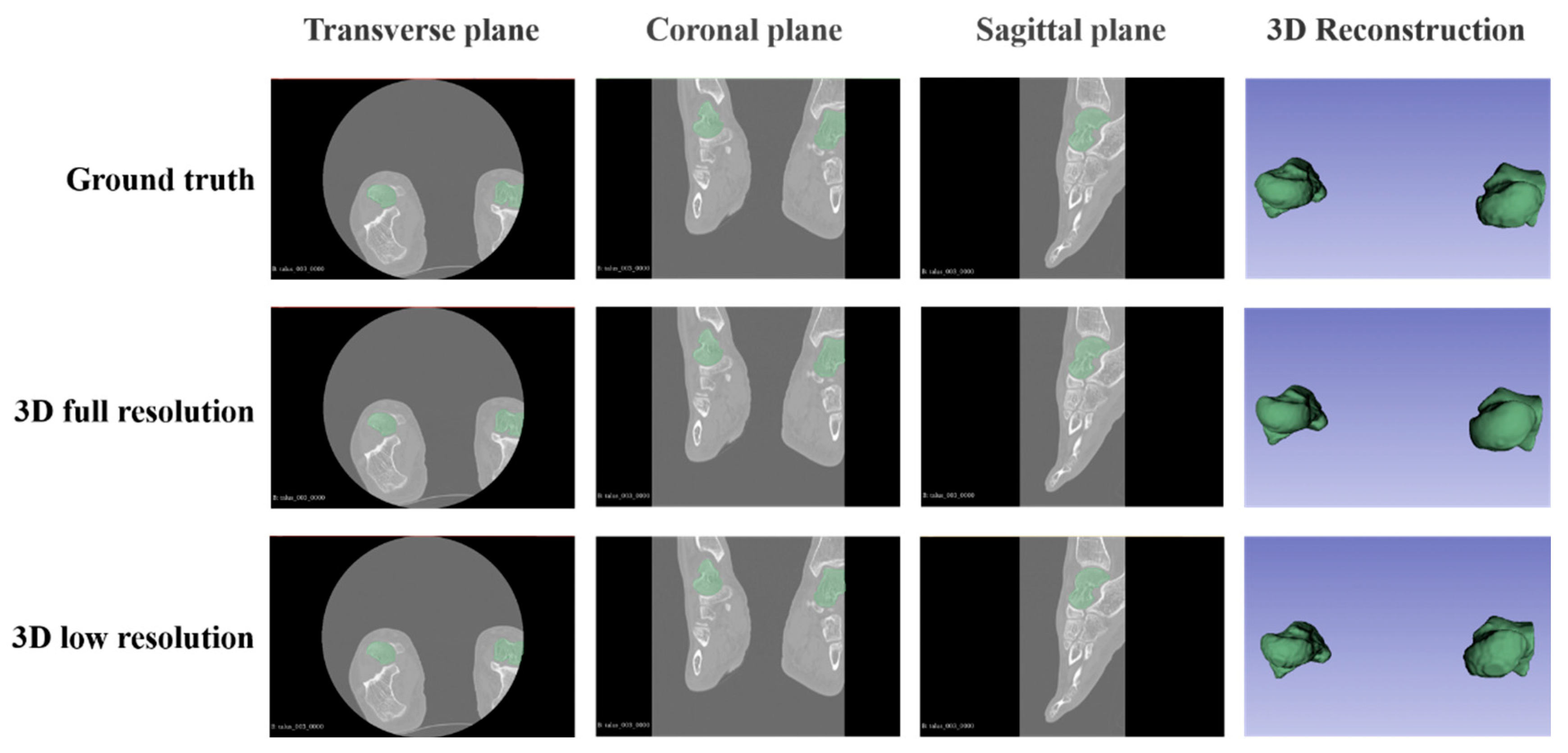
Figure 2 shows that all configurations returned perfect masks and 3D models and made it easy to distinguish the bone. According to the results, both configurations had almost the same performance, resulting in a perfect and robust model. The talus was automatically characterized by U-Net and included the results of DSC and HD95. The 3D full-resolution U-Net also proved its high accuracy in the detection of small bone spurs. The performance results of all network architectures met the requirements of automatic talus segmentation. And the automatically segmented 3D models of talus will be applied in the pre-planning of individualized talus implants as an approach to prevent scoliosis deformity.
In the future, more types of talus fracture CT scan images with different features will be used to evaluate the performance of U-Net. The improved accuracy of automatic segmentation will facilitate the development of talus fracture classification, which can be extended to diagnosing suspicious fractures and designing talus implants in the clinical trial. It will expand the conditional strategy of talus and calcaneus segmentation and develop a complete segmentation system for at-risk organs in foot treatment planning. This is a challenging task due to heterogeneity. Furthermore, a statistical shape model of the talus will be generated to adapt talus implants for Chinese patients, which could be an approach to prevent scoliosis deformity. This work will focus on talus anatomic variations and evaluate the statistical shape model, which suggests that the framework could be used in the early prevention of spine disorders.
Author Contributions
Jie He and Zhexiao Guo contributed equally.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
- Zang, J., et al., The treatment of neurotrophic foot and ankle deformity of spinal bifida: 248 cases in single center. Journal of Neurorestoratology, 2019. 7(3): p. 153-160.
- Carroll, L.A., S. Paulseth, and R.L. Martin, Forefoot Injuries in Athletes: Integration of the Movement System. International Journal of Sports Physical Therapy, 2022. 17(1): p. 81.
- Rozis, M., et al., Results and outcomes of combined cross screw and ilizarov external fixator frame in ankle fusion. The Journal of Foot and Ankle Surgery, 2020. 59(2): p. 337-342.
- Knupp, M. , Diffuse Ankle Osteoarthritis, in Foot and Ankle Disorders. 2022, Springer. p. 723-742.
- Kadakia, R.J. , et al., 3D printed total talus replacement for avascular necrosis of the talus. Foot & Ankle International, 2020. 41(12): p. 1529-1536.
- Scott, D.J., et al., Early outcomes of 3D printed total talus arthroplasty. Foot & Ankle Specialist, 2020. 13(5): p. 372-377.
- West, T.A. and S.M. Rush, Total talus replacement: case series and literature review. The Journal of Foot and Ankle Surgery, 2021. 60(1): p. 187-193.
- Strand, G., C. Juels, and J. Nowak, Custom total talus replacement as a salvage option for failed total ankle arthroplasty: A prospective report of two cases. Foot & Ankle Surgery: Techniques, Reports & Cases, 2022. 2(1): p. 100113.
- Lullini, G., et al., Custom-Made Total Talonavicular Replacement in a Professional Rock Climber: Functional Evaluation With Gait Analysis and 3-Dimensional Medical Imaging in Weightbearing at 5 Years’ Follow-Up. The Journal of Foot and Ankle Surgery, 2020. 59(5): p. 1118-1127.
- Grau, D., et al., A 3D-Printed Model of a Titanium Custom-Made Talus for the Treatment of a Chronic Infection of the Ankle. The Journal of Foot and Ankle Surgery, 2022. 61(1): p. 212-217.
- Wang, G., et al., Automatic Detection of Osteochondral Lesions of the Talus via Deep Learning. Frontiers in Physics, 2022: p. 113.
- Engström Messén, M. and E. Moser, Pre-planning of Individualized Ankle Implants Based on Computed Tomography-Automated Segmentation and Optimization of Acquisition Parameters. 2021.
- Liu, X., et al., A review of deep-learning-based medical image segmentation methods. Sustainability, 2021. 13(3): p. 1224.
- Shadid, W.G. and A. Willis, Bone fragment segmentation from 3D CT imagery. Computerized Medical Imaging and Graphics, 2018. 66: p. 14-27.
- Ang, I.C., et al., An algorithm for automated separation of trabecular bone from variably thick cortices in high-resolution computed tomography data. IEEE Transactions on Biomedical Engineering, 2019. 67(3): p. 924-930.
- Ma, J., et al. A novel bayesian model incorporating deep neural network and statistical shape model for pancreas segmentation. in International Conference on Medical Image Computing and Computer-Assisted Intervention. 2018. Springer.
- Rahmaniar, W. and W.-J. Wang, Real-time automated segmentation and classification of calcaneal fractures in CT images. Applied Sciences, 2019. 9(15): p. 3011.
- Boutillon, A., et al., Multi-Task, Multi-Domain Deep Segmentation with Shared Representations and Contrastive Regularization for Sparse Pediatric Datasets. arXiv preprint arXiv:2105.10310, 2021.
- Courtis, P., et al. ACCURACY EVALUATION OF AN X-RAY-BASED 2D/3D KNEE SEGMENTATION SYSTEM. in Orthopaedic Proceedings. 2017. The British Editorial Society of Bone & Joint Surgery.
- Isensee, F., et al. nnU-Net for brain tumor segmentation. in International MICCAI Brainlesion Workshop. 2020. Springer.
- Fick, T., et al., Fully automatic brain tumor segmentation for 3D evaluation in augmented reality. 2021. 51(2): p. E14.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).