Introduction
The multi-functional scaffold IQGAP1 is a 190 kDa scaffold protein expressed in several cell types 1, 2, 3, 4, 5. There are three members in the IQGAP family, conserved in humans and mice 6. IQGAP1 is ubiquitously expressed 7, while IQGAP2 is found to be expressed in the liver, platelets, kidney, stomach, prostate, thyroid, and salivary glands 8, 9, 10, 11. In contrast, the expression of IQGAP3 is different from the other two isoforms, primarily found in the brain, lung, testis, small intestine, and colon 8, 12. Murine IQGAP1 and IQGAP2 isoforms share 52% of their amino acid identities with a 68% overall protein homology. IQGAP1 and IQGAP3 isoforms display 57% amino acid and 74% overall protein homology 13. IQGAP1 possesses six well-defined interactive domains (Figure 1A), which interact with numerous cellular protein partners 14. Among lymphocytes, murine NK, B, and T cells express IQGAP115, 16. Earlier study has characterized the role of IQGAP1 in murine B cells in the context of Rap1-cofilin-1 pathway in actin reorganization and MTOC polarization17. However, its role in B cell development has not been explored.
Figure 1.
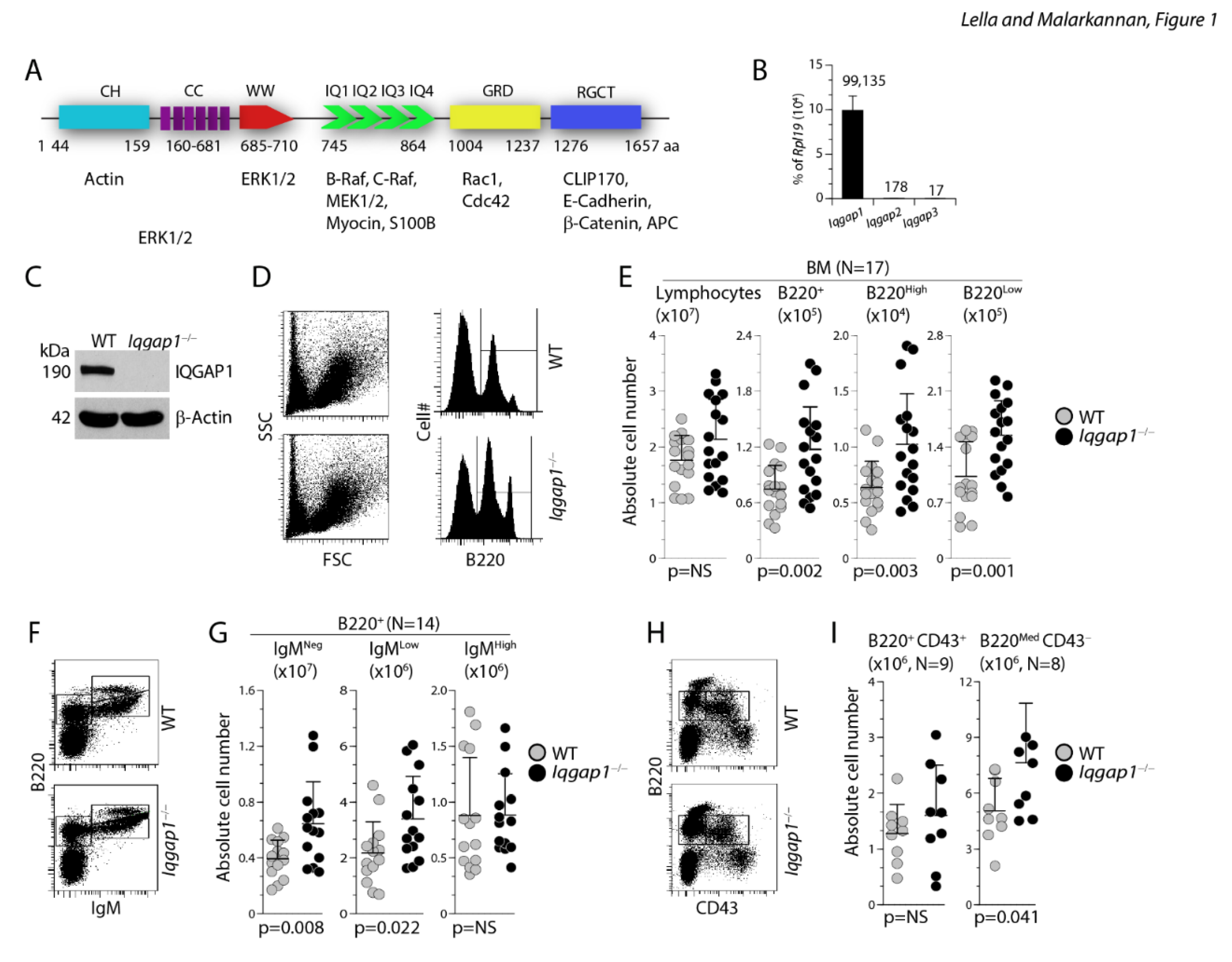
Lack of IQGAP1 results in dysregulated early B cell development in the BM. A) Calponin homology (CH) domain regulates the cytoskeleton by binding to N-WASp and F-actin. Coiled-coil (CC) regions, comprised of conserved amino acid repeats, bind to Ezrin. WW domain is involved in recruiting Erk1/2 to facilitate a sequential phosphorylation cascade. Isoleucine/glutamine-containing (IQ) domains recruit Rap1a, Rap1b, B-Raf, C-Raf, Mek1, and Mek2 to promote phosphoinositide (PIPKIγ) and calcium signaling. The Ras-GAP domain (GRD) interacts with small GTPases Cdc42, Rac1, and TC10. The Ras-GAP C-terminus (RGCT) domain interacts with CLIP-170, Clasp2, β-catenin, E-cadherin, and APC. B) Sorted total B220+ resting B cells from the BM were analyzed for the expression levels of Iqgap1, Iqgap2, and Iqgap3 transcripts using RTqPCR. Relative expression of these transcripts is shown as percent of Rpl19 house-keeping gene. The data shown are the average of three independent experiments. C) Expression of IQGAP1 protein in the WT and Iqgap1−/− mice were analyzed by western blot using an anti-IQGAP1 antibody. Level of β-actin was used as loading controls. Blots are a representative set from three different experiments. D, E) Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells. Staining with anti-B220 antibody indicated the presence of both B220High and B220Low B cells in both the WT and Iqgap1−/− mice. F, G) Absolute numbers of B220+IgM− pro/pre-B, B220+IgM+ immature B, and B220Hi recirculating B cells were defined using anti-B220 and anti-IgM antibodies, and their numbers were calculated from live lymphocyte gates. H, I) The absolute numbers of pro-B cells were further identified with anti-B220 and anti-CD43 antibody staining. Pro-B cells are B220MedCD43+, and pre-B cells are B220+CD43−. Data shown in C, E, and G are representative panels. Open and filled circles in D, F, and H are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 1.
Lack of IQGAP1 results in dysregulated early B cell development in the BM. A) Calponin homology (CH) domain regulates the cytoskeleton by binding to N-WASp and F-actin. Coiled-coil (CC) regions, comprised of conserved amino acid repeats, bind to Ezrin. WW domain is involved in recruiting Erk1/2 to facilitate a sequential phosphorylation cascade. Isoleucine/glutamine-containing (IQ) domains recruit Rap1a, Rap1b, B-Raf, C-Raf, Mek1, and Mek2 to promote phosphoinositide (PIPKIγ) and calcium signaling. The Ras-GAP domain (GRD) interacts with small GTPases Cdc42, Rac1, and TC10. The Ras-GAP C-terminus (RGCT) domain interacts with CLIP-170, Clasp2, β-catenin, E-cadherin, and APC. B) Sorted total B220+ resting B cells from the BM were analyzed for the expression levels of Iqgap1, Iqgap2, and Iqgap3 transcripts using RTqPCR. Relative expression of these transcripts is shown as percent of Rpl19 house-keeping gene. The data shown are the average of three independent experiments. C) Expression of IQGAP1 protein in the WT and Iqgap1−/− mice were analyzed by western blot using an anti-IQGAP1 antibody. Level of β-actin was used as loading controls. Blots are a representative set from three different experiments. D, E) Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells. Staining with anti-B220 antibody indicated the presence of both B220High and B220Low B cells in both the WT and Iqgap1−/− mice. F, G) Absolute numbers of B220+IgM− pro/pre-B, B220+IgM+ immature B, and B220Hi recirculating B cells were defined using anti-B220 and anti-IgM antibodies, and their numbers were calculated from live lymphocyte gates. H, I) The absolute numbers of pro-B cells were further identified with anti-B220 and anti-CD43 antibody staining. Pro-B cells are B220MedCD43+, and pre-B cells are B220+CD43−. Data shown in C, E, and G are representative panels. Open and filled circles in D, F, and H are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Due to its ability to directly bind and cross-link F-actin, IQGAP1 plays an essential role in cytoskeletal maintenance and cell integrity, filopodia formation, and directional movement 18, 19. Our earlier study identified an IQGAP1-based master signalosome upon receptor-mediated activation. This signalosome localized in the perinuclear region of murine NK cells coordinated the sequential phosphorylation of Raf→MEK1/2→ERK1/2 15, 16. The functional domains of IQGAP1 regulate several vital cellular processes, including actin/tubulin-mediated cytoskeletal remodeling, Erk1/2-mediated c-Fos, c-Jun, and Elk1 activation β-catenin-mediated LEF and TCF activation, and calmodulin-mediated Ca2+ signaling. Irrespective of these findings, the precise functions of IQGAP1 in B cells are yet to be determined.
Here, using wild-type (WT) and Iqgap1−/− mice, we found that lack of IQGAP1 significantly altered the development and function of different B cell subsets. The bone marrow (BM) of Iqgap1−/− mice contained significantly increased numbers of B220+ B, B220+IgM− pro/pre-B, and B220+IgM+ immature-B cells. The new-forming (NF) and follicular (FO) B cell numbers were increased, while the marginal zone (MZ) B cell numbers were significantly reduced in the spleen of the Iqgap1−/− mice. These developmental defects are B cell-intrinsic, and the absence of IQGAP1 significantly reduced T-dependent (TD) and T-independent (TI) humoral responses. Mechanistically, lack of IQGAP1 resulted in the reduction of Mek1/2, Erk1/2, and Jnk1/2 phosphorylation following anti-IgM mAb-mediated activation. In addition, lack of IQGAP1 resulted in a substantial reduction in Irf4 in both transcript and protein levels. B cells from Iqgap1−/− mice failed to suppress IL-7R-mediated activation of Stat5a and Stat5b, which resulted in a higher number of pro and pre-B cells. Analyses of recombining sequence (RS) rearrangement frequency demonstrate a moderate reduction in the absence of IQGAP1. Our study provides the first evidence that IQGAP1-based signalosome is essential for the development and functions of B cells.
Results
B cell development in the BM of Iqgap1−/− mice is altered
To investigate the role of IQGAP1 in B cell development, we analyzed the absolute numbers of B cells in the BM. The absolute numbers of total lymphocytes in the BM of Iqgap1−/− mice were comparable to that of WT (Figure 1D, E). Irrespective of this, the absolute numbers of B220+ B cells were significantly increased in the BM of Iqgap1−/− mice compared to that of WT (Iqgap1−/−: 11.74 ± 4.72×104; WT: 7.46 ± 2.62×104; n=17, 17; and p=0.002) (Figure 1D, E). B220+ B cells can be further divided into B220High and B220Low subsets. Iqgap1−/− mice exhibited significant increases in both B220High (Iqgap1−/−: 10.2 ± 4.5×105; WT: 6.4 ± 2.3×105; n=17, 17; and p=0.003) and B220Low subsets (Iqgap1−/−: 1.55 ± 0.44×104; WT: 1.02 ± 0.43×104; n=17, 17; and p=0.001). Based on B220 and IgM expression, BM B cells are classified into pro/pre-B (B220+IgM−), immature B (B220+IgM+), and recirculating mature B cells (B220Hi). The absolute numbers of B220+IgM− pro/pre (Iqgap1−/−: 6.45 ± 3.0×104; WT: 3.94 ± 1.3×104; n=14, 14; and p=0.008) and recirculating mature B220+IgMLow population (Iqgap1−/−: 3.4 ± 1.5×104; WT: 2.17 ± 1.11×104; n=14, 14; and p=0.022) were significantly increased in Iqgap1−/− mice compared to WT (Figure 1F, G). In contrast, the number of B220+IgM+ immature B cells was comparable between Iqgap1−/− and WT mice (Iqgap1−/−:0.89 ± 0.37×104; WT: 0.88 ± 0.52×104; n=14, 14; and p=NS) (Figure 1G). B cell development in BM can be further characterized through the expression of CD43 (Leukosialin), CD117 (c-Kit), and CD25 (IL-2Rα). Early Pro-B cells are B220+CD43+, which develop into B220+CD43− B cells. The absolute B220+CD43+ pro-B cell numbers did not differ significantly between Iqgap1−/− and WT (Iqgap1−/−: 1.6 ± 0.89×106 and WT: 1.3 ± 0.52×106; n=9, 9; p=0.36) (Figure 1H, I).
Lack of IQGAP1 forms a blockade at early B cell development, and it is cell-autonomous
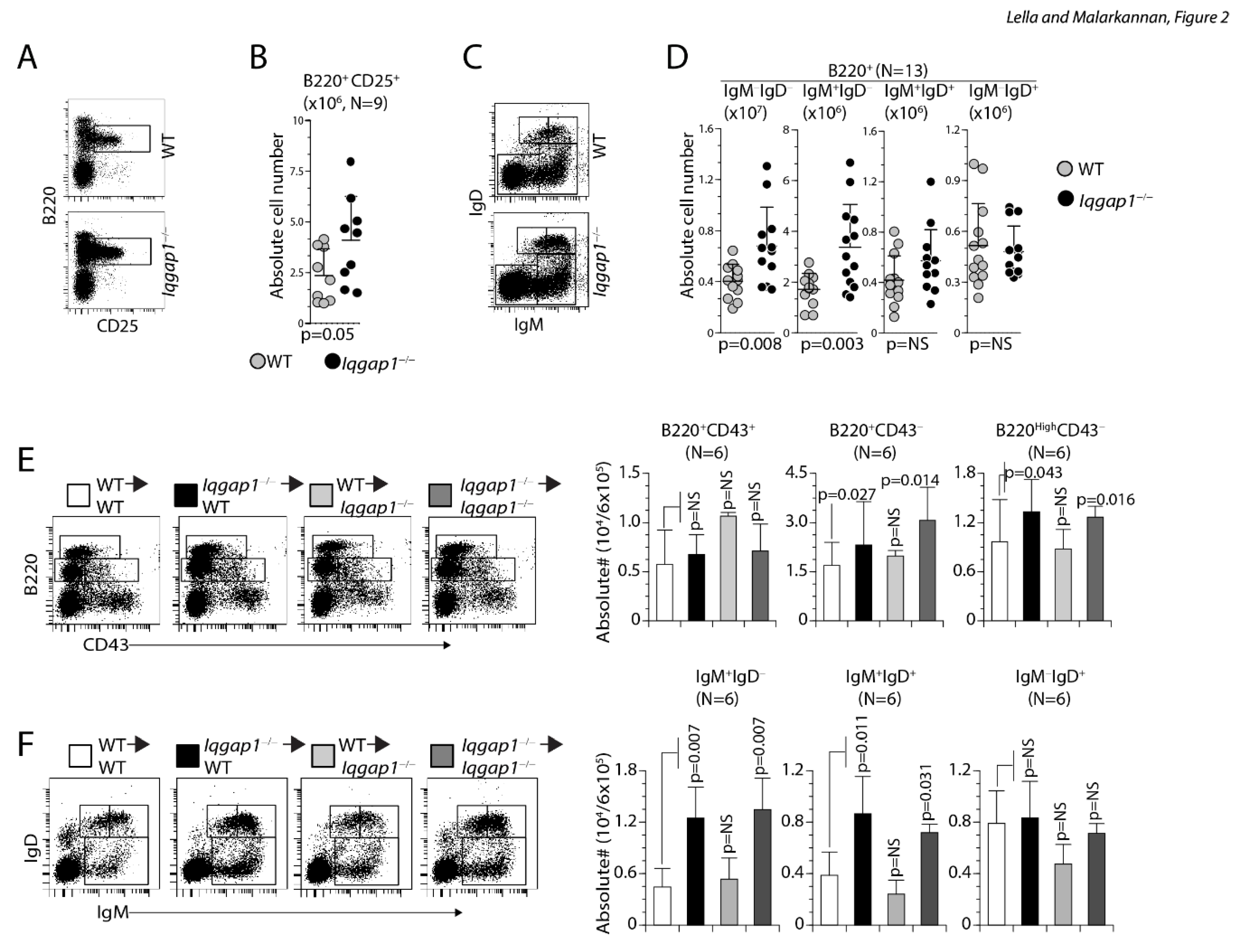
However, B220MedCD43− pre-B cells were significantly higher in Iqgap1−/− (Iqgap1−/−: 7.6 ± 3.2×106 and WT: 5.0 ± 1.7×106; n=8, 8; p=0.041), which is consistent with the increased B220+IgM− pro/pre-B cells populations. As pro-pre-B-I cells mature into pre-B-II (large and small), c-kit expression decreases and CD25 level increases. In this context, we also observed a significantly increased B220+CD25+ pre-B-II population (Iqgap1−/−: 4.10 ± 2.16×106 and WT: 2.35 ± 1.31×104; n=9, 9; p=0.05) in Iqgap1−/− mice (Figure 2A, B). BM-derived cells were also stained with anti-B220, anti-IgM, and anti-IgD Abs and analyzed. The absolute numbers of IgM+IgD+ (Iqgap1−/−: 0.57 ± 0.25×104and WT: 0.42 ± 0.19×104; n=13, 13; p=NS) and IgM−IgD+ (Iqgap1−/−: 0.48 ± 0.15×104 and WT: 0.52 ± 0.25×104; n=13, 13; p=NS) B cells were comparable between Iqgap1−/− and WT mice (Figure 2C, D). However, both the immature IgM−IgD− (Iqgap1−/−: 6.8 ± 3.0×104 and WT: 4.0 ± 1.3×104; n=13, 13; p=0.008) and IgM+IgD− (Iqgap1−/−: 3.38 ± 1.68×104 and WT: 1.71 ± 0.62×104; n=13, 13; p=0.003) B cells were increased in Iqgap1−/− mice compared to that of WT (Figure 2C, D), indicating a loss of developmental regulation during the early stages of B cell maturation.
Figure 2.
Lack of IQGAP1 leads to a potential blockade at pre-B cell stages in the BM and the defects in the transition of pro-B to pre-B cells in the BM of Iqgap1−/− mice is cell-intrinsic. A, B) Absolute numbers of B220+CD25+ large and small pre-B-II stage. C, D) Absolute numbers of B220+IgM−IgD− or B220+IgM+IgD− immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. BM cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies, and the absolute numbers were calculated per million total gated lymphocytes. Data shown in A, C, and E are one representative panel from a minimum of three independent experiments. Open and filled circles in B and D are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. E) Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host BM were harvested and analyzed. The absolute numbers of reconstituted pro-B cells were identified with anti-B220 and anti-CD43 antibody staining. Pro-B cells (B220MedCD43+) and pre-B cells (B220+CD43−) were quantified from six host mice, and the average absolute numbers are presented as bar graphs. F) Absolute numbers of reconstituted B220+IgM−IgD− or B220+IgM+IgD− immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. BM cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies, and the absolute numbers were calculated per million total gated lymphocytes. Data shown on the left are representative panels. The average absolute numbers are presented as bar graphs. Data shown are from two independent experiments with three mice each. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 2.
Lack of IQGAP1 leads to a potential blockade at pre-B cell stages in the BM and the defects in the transition of pro-B to pre-B cells in the BM of Iqgap1−/− mice is cell-intrinsic. A, B) Absolute numbers of B220+CD25+ large and small pre-B-II stage. C, D) Absolute numbers of B220+IgM−IgD− or B220+IgM+IgD− immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. BM cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies, and the absolute numbers were calculated per million total gated lymphocytes. Data shown in A, C, and E are one representative panel from a minimum of three independent experiments. Open and filled circles in B and D are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. E) Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host BM were harvested and analyzed. The absolute numbers of reconstituted pro-B cells were identified with anti-B220 and anti-CD43 antibody staining. Pro-B cells (B220MedCD43+) and pre-B cells (B220+CD43−) were quantified from six host mice, and the average absolute numbers are presented as bar graphs. F) Absolute numbers of reconstituted B220+IgM−IgD− or B220+IgM+IgD− immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. BM cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies, and the absolute numbers were calculated per million total gated lymphocytes. Data shown on the left are representative panels. The average absolute numbers are presented as bar graphs. Data shown are from two independent experiments with three mice each. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
IQGAP1 is expressed in most of the hematopoietic cell lineages 21. Since IQGAP1 has been globally deleted 20 in the mice we used, the defects we observed in absolute B cell numbers can be cell-intrinsic or due to an abnormality in the stromal microenvironment. To distinguish between these two possibilities, we performed bone marrow (BM) transplantation (WT→WT; Iqgap1−/−→WT; WT→Iqgap1−/−; Iqgap1−/−→Iqgap1−/−). After eight weeks, B cell development was analyzed in the reconstituted host mice. Lymphocyte reconstitution was successfully observed in all hosts. Compared to WT, the transfer of Iqgap1−/− BM cells into WT or Iqgap1−/− hosts resulted in an increased absolute number of lymphocytes in the BM (Supplementary Figure S1A). Absolute numbers of B220+ B, B220High, and B220Low cell numbers were significantly increased in host mice that received IQGAP1−/− BM cells (Supplementary Figure S1B). Also, although B220+IgMNeg cells did not show any considerable change, B220+IgMHigh and B220+IgMLow cells were significantly increased in the recipient mice that were transplanted with Iqgap1−/− BM cells compared to that of WT (Supplementary Figure S1C). Analyzing Pro/Pre-B cells (B220+CD43+), Pro-B cells (B220MedCD43−), and Pre-B cells (B220HighCD43−) in reconstituted mice confirmed accumulation of Pro-B cells (Figure 2E). Staining for immature (IgM+IgD−) and mature (IgM+IgD+ and IgM−IgD+) B cell populations in the reconstituted mice indicated a significant increase in the immature but not the mature B cells (Figure 2F). In the spleens from the BM reconstituted mice, we did not observe an increase in the absolute number of lymphocytes or B220+ B cells (Supplementary Figure S2A). However, the transfer of Iqgap1−/− BM cells into the WT or Iqgap1−/− mice lead to a significant increase in the absolute numbers of immature (IgM+IgD−) but not mature (IgM+IgD+ and IgM−IgD+) B cell populations (Supplementary Figure S2B). These results demonstrate that the lack of IQGAP1 in the hematopoietic compartment but not the stromal environment is primarily responsible for the B cell developmental defects.
Lack of IQGAP1 significantly increases IgMHighCD23− T1 B cells in the spleens
Next, we analyzed the B cell maturation in the spleen. Lack of IQGAP1 did not alter the total lymphocyte numbers in the Iqgap1−/− mice (Iqgap1−/−: 39.0 ± 8.2×106 and WT: 40.1 ± 8.4×106; n=16, 16; p=NS) (Figure 3A). Moreover, similar to the BM, the absolute numbers of B220+ B cells in the spleen were moderately but significantly increased in Iqgap1−/− compared to that of the WT mice (Iqgap1−/−: 16.58 ± 6.26×106 and WT: 13.17 ± 5.16×106; n=31, 31; p=0.022) (Figure 3B). To further analyze the role of IQGAP1 in splenic B cell development, we stained them with anti-IgM and anti-IgD Abs. Although there were moderate increases in immature IgM+IgD− B (Iqgap1−/−: 2.91 ± 2.17×106 and WT: 1.85 ± 1.37×106; n=12, 12; p=NS) and mature IgM+IgD+ (Iqgap1−/−: 5.3 ± 2. 95×106 and WT: 3.46 ± 2.01×106; n=12, 12; p=NS) or IgM−IgD+ (Iqgap1−/−: 5.08 ± 2.49×106 and WT: 3.88 ± 1.82×106; n=12, 12; p=NS) B cells in the spleen of Iqgap1−/− mice, they did not differ significantly (Figure 3C).
Figure 3.
Lack of IQGAP1 leads to increased T1 B cells in the spleen. A) Absolute number of total lymphocytes within the spleens of WT and Iqgap1−/− mice were comparable. Absolute numbers were calculated from live gating of the lymphocyte population. Data shown represent the total lymphocytes per spleen from both strains. B) Absolute total splenic B cells were comparable between the WT and Iqgap1−/− mice. Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells using anti-B220 antibody. Data shown represent the B220+ B cells per spleen from both strains. C) Absolute numbers of immature and mature B cells do not differ between the WT and Iqgap1−/− mice. Total splenic B cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies. Data shown represent the B220+IgM+ immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. Data represent absolute numbers of immature or mature B cells per spleen from both strains. D) Absolute numbers of T1-B but not T2-B or T3-B are altered in Iqgap1−/− mice. Total splenocytes were stained with anti-B220, and anti-CD93 antibodies and the double-positive cells were gated and quantified. E) B220+CD93+ B cells were further sub-divided into B220+CD93+IgM+ T1-B, B220+CD93+IgM+CD23+ T2-B, and B220+CD93+IgM−CD24+ T3-B cells. Open and filled circles in A, B, C, and E are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 3.
Lack of IQGAP1 leads to increased T1 B cells in the spleen. A) Absolute number of total lymphocytes within the spleens of WT and Iqgap1−/− mice were comparable. Absolute numbers were calculated from live gating of the lymphocyte population. Data shown represent the total lymphocytes per spleen from both strains. B) Absolute total splenic B cells were comparable between the WT and Iqgap1−/− mice. Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells using anti-B220 antibody. Data shown represent the B220+ B cells per spleen from both strains. C) Absolute numbers of immature and mature B cells do not differ between the WT and Iqgap1−/− mice. Total splenic B cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies. Data shown represent the B220+IgM+ immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. Data represent absolute numbers of immature or mature B cells per spleen from both strains. D) Absolute numbers of T1-B but not T2-B or T3-B are altered in Iqgap1−/− mice. Total splenocytes were stained with anti-B220, and anti-CD93 antibodies and the double-positive cells were gated and quantified. E) B220+CD93+ B cells were further sub-divided into B220+CD93+IgM+ T1-B, B220+CD93+IgM+CD23+ T2-B, and B220+CD93+IgM−CD24+ T3-B cells. Open and filled circles in A, B, C, and E are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
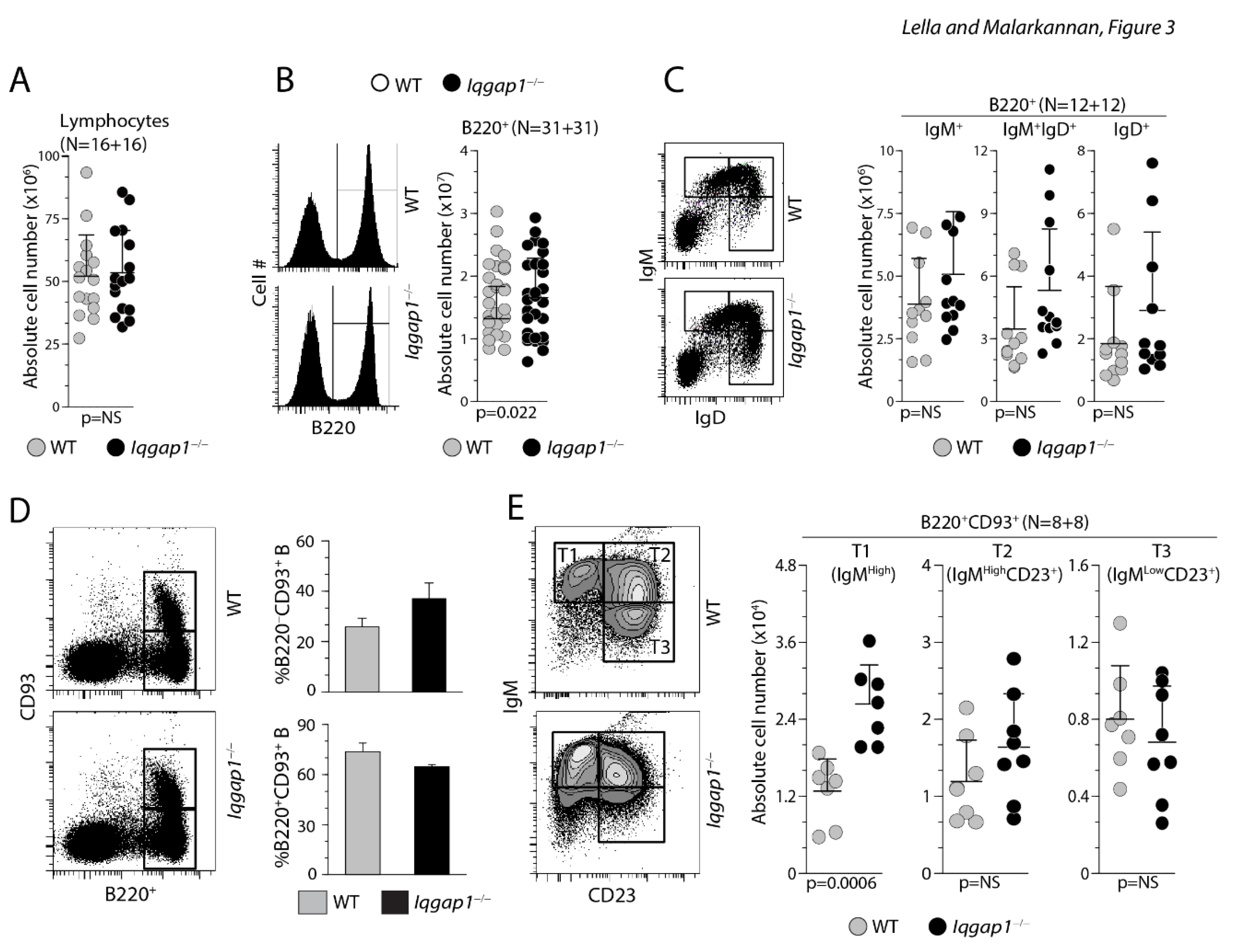
Next, we identified the transitional B cells among the total B220+ splenocytes based on their CD93 positivity. Transitional T1, T2, and T3 B cells are defined as IgMHighCD23−, IgMHighCD23+, and IgMLowCD23+, respectively. We gated the B220+ splenocytes into CD93+ and CD93− B cells (Figure 3D). We selected the CD93+ B cells and subdivided them based on IgM and CD23 positivity. Total percentages of B220+CD93+ B cells did not vary between the WT and the Iqgap1−/− splenocytes. However, the absolute number of IgMHighCD23− T1 B cells was significantly increased among the splenocytes from Iqgap1−/− mice (Figure 3E). Absolute numbers of T2 and T3 B cells were comparable between the WT and the Iqgap1−/− mice.
Lack of IQGAP1 differentially affects FO B and MZ B cells in the spleens
B220+ splenic B cells can also be separated into new-forming (NF B) (CD21Low/−CD23Low/−), follicular (FO B) (CD23HighCD21Low/−) and marginal zone (MZ B) (CD21HighCD23Low/−) cells. Therefore, we next analyzed the numbers FO, NF, and MZ B cells (Figure 4A). The absolute numbers of B220-gated CD21Low/−CD23High FO B (Iqgap1−/−: 11.7 ± 3.5×106 and WT: 8.92 ± 3.82×106; n=14, 14; p=0.026) and the CD21Low/−CD2 3Low/− NF B cells (Iqgap1−/−: 6.37 ± 2.71×106 and WT: 4.33 ± 1.18×106; n=14, 14; p=0.018) were significantly increased in Iqgap1−/− mice. However, the absolute number of MZ B was significantly reduced in the spleens of IQGAP1−/− mice (Iqgap1−/−: 1.18 ± 0.6×106 and WT: 1.59 ± 0.87×106; n=14, 14; p=0.05). In addition to these changes, the B cell follicles in the spleens of Iqgap1−/− mice were extended and multi-centered compared to that of WT. MZ B cells can also be defined by their exclusive high-level expression of IgM, CD21, and CD1d 22. Staining for IgM and CD21 (Iqgap1−/−: 0.93 ± 0.44×106 and WT: 1.56 ± 0.0.53×106; n=11, 11; p=0.0049) or IgM and CD1d (Iqgap1−/−: 0.75 ± 0.32×106 and WT: 1.18 ± 0.31×106; n=9, 9; p=0.0364) confirmed the significant decrease in the absolute number of MZ B cells (Figure 4B, C). We immune-stained spleen cryosections to establish further the exclusive reduction in MZ B cells. Iqgap1−/− mice displayed normal B cell follicle architecture (Figure 4D), as revealed by the anti-CD3ε and anti-B220 mAb staining. Since the metallophilic macrophages form a boundary between the follicular and marginal zones, we used MOMA-1 mAb to determine the changes in the marginal zone. The spleen sections from the WT mice revealed a well-defined band of B220+ MZ area outside of the MOMA-1+ metallophilic macrophages. However, similar regions were greatly reduced and less contiguous in Iqgap1−/− mice, which further confirmed a reduction in the number of MZ B cells (Figure 4E).
Figure 4.
Lack of IQGAP1 leads to increased NF-B, FO-B but decreased MZ-B cells in the spleen and the dysregulated early B cell development in the spleen of Iqgap1−/− mice is B cell-intrinsic. A) Absolute numbers of CD21Low/−CD23Low/− NF-B, CD21LowCD23High FO-B, and CD21HighCD23Low/− MZ-B cells are shown. Total splenic B cells were stained with anti-B220, anti-CD21, and anti-CD23 antibodies, and live cells were gated and compared between the WT and Iqgap1−/− mice. Data shown represent absolute numbers per million live B220+ B cells. B) Reduction in MZ-B cells was validated using anti-IgM and anti-CD21 antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. C) Reduction in MZ-B cells was validated using anti-IgM and anti-CD1d antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. Open and filled circles in A, B, and C are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. D, E) Immunohistochemical analyses of splenic sections from WT and Iqgap1−/− mice. Spleens were isolated, embedded in paraffin, 7-micron thick sections were cut, and stained for total B cells (anti-B220), T cells (anti-CD3ε), and metallophilic macrophages (anti-MOMA antibody). Panels show the locations of T and B cells as part of the B cell follicles and metalophillic antigen-1+ macrophages surrounding the follicles. F) Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host BM were harvested and analyzed. Absolute numbers of reconstituted CD21Low/−CD23Low/− NF-B, CD21LowCD23High FO-B, and CD21HighCD23Low/− MZ-B cells are shown. Total splenic B cells were stained with anti-B220, anti-CD21, and anti-CD23 antibodies, and live cells were gated and compared between the WT and Iqgap1−/− mice. Data shown represent absolute numbers per million live B220+ B cells.
Figure 4.
Lack of IQGAP1 leads to increased NF-B, FO-B but decreased MZ-B cells in the spleen and the dysregulated early B cell development in the spleen of Iqgap1−/− mice is B cell-intrinsic. A) Absolute numbers of CD21Low/−CD23Low/− NF-B, CD21LowCD23High FO-B, and CD21HighCD23Low/− MZ-B cells are shown. Total splenic B cells were stained with anti-B220, anti-CD21, and anti-CD23 antibodies, and live cells were gated and compared between the WT and Iqgap1−/− mice. Data shown represent absolute numbers per million live B220+ B cells. B) Reduction in MZ-B cells was validated using anti-IgM and anti-CD21 antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. C) Reduction in MZ-B cells was validated using anti-IgM and anti-CD1d antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. Open and filled circles in A, B, and C are values obtained from an individual mouse from four independent experiments. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. D, E) Immunohistochemical analyses of splenic sections from WT and Iqgap1−/− mice. Spleens were isolated, embedded in paraffin, 7-micron thick sections were cut, and stained for total B cells (anti-B220), T cells (anti-CD3ε), and metallophilic macrophages (anti-MOMA antibody). Panels show the locations of T and B cells as part of the B cell follicles and metalophillic antigen-1+ macrophages surrounding the follicles. F) Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host BM were harvested and analyzed. Absolute numbers of reconstituted CD21Low/−CD23Low/− NF-B, CD21LowCD23High FO-B, and CD21HighCD23Low/− MZ-B cells are shown. Total splenic B cells were stained with anti-B220, anti-CD21, and anti-CD23 antibodies, and live cells were gated and compared between the WT and Iqgap1−/− mice. Data shown represent absolute numbers per million live B220+ B cells.
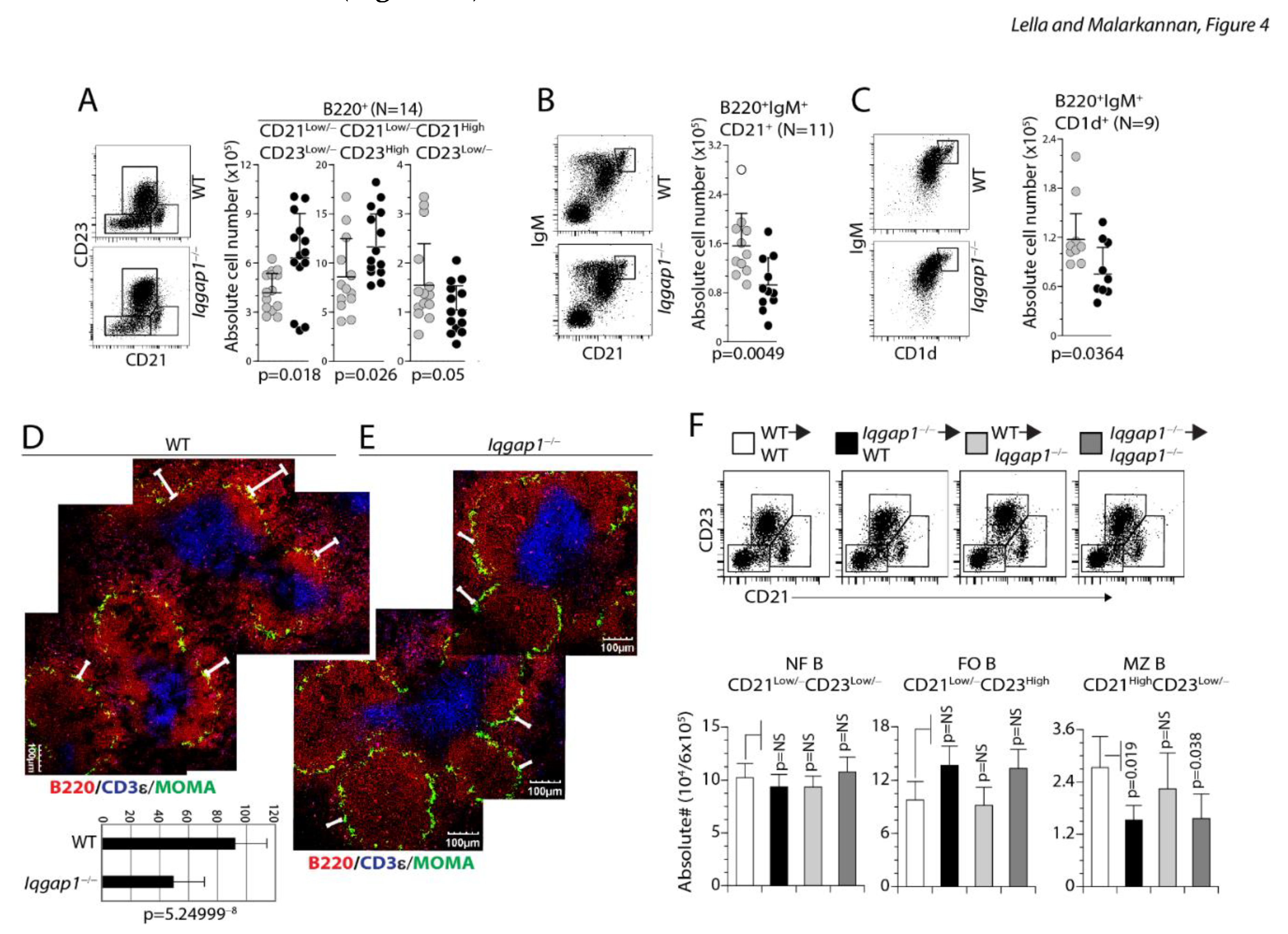
These reductions were B cell-intrinsic as reconstitution with Iqgap1−/− BM cells either into WT or Iqgap1−/− mice confirmed these defects. In the spleen of the reconstituted mice, we did not observe an increase in the absolute number of lymphocytes or B220+ B cells. However, the transfer of Iqgap1−/− BM cells into the WT or Iqgap1−/− mice lead to a significant increase in the absolute numbers of immature (IgM+IgD−) but not mature (IgM+IgD+ and IgM−IgD+) B cell populations. Reconstitution with Iqgap1−/− BM cells either into WT or Iqgap1−/− mice showed an increase in the CD23HiCD21Low/− FO B and a decrease in CD21HiCD23Low/− MZ B cells (Figure 4F). A decrease in the MZ B cells was further validated by a reduction in IgM+CD21High or IgM+CD1dHigh B cells in the reconstituted mice (Figure 5A, B). Our results demonstrate that the B cell defects in the BM and spleen of Iqgap1−/− mice were cell-intrinsic.
Figure 5.
Dysregulated early B cell development in the spleen of Iqgap1−/− mice is B cell-intrinsic. A) Reduction in reconstituted MZ-B cells was validated using anti-IgM and anti-CD1d antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. B) Reduction in reconstituted MZ-B cells was validated using anti-IgM and anti-CD21antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 5.
Dysregulated early B cell development in the spleen of Iqgap1−/− mice is B cell-intrinsic. A) Reduction in reconstituted MZ-B cells was validated using anti-IgM and anti-CD1d antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. B) Reduction in reconstituted MZ-B cells was validated using anti-IgM and anti-CD21antibody staining of total B220+ splenic B cells. Data shown represent absolute numbers per million live B220+ B cells. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Lack of IQGAP1 impairs T-independent and T-dependent antibody production
The significant impairment in B cell response in Iqgap1−/− mice could be due to either a developmental or a functional defect. To narrow it down, we first analyzed the basal immunoglobulin levels in the pre-immune sera of WT and Iqgap1−/− mice. Then, we quantified each isotype in the pre-immune sera. Figure 6A shows that IgG1, IgG2a, IgG2b, and IgA levels were comparable between WT and Iqgap1−/− mice. However, the levels of IgM were significantly reduced, and IgG3 was significantly increased in the sera of Iqgap1−/− mice (Figure 6A). Secreted ‘natural’ IgM is central to neutralizing pathogens and self-antigens. Thus, the presence of multiple Ig isotypes in the sera of Iqgap1−/− mice indicated that the lack of IQGAP1 did not result in a global hypo-responsiveness of B cells. IgG3 subtype possesses an extended hinge region, superior molecular flexibility, broad polymorphisms, and unique glycosylation sites compared to other IgG subclasses.
Figure 6.
Lack of IQGAP1 results in significantly reduced IgM and IgG3 levels in the sera of Iqgap1−/− mice and results in significantly reduced T-independent and T-dependent humoral responses. A) Pre-immune sera were collected from 20 WT or Iqgap1−/− mice each and were analyzed for the levels of soluble IgM, IgA, IgG1, IgG2a, IgG2b, and IgG3 using isotype-specific capture and detection antibodies-based ELISA. Data presented were an average of duplicates for each isotype of each serum from multiple experiments. A total of 20 sera were used for each strain. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. B) T-independent B cell responses were measured following the TNP-AECM-Ficoll antigen challenge. Four WT or four Iqgap1−/− mice were used to collect pre-immune or immune sera. ELISA was performed with titrated concentrations of sera (1:4, 1:8, and 1:16). IgM, IgG1, IgG2a, and IgG3 isotypes were tested and shown. C) T-dependent B cell responses were measured following the TNP-KLH antigen challenge. Four WT or four Iqgap1−/− mice were used to collect pre-immune or immune sera. ELISA was performed with sera collected at different time points before (pre-immune) and after antigen challenge (post 7d, 21d, and following 7d after booster challenge). IgM, IgG1, IgG2a, and IgG2b isotypes were tested and shown. Open and filled circles in A and B are values obtained from an individual mouse. Black and grey bars represent, respectively. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 6.
Lack of IQGAP1 results in significantly reduced IgM and IgG3 levels in the sera of Iqgap1−/− mice and results in significantly reduced T-independent and T-dependent humoral responses. A) Pre-immune sera were collected from 20 WT or Iqgap1−/− mice each and were analyzed for the levels of soluble IgM, IgA, IgG1, IgG2a, IgG2b, and IgG3 using isotype-specific capture and detection antibodies-based ELISA. Data presented were an average of duplicates for each isotype of each serum from multiple experiments. A total of 20 sera were used for each strain. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. B) T-independent B cell responses were measured following the TNP-AECM-Ficoll antigen challenge. Four WT or four Iqgap1−/− mice were used to collect pre-immune or immune sera. ELISA was performed with titrated concentrations of sera (1:4, 1:8, and 1:16). IgM, IgG1, IgG2a, and IgG3 isotypes were tested and shown. C) T-dependent B cell responses were measured following the TNP-KLH antigen challenge. Four WT or four Iqgap1−/− mice were used to collect pre-immune or immune sera. ELISA was performed with sera collected at different time points before (pre-immune) and after antigen challenge (post 7d, 21d, and following 7d after booster challenge). IgM, IgG1, IgG2a, and IgG2b isotypes were tested and shown. Open and filled circles in A and B are values obtained from an individual mouse. Black and grey bars represent, respectively. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
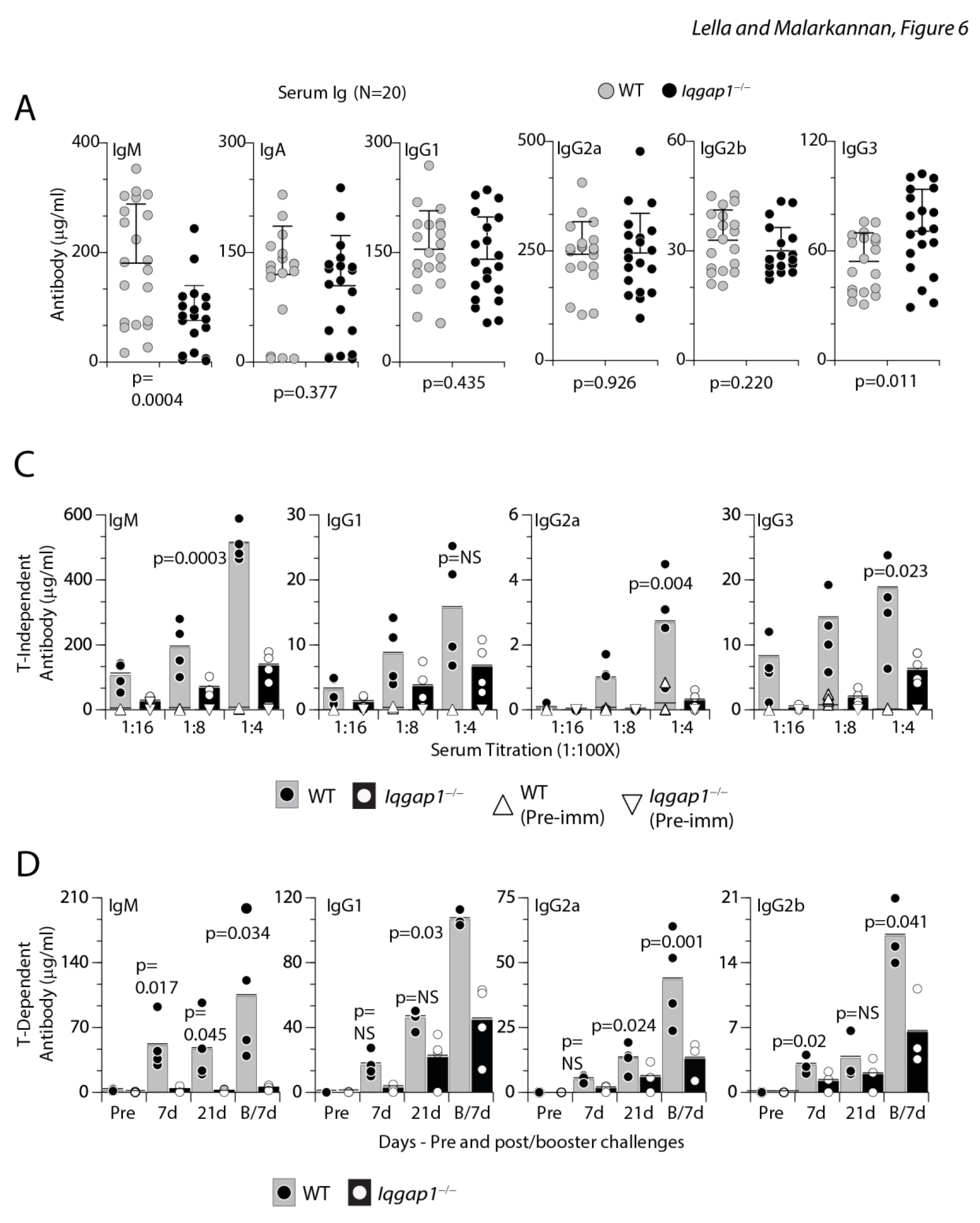
To test whether IQGAP1 plays a role in humoral immune responses, we challenged age and sex-matched WT and Iqgap1−/− mice with T-independent (TI) antigen TNP-AECM-Ficoll (TNP (2,4,6-Trinitrophenyl), where the hapten is conjugated to Amino Ethyl Carboxy Methyl-Ficoll and collected sera to analyze the presence of antigen-specific IgM, IgG1, IgG2a, or IgG3. Our phenotypic analyses indicated an increased number of naïve B cells in Iqgap1−/− mice expressed membrane-bound IgM and IgD in both the BM and spleen. Irrespective of this and an increase in the overall FO B cell number, Iqgap1−/− mice could not produce similar amounts of TNP-specific antibodies during TI responses as WT mice (Figure 6B). Next, we examined the T-dependent (TD) immune responses by intraperitoneal injection of TNP-KLH. In the primary response, the generation of TNP-specific IgM, IgG1, IgG2a, or IgG2b was significantly reduced in Iqgap1−/− mice when compared to that of WT mice (Figure 6C). Further, after the antigen boost, the KLH-specific IgG1 did not recover and remained significantly low in the Iqgap1−/− mice compared to WT (Figure 6C). These results demonstrate that IQGAP1 plays a crucial role in the activation and functions of B cells.
Receptor editing at κ the locus is moderately defective in B cells lacking IQGAP1
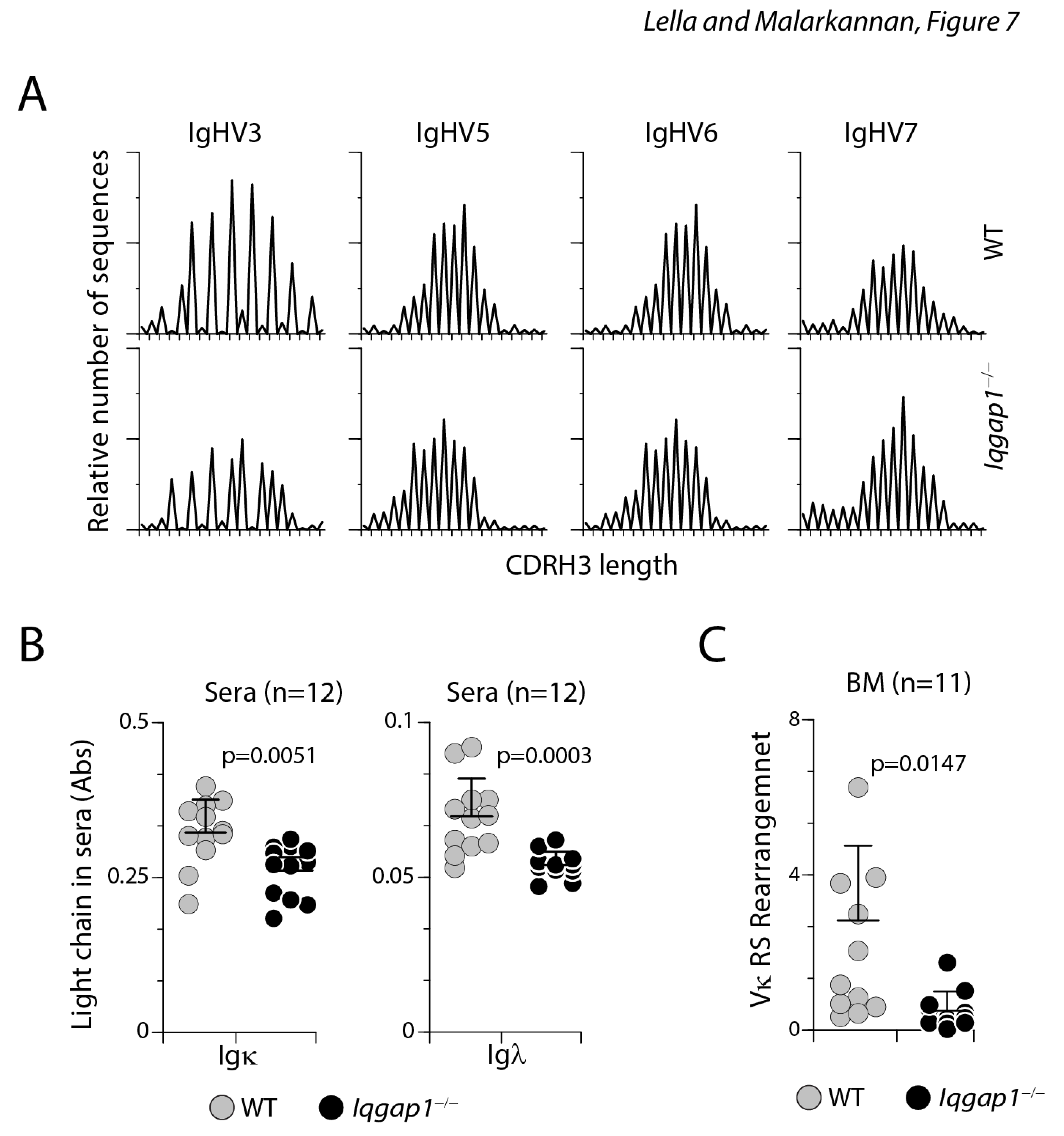
The diversity in the length of hypervariable complementarity-determining region 3 (CDR3) of the heavy chain (CDRH3) indicates a repertoire of rearranged VH genes and polyclonality 23. Using PCR-based spectrotyping, we analyzed the fragment size and peak height for IgHV3, IgHV5, IgHV6, and IgHV7 in B cells from the BM. Our results show that the length of the variable regions and the relative number of sequence-occurrences of the heavy chains in B cells from the WT and Iqgap1−/− mice followed an expected normal Gaussian distribution indicative of diverse and comparable repertoire (Figure 7A). Thus, the lack of IQGAP1 did not affect the B cell repertoire diversity at the heavy chain rearrangement. Furthermore, the preserved heavy chain rearrangement in B cells from Iqgap1−/− mice indicated that lack of IQGAP1 did not affect the B cell repertoire diversity at the heavy chain rearrangement.
Figure 7.
Light chain but not heavy recombination is affected by the lack of IQGAP1. A) PCR-based spectrotyping was employed to quantify the diversity in the length of hypervariable CDR3 of the heavy chain (CDRH3) in B cells from the BM of WT and Iqgap1−/− mice. The fragment size and peak height for IgHV3, IgHV5, IgHV6, and IgHV7 indicated that the repertoire of rearrangement VH genes and polyclonality between these two strains are comparable and followed the expected normal Gaussian distribution indicative of diverse and comparable repertoire. B) Igκ and Igλ light chains in the sera are reduced in Iqgap1−/− mice. C) RS rearrangement at the k-locus is reduced in the absence of IQGAP1. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Figure 7.
Light chain but not heavy recombination is affected by the lack of IQGAP1. A) PCR-based spectrotyping was employed to quantify the diversity in the length of hypervariable CDR3 of the heavy chain (CDRH3) in B cells from the BM of WT and Iqgap1−/− mice. The fragment size and peak height for IgHV3, IgHV5, IgHV6, and IgHV7 indicated that the repertoire of rearrangement VH genes and polyclonality between these two strains are comparable and followed the expected normal Gaussian distribution indicative of diverse and comparable repertoire. B) Igκ and Igλ light chains in the sera are reduced in Iqgap1−/− mice. C) RS rearrangement at the k-locus is reduced in the absence of IQGAP1. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
CD43+ B cell progenitors in mice rearrange immunoglobulin Igκ genes before the assembly of a productive VHDHJH joint. Thus, μ chain rearrangement and expression are not prerequisites to Igκ light chain gene rearrangements in normal development 24. Therefore, we next analyzed the quantity of κ and λ light chains in the sera of non-challenged healthy WT and Iqgap1−/− mice using an ELISA-based assay. We found a significant reduction in both the κ and λ light chains in Iqgap1−/− mice compared to the WT (Figure 7B). This demonstrates that the defect we see in the antibody production in challenged mice could be related to the production of the light but not the heavy chains. Indeed small GTPase, Ras, plays an essential role via Mek1/2 and Erk1/2 in silencing Ccnd3 transcription, exiting from the cell cycle, and initiating Igκ recombination and light chain editing 25. Receptor editing in the light chain loci forms a critical basis for high-affinity maturation that is a continuous process in an ongoing immune response 26, 27, and impairment in κ chain rearrangement could account for the functional defects in T-dependent and independent antibody responses. Therefore, we next analyzed the light chain rearrangement. The orderly recombination of genes at the κ locus is the first step in the light chain rearrangement. The successful recombination of genes at the κ locus is proceeded by rearranging non-coding gene segments defined as ‘recombining sequence’ (RS) 28. RS rearrangements occur higher in the most highly edited B cells. Thus, quantifying RS provides a readout for successful κ chain rearrangement 29. Our results show that the levels of amplified RS products were moderately lower in the B cells derived from Iqgap1−/− mice (Figure 7C), indicating that the early pre-B cell development is largely intact in Iqgap1−/− mice. Thus, the functional defects observed in the B cells from Iqgap1−/− mice are due to an inability to rearrange the light chains appropriately.
B cell activation requires IQGAP1-Mek1/2-Erk1/2 signalosome
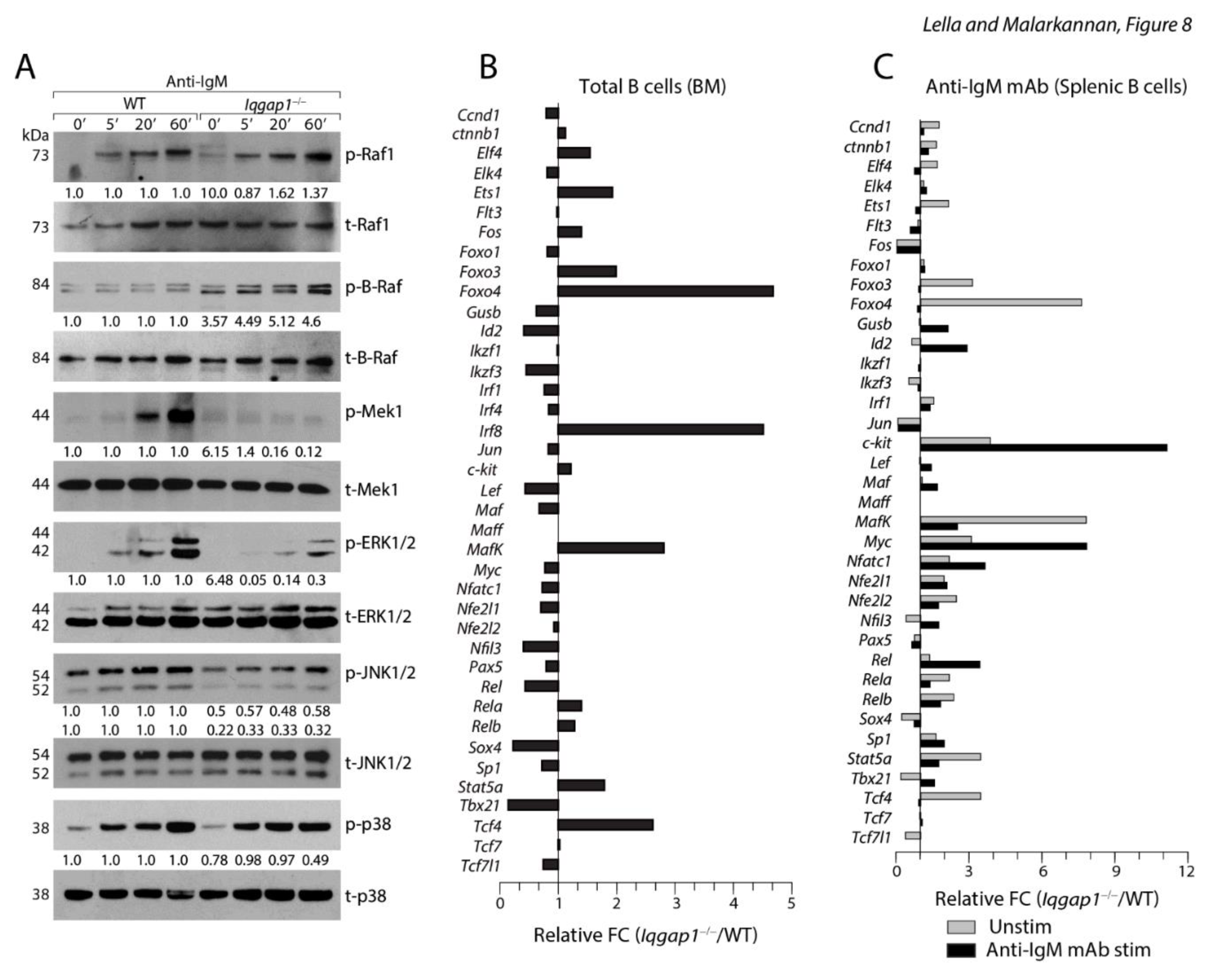
IQGAP1 is an essential scaffold in organizing Erk1/2 activation and phosphorylation 30. Four tandem Isoleucine-Glutamine (IQ) domains are located between 745-864 amino acids. These IQ domains primarily mediate interactions with Rap1 31, B-Raf or C-Raf 32, and Mek1/2 30. Therefore, we hypothesized that IQGAP1 regulates the sequential activation of the Raf/Mek1/2/Erk1/2 cascade. To test this, we activated purified total splenic B cells with soluble anti-IgM mAb. Rac1/Cdc42 activates p21-activated kinase (Pak) that connects the upstream signaling to Raf by phosphorylating S445 in B-Raf and S338 in C-Raf 33. We found comparable amounts of phosphorylated B-Raf (Serine 445), C-Raf (Serine 338), and total B-Raf and C-Raf were present in Iqgap1−/− and WT B cells (Figure 8A). B-Raf or C-Raf catalyzes the phosphorylation of Mek1/2, which in turn phosphorylates Erk1/2 34. Since IQ domains recruit Mek1/2, we analyzed their activation status. and found the phosphorylation of Mek1/2 were considerably reduced in B cells lacking IQGAP1. The WW domain of IQGAP1 (685-710 amino acids) contains two conserved tryptophan, positioned about 20-22 amino acids apart (Wx20-22W) that function as an interaction module for proline-rich ligands 35. This domain is also responsible for recruiting Erk1/2 via a polyproline (APPPxxPY) motif 36. Therefore, we next analyzed the phosphorylation status of Erk1/2 along with the two other MAPKs, Jnk1/2, and p38. All three kinases were active in the WT B cells. p38 phosphorylation was not affected in any of the Iqgap1−/− B cells. However, the levels of Erk1/2 and Jnk1/2 phosphorylation were considerably reduced in Iqgap1−/− B cells (Figure 8A).
Figure 8.
IQGAP1 orchestrates ERK1/2 activation in B cells. A) Sorted splenic B cells were activated with soluble anti-IgM antibody for indicated time points and lysed, protein quantities were estimated and analyzed by western blots. Lysates were analyzed for the total and phosphorylated forms of Raf1, B-Raf, Mek1, Erk1/2, JNK1/2, and p38. Numbers below each blot represent fold change calculated following normalized values against their respective total and based on the phosphorylated protein levels of the WT. B) PCR array results. C) PCR array results. Data shown are one representative panel of three independent experiments. Data shown are obtained by repeated probing of the same membrane for indicated proteins.
Figure 8.
IQGAP1 orchestrates ERK1/2 activation in B cells. A) Sorted splenic B cells were activated with soluble anti-IgM antibody for indicated time points and lysed, protein quantities were estimated and analyzed by western blots. Lysates were analyzed for the total and phosphorylated forms of Raf1, B-Raf, Mek1, Erk1/2, JNK1/2, and p38. Numbers below each blot represent fold change calculated following normalized values against their respective total and based on the phosphorylated protein levels of the WT. B) PCR array results. C) PCR array results. Data shown are one representative panel of three independent experiments. Data shown are obtained by repeated probing of the same membrane for indicated proteins.
To further define the status of specific transcription factors altered in the early B cells from Iqgap1−/− mice, we performed a PCR array analysis. We sorted total B cells from the BM or spleen of three mice each and pooled the mRNA for each strain. cDNA preparations were analyzed in triplicates, and average expression was presented as fold change (Fc) between Iqgap1−/− B cells over WT (Figure 8B). Total naïve BM B cells were analyzed for the expression levels of transcription factors that play an essential role in early B cell commitment and maturation. Expressions of Foxo3, Foxo4, MafK, Stat5a, and Tcf4 were increased two-fold in Iqgap1−/− B cells compared to WT. Id2, Ikzf3, Lef, Nfil3, Rel, Sox4, and Tbx21 were reduced in the Iqgap1−/− B cells. It is important to note that the expression of Id2 is regulated by Erk/Jnk/Cdk2 pathway. Ikzf3 (Ailos) is essential for the heterochromatic sequestration of the Igκ gene. Next, we stimulated the sorted splenic B cells with anti-IgM mAb and compared the expression of transcription factors in between unstimulated and stimulated conditions from both strains. Averages from triplicates were used to calculate the fold change (Fc) between Iqgap1−/− B cells over WT under both the experimental conditions (Figure 8C). Similar to the total B cells from the BM, the levels of Foxo3, Foxo4, MafK, Stat5a, and Tcf4 have increased two or more folds in non-stimulated Iqgap1−/− B cells compared to WT. The expression levels of all these factors were considerably reduced in Iqgap1−/− B cells following anti-IgM mAb-mediated activation. However, the levels of c-Fos, Jun, Sox4, and Tbx21 were reduced in both unstimulated and stimulated conditions. Interestingly, levels of Id2, c-Kit, Myc, Nfatc1, and Rel were increased in Iqgap1−/− B cells following anti-IgM mAb-mediated activation (Figure 8C). Expression of c-Kit is limited only to the pre-lymphoid progenitors, pro-B, and pre-B-I cells. Therefore, a high level of c-Kit expression in Iqgap1−/− B cells at both unstimulated and stimulated conditions strongly validates our phenotypic characterization and supports our notion that lack of IQGAP1 results in a defect in the transition of pre-B-I into large pre-B-II cells.
IQGAP1 is essential for suppressing Stat5a and Stat5b
Earlier studies have shown that pre-BCR-mediated activation counters the IL-7R pathway 25. Signaling via IL-7R sustains the survival and proliferation of pro-B cells 37. This is primarily mediated by cell-cycle-related genes, including Ccnd3. In addition, phosphorylated Stat5A and Stat5B, downstream of IL-7R, translocated into the nucleus and form a complex with PRC2/EZH2 to bind to 5′ intronic enhancer of the κ gene (iEκ) to actively suppress its transcription by limiting the access to E2A, which is induced by the pre-BCR signaling. In addition, the pre-BCR-mediated Ras-Mek1/2-Erk1/2 pathway silences Ccnd3 to orchestrate the exit from the cell cycle and to initiate κ chain recombination 25, 38. First, we investigated the status of Ccnd3 transcripts. For this, we purified the relatively immature T1 B cells from the spleens of WT and Iqgap1−/− mice. Expression of cell cycle-related genes such as β-catenin, Bcl-xl, Ccnd3, Ccnd1, and Bim were analyzed. We found that the levels of β-catenin and Ccnd3 transcripts were significantly and moderately reduced, respectively (Supplementary Figure S3). Next, we investigated the status of E2A, Ezh2, Stat3, Stat5A, and Stat5B in WT and Iqgap1−/− B cells. First, we analyzed the protein levels of these transcription factors in the total B cells derived from the BM of both strains (Figure 9A). The limited numbers of pro-and pre-B cells excluded our ability to use them in these assays. Compared to the WT, Iqgap1−/− B cells possessed moderately higher levels of E2A, Stat5A, Stat5B, and Ezh2.
Figure 9.
IQGAP1 is required to suppress Stat5a and Stat5b and to initiate the transcription of IRF4 and IRF8. A) Total sorted BM B cells were used to analyze the protein levels of E2A, Stat3, Stat5A, Stat5B, IRF4, and β-actin. Data shown are one representative of three experiments. B) Total sorted BM B cells were activated with anti-IgM mAb, and the cytoplasmic and nuclear protein fractions were separated and analyzed for E2A, Stat3, Stat5A, Stat5B, and β-actin. Data shown are one representative of three experiments. C) mRNA from sorted BM B cells was used to quantify the expression of Irf4 and Irf8 transcripts. Data shown are the average relative expression of these transcripts from four mice. D) mRNA from sorted splenic B cells was used to quantify the expression of Irf4 and Irf8 transcripts. Data shown are the average relative expression of these transcripts from four mice. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. E) Cartoon summarizing the role of IQGAP1 during early B cell development. IL-7R transduces its signal via Stat3, Stat5A, and Stat5B. Signals via pre-BCR initiate two distinct pathways, One via SLP65-mediated IRF4 and IRF8 activation and the second via the Ras-Raf-Mek-Erk/Jnk pathway mediated by IQGAP1. Four tandemly organized IQ domains can recruit C-Raf or B-Raf. IQ domains also have the specificity to Mek1/2. Rafs mediate the activation of Mek1/2 on the IQGAP1 scaffold. The WW domain of IQGAP1 recruits Erk1/2, which is phosphorylated by Mek1/2. Phosphorylated Erk1/2 activates several transcription factors, including E2A, Elk1, and CREB. E2A competes with Stat5 dimers for the 5′ intronic Eκ enhancer accessibility. Successful displacement of Stat5 dimers initiates the recombination process of Igκ gene. E2A also relieves the Stat5-mediated repression of Rag1 and Rag2 genes. Stat5-mediated transcription of Ccnd3 is relieved by the phosphorylated Ras-Mek1/2-Erk1/2 pathway. The potential candidates that repress the transcription of Ccnd3 are Cdkn2a or p21. The mechanism by which IQGAP1 regulates the expression of Irf4 and Irf8 genes are not known. IRF4 binds to the 3′ Eκ in further promoting light chain recombination. IRF4 also binds to the 5′ Eλ to initiate the transcription of Igλ gene. IRF4, along with Ikaros3 (Ailos) represses the surrogate light chain component λ5-encoding gene. IRF4 also plays an important role in initiating the transcription of Cxcr4, which is essential for the pre-B cells to traffic from their IL-7-producing stromal niche to IL-7-deficient niche to transition into immature B cells. Repressing Id2 and Id3 is also needed for the pre-B cells that is regulated by Ebf1, a potential target downstream of Erk1/2.
Figure 9.
IQGAP1 is required to suppress Stat5a and Stat5b and to initiate the transcription of IRF4 and IRF8. A) Total sorted BM B cells were used to analyze the protein levels of E2A, Stat3, Stat5A, Stat5B, IRF4, and β-actin. Data shown are one representative of three experiments. B) Total sorted BM B cells were activated with anti-IgM mAb, and the cytoplasmic and nuclear protein fractions were separated and analyzed for E2A, Stat3, Stat5A, Stat5B, and β-actin. Data shown are one representative of three experiments. C) mRNA from sorted BM B cells was used to quantify the expression of Irf4 and Irf8 transcripts. Data shown are the average relative expression of these transcripts from four mice. D) mRNA from sorted splenic B cells was used to quantify the expression of Irf4 and Irf8 transcripts. Data shown are the average relative expression of these transcripts from four mice. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. E) Cartoon summarizing the role of IQGAP1 during early B cell development. IL-7R transduces its signal via Stat3, Stat5A, and Stat5B. Signals via pre-BCR initiate two distinct pathways, One via SLP65-mediated IRF4 and IRF8 activation and the second via the Ras-Raf-Mek-Erk/Jnk pathway mediated by IQGAP1. Four tandemly organized IQ domains can recruit C-Raf or B-Raf. IQ domains also have the specificity to Mek1/2. Rafs mediate the activation of Mek1/2 on the IQGAP1 scaffold. The WW domain of IQGAP1 recruits Erk1/2, which is phosphorylated by Mek1/2. Phosphorylated Erk1/2 activates several transcription factors, including E2A, Elk1, and CREB. E2A competes with Stat5 dimers for the 5′ intronic Eκ enhancer accessibility. Successful displacement of Stat5 dimers initiates the recombination process of Igκ gene. E2A also relieves the Stat5-mediated repression of Rag1 and Rag2 genes. Stat5-mediated transcription of Ccnd3 is relieved by the phosphorylated Ras-Mek1/2-Erk1/2 pathway. The potential candidates that repress the transcription of Ccnd3 are Cdkn2a or p21. The mechanism by which IQGAP1 regulates the expression of Irf4 and Irf8 genes are not known. IRF4 binds to the 3′ Eκ in further promoting light chain recombination. IRF4 also binds to the 5′ Eλ to initiate the transcription of Igλ gene. IRF4, along with Ikaros3 (Ailos) represses the surrogate light chain component λ5-encoding gene. IRF4 also plays an important role in initiating the transcription of Cxcr4, which is essential for the pre-B cells to traffic from their IL-7-producing stromal niche to IL-7-deficient niche to transition into immature B cells. Repressing Id2 and Id3 is also needed for the pre-B cells that is regulated by Ebf1, a potential target downstream of Erk1/2.
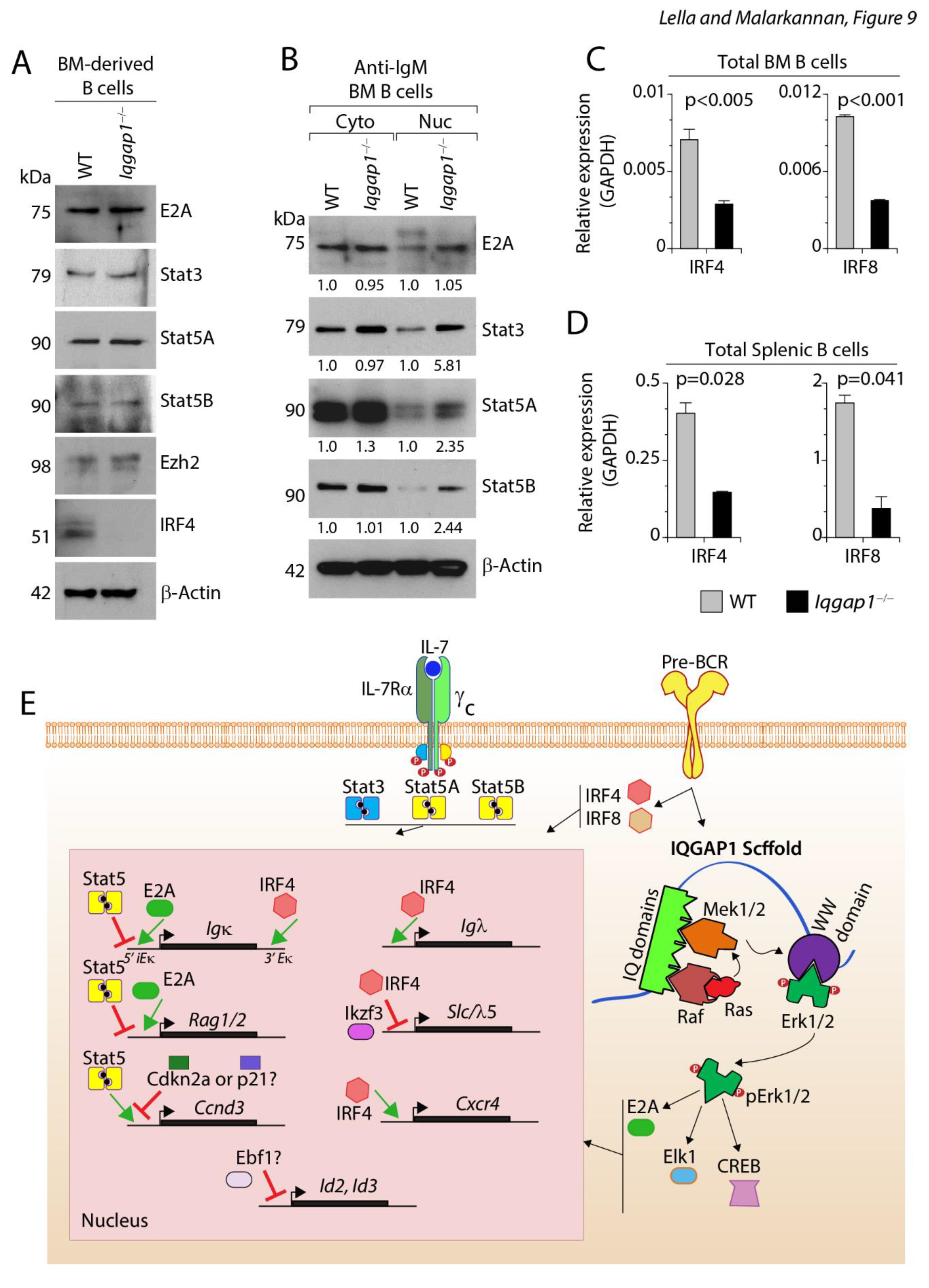
Interestingly, no protein band was found for IRF4 in Iqgap1−/− B cells (Figure 9A). Next, we evaluated the levels of these transcription factors translocated into the nucleus following anti-IgM mAb-mediated activation. The B cells activated by this method are not pro- or pre-B cells; but immature and mature B cells. However, it served as an indicator of the status of these transcription factors. We isolated total BM B cells and activated them with anti-IgM mAb for two hours. Nuclear and cytoplasmic fractions were isolated and analyzed for total E2A, Stat3, Stat5A, Stat5B, and β-actin (Figure 9B). Stat3, Stat5A, and Stat5B were present at comparable levels in the cytoplasmic fractions of B cells in both the WT and Iqgap1−/− mice. However, following anti-IgM mAb-mediated activation, the levels of these transcription factors were higher in Iqgap1−/− B cells compared to that of the WT. These results indicate a sustained presence of E2A, Stat3, Stat5A, and Stat5B in the nucleus of Iqgap1−/− B cells. In addition to these transcription factors, IRF4 and IRF8 are obligatory for the cycling pre-B to initiate light chain rearrangement and transition into immature B cells 39, 40. Therefore, next, we quantified the levels of IRF4 and IRF8 in total BM- and spleen-derived B cells. Both IRF4 and IRF8 were significantly reduced in B cells from Iqgap1−/− compared to the WT mice (Figure 9C, D). These results demonstrate that the lack of IQGAP1 scaffold significantly impairs early B cell development due to the reduced levels of IRF4 and IRF8 transcription factors.
Discussion
In this report, we define that IQGAP1 functions as an essential scaffold regulating the early development and maturation of B cells. Earlier studies have shown that the MAPK/Erk1/2 pathway plays a vital role in most B cell developmental stages and functions 25, 37, 38, 41. However, the identity of the scaffold proteins and their stage-specific roles were not defined. We found lack of IQGAP1 significantly blocked the transition of pre-B-I cells into large pre-B-II cells in the BM. In conjunction with this blockade, recombination of Igκ and the expression of Igλ chains were moderately reduced.
Furthermore, the lack of IQGAP1 resulted in a significant reduction in the transcription of IRF4 and IRF8, two essential transcription factors for light chain recombination. In the periphery, the splenic NF B and follicular FO B cell numbers were increased, while the MZ B cell numbers were significantly reduced in the Iqgap1−/− mice. Our findings demonstrate that these developmental defects are B cell-intrinsic. In addition, the lack of IQGAP1 significantly reduced TD and TI humoral responses. Mechanistically, the lack of IQGAP1 resulted in the reduction of Mek1/2, Erk1/2, and Jnk1/2 phosphorylation, suggesting a basis for the impaired B cell development and function.
Pre-BCR signaling initiated two major activation pathways. First, one involves the Ras-Raf-Mek-Erk/Jnk pathway. The second pathway involves the PLC-γ2/SLP65/Btk/IRF4/IRF8 signaling. These two pathways are obligatory and converge to attenuate the survival and proliferation signaling regulated by the IL-7R in pro-pre B cells. Earlier studies have shown that the absence of both Erk1 and Erk2 in murine pro-B cells resulted in defective pre-BCR signaling, proliferation, and their transition into pre-B cells 38. Thus, if the IQGAP1 is an obligatory scaffold for organizing the Ras-Raf-Mek-Erk1/2 pathway, it should impair the proliferation of pro-B cells and their transition into pre-B cells. Although a primitive GTPase-activation-related domain (GRD) is present in IQGAP1 between 1004-1237 amino acids, it lacks GTPase-activating protein (GAP) activity. It thereby is unable to regulate Ras-GTP 42 or Rap-GTP hydrolysis 43. The crystal structure of the GRD domain demonstrates the presence of a conserved threonine in place of the catalytic ‘arginine finger’ essential for GTP hydrolysis 44. Irrespective of lacking a GAP function, IQGAP1 possesses six highly conserved domains, with which it recruits and sequentially activates B-Raf 15, 32, 45, Mek1/2 30, and Erk1/2 12, 30, 36, 46. These earlier findings strongly suggest that IQGAP1 can function as an essential scaffold in developing B cells.
Receptor-mediated stimulations result in Vav1-Cdc42-Pak1 activation, and Pak1 binds to Rafs to induce conformational changes and phosphorylate serine445 of B-Raf or serine338 of C-Raf 47, 48. IQGAP1, via its Ras-GAP C-terminus (RGCT) domain (1276-1657 amino acids), recruits the Cdc42-GTP 49 and activates Pak1. Ras is an essential GTPase for the activation of Rafs 50, 51. B-Raf and C-Raf possess an N-terminal Ras-binding domain (RBD), a ubiquitin-like fold, which functions as an auto-inhibitory domain. Upon Ras-GTPase binding, Rafs are activated to phosphorylate downstream substrates such as Mek1/2. Four tandem Isoleucine-Glutamine (IQ) domains located between 745-864 amino acids contain repeats that are 25 amino acids long and have [I/V/L]Qxxx[RG]xxx[RK] motif, which primarily interacts with Rap1 31, B-Raf or C-Raf 32, Mek1/2 30. Two highly conserved tryptophan within the WW domain (685-710 amino acids) positioned about 20-22 amino acids apart (Wx20-22W) function as an interaction module for proline-rich ligands 35. This WW domain is responsible for recruiting Erk1/2 via a polyproline (APPPxxPY) motif 36. Thus, IQGAP1 can orchestrate the Ras→Raf→Mek→Erk pathway in B cells.
We found that the overall absolute number of B cells in terms of B220+ (both B220High and B220Low B cells) were significantly increased in the BM of Iqgap1−/− mice compared to the WT. Early B cell development in BM is characterized by the expression of CD43 (Leukosialin), CD117 (c-Kit), and CD25 (IL-2Rα). B220+CD43+ Pro-B cells develop into B220+CD43− pre-B cells. The absolute cell numbers of Pro-B cells did not differ between Iqgap1−/− and WT. However, B220MedCD43− B cells were much higher in Iqgap1−/−, which is consistent with the increased B220+IgM− pro/pre-B cells populations. Maturing pro-pre-B cells decrease the expression of c-kit and increase the level of CD25. Therefore, we used CD25 (IL-2Rα) as a marker to specify large pre-B (stage-II, early pre-B) and small pre-B (stage-II, late pre-B). Pro-pre-B and pre-B stage-I cells do not express CD25. Immature and mature B cells stop expressing CD25. We found a significant increase in the numbers of B220+CD25+ B cells in the BM of Iqgap1−/− mice. These findings are validated independently with a PCR array analysis. We found significantly increased levels of c-Kit expression in Iqgap1−/− total splenic B cells at both unstimulated and anti-IgM mAb-stimulated conditions. These results demonstrate that lack of IQGAP1 results in a blockade in the transition of pre-B-I into large pre-B-II cells.
As expected, with the increase in the B220+CD43+ pro-B cells or B220+CD25+ large/small pre-B cells, the B220+IgM− B cells also significantly increased. Notably, the B220+IgM+IgD− immature B cells but not the B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells were increased in the Iqgap1−/− mice. Thus, our data from the BM of non-challenged WT and Iqgap1−/− mice imply that IQGAP1 is essential during the early developmental stages of B cells; however, its absence does not negatively impact the commitment to B cell lineage. The light chain rearrangement is moderately but significantly reduced in Iqgap1−/− B cells. This may be because other MAPK scaffolds, such as KSR1, may compensate for the absence of IQGAP1. The early developmental progression of B cells is regulated by multiple soluble factors, including Flt3 ligand, SCF, and IL-7, produced by BM stromal cells. We performed BM chimera experiments to distinguish between the B cell-intrinsic and extrinsic effects. Our findings strongly indicate that the impairments we see in B cells are due to the lack of IQGAP1 within the B cells and the minimal role of cell-extrinsic factors.
BM B cells continue to mature in the spleen 52, 53. Total B220+ B cells were considerably higher in mice lacking IQGAP1 than the WT, irrespective of comparable numbers of total lymphocytes in both strains. Classification of immature and mature B cells based on their IgM and IgD expression did not show any changes in cell numbers due to the lack of IQGAP1. However, identification of T1, T2, and T3 splenic B cells 53 based on their expression of CD220, CD93 (AA4), IgM, and CD23 revealed a significant increase in the absolute numbers of T1 (CD220+CD93+IgM+) B cells. IgMhighIgD−CD21−CD23− T1 B cells in the spleen are the continuation of IgM+ immature B cells in the BM. The lack of IQGAP1 did not alter the numbers of T2 and T3 B cells. One possibility for the higher number of T1 B cells may be due to an impairment in deleting autoreactive B cells in the spleen during their maturation process. The mature T2 and T3 B cells can be further classified into NF B (B220+CD21LowCD23Low), FO B (B220+CD21LowCD23High), and MZ B (B220+CD21HighCD23Low) cells. Lack of IQGAP1 moderately increased the numbers of NF B and FO B cells and significantly reduced the numbers of MZ B cells. One possibility for the decrease in the MZ B, but not NF B or FO B, could be the requirement of differing levels of tonic BCR signaling in these subsets. These results indicate that the developmental defects of the B cells in BM extend and persist in the periphery.
Challenging mice with TI and TD antigens in vivo indicated that lack of IQGAP1 significantly reduced the ability of B cells to respond and generate IgM, IgG1, IgG2a, IgG2b, and IgG3 antibodies. These functional defects could occur due to two reasons. One, an inherent developmental defect might have resulted in the inability of Iqgap1−/− B cells to respond. However, it is also possible that a defective Erk1/2/MAPK pathway leads to functional impairments in partially matured B cells in the Iqgap1−/− mice.
Differential expression of cell surface IgM and IgD define immature and mature B cells. The pre-B cells at Stage-II level are cytoplasmic IgM-positive; but stop the expression of surrogate light chains (SLC) composed of λ5 and VpreB 54, 55. Expression of Rag1 and Rag2 at the late pre-B stage-II (BP-1+ Fraction-D) initiates the rearrangement of Vκ chains 56. Thus, expression of cell surface IgM marks the transition of late pre-B into mature B cells. The unique recombining sequence (RS) is a marker of ongoing light chain rearrangement, and it occurs at the time of late κ recombination 29. Most RS rearrangements occurred in small pre-B cells that possess cytoplasmic Igμ+; but do not express surrogate light chains. At this stage, the pre-B cells start expressing Rag1 and Rag2 genes, which are essential for the κ or λ rearrangement 29. We find that the lack of IQGAP1 resulted in both a reduced RS occurrence and a significantly reduced Igκ and Igλ chains in the sera. Earlier studies have shown that the Ras-mediated MEK-Erk1/2 pathway is crucial for the exit from sustained IL-7-driven cell division and the initiation of light-chain recombination 25, 37. Our analyses demonstrated that the phosphorylated Mek1, Erk1/2, and Jnk1/2 were considerably reduced in the Iqgap1−/− B cells. However, we did not observe any reductions in phosphorylated Raf1 or B-Raf levels. We do not know the precise reasons for these observations. Earlier studies have predicted the hierarchical interplay between IQGAP1 and caveolin scaffolds; however, whether such a mechanism operates in lymphocytes is unknown 51.
The phenotypic and functional outcome of Iqgap1−/− B cells provides a model (Figure 9E) that concurs with earlier observations. Erk1/2-mediated activation of E2A plays an essential in relieving the Stat5-mediated suppression of Igκ chain recombination 25. The levels of E2A under unstimulated and anti-IgM mAb-activation did not change in the BM-derived B cells from the WT. Albeit still lower than the WT, its level was increased only after anti-IgM mAb-stimulation in B cells from the BM of Iqgap1−/− mice. This demonstrates that under homeostatic conditions, the level of E2A was significantly lower in the BM-derived B cells of Iqgap1−/− mice. In addition, our data indicate an increase in the levels of nuclear Stat3, Stat5A, and Stat5B in the BM-derived B cells from Iqgap1−/− mice. Thus, a significant increase in the pre-B-I cells and immature B cells could be due to the disruption of Erk1/2 activation and persistence of Stat5 functions. These findings imply that the loss of IQGAP1 leads to a failure to suppress cell-cycle-regulated genes, and this could be due to impaired activation of the ERK pathway by the pre-BCR. Importantly, levels of c-Fos and c-Jun, two crucial AP1 transcription factors regulated by Erk1/2 and Jnk1/2, respectively, were decreased in the Iqgap1−/− splenic B cells. Through a second pathway, activation via pre-BCR elevates the transcription of Irf4 and Irf8 40. This study showed that IRF4 bound to the 3′Eκ and 3′Eλ enhancers and positioned the alleles away from the pericentric chromatin. This, along with the attenuation of the IL-7R-mediated signaling, is required to displace Stat5, followed by the binding of E2A for the activation of iEκ enhancer. Our data indicate a significant reduction in both Irf4 and Irf8 transcripts in the BM and splenic B cells derived from Iqgap1−/− mice. The reductions in Irf4 and Irf8 transcripts are substantial and striking; however, currently, we do not know where and how IQGAP1 links and coordinates the pre-BCR-mediated SLP65 activation. Furthermore, no direct association between SLP65 and IQGAP1 is known. Future studies should focus in defining these molecular interactions.
In summary, we define IQGAP1 as an essential scaffold protein for the development and functions of B cells. IQGAP1 regulates the Ras/Raf→Mek1/2→Erk1/2/Jnk1/2 pathway in pre-B cells, leading to the exit from IL-7R/Stat5-dependent cell cycle and the entry into Igκ light chain recombination. In addition, through an unknown mechanism, IQGAP1 is involved in initiating the transcription of IRF4 and IRF8 that are required for the transition of pre-B into immature B cells. Lack of IQGAP1 significantly reduced the humoral responses against both TD and TI B cell responses. Future studies should focus on further delineating the molecular mechanisms regulated by IQGAP1 and other scaffold proteins.
Materials and Methods
Mice
Iqgap1−/− mice in the C57BL/6 background 20 were used along with the C57BL/6 (WT, Jackson Laboratory, Bar Harbor, Maine) as controls. Mice used for this study were maintained in the Biology Resource Center (BRC) at the Medical College of Wisconsin (MCW) in their pathogen-free facility and were used for experiments between the ages of 6 and 12 weeks of age. All animal protocols were approved by Institutional Animal Care and Use Committees of the IACUC at the Medical College of Wisconsin, Milwaukee, WI. In addition, medical College of Wisconsin is formally accredited by AAALAC, and all the animal care and use-protocols used in this study fully adhere to the specified guidelines of AAALAC.
Flow cytometry
Single-cell suspensions were prepared by gently mincing the dissected organs through 70-µm cell strainers. Single-cell preparations were stained with fluorescent-labeled mAbs in a FACS buffer (PBS containing 5% BSA and 0.5% Sodium Azide solution) for half an hour and washed with the FACS buffer twice. Flow cytometry analyses were conducted in LSR-II (BD Biosciences, San Jose, CA) or MACSQuant Analyzer 10 (Miltenyi Biotec, Bergisch Gladbach, Germany) and analyzed with FlowJo software (FlowJo LLC, Ashland, OR). We used the following antibodies purchased from eBioscience at Thermo-Fischer (Waltham, MA) for flow cytometry: rat anti-mouse B220 (RA3-6B2), rat anti-mouse IgM (R6-60.2), rat anti-mouse IgD (11-26c.2a), rat anti-mouse CD19 (1D3), rat anti-mouse CD1d (1B1), rat anti-mouse CD21/CD35 (4E3), rat-anti-mouse CD23 (B3B4), Armenian hamster anti-moue CD3ε (145-2C11), rat anti-mouse CD16/CD32(93), rat anti-mouse CD43, rat anti-mouse CD23 (B3B4), rat anti-mouse CD93 (AA4.1), rat anti-mouse CD25 and IL-4 (eBioscience, San Diego, CA); rat anti-mouse c-Kit (BD Pharmingen, San Jose, CA); MOMA-1-FITC (Serotec, Raleigh, NC); goat anti-mouse IgM F(ab′)2 (Jackson ImmunoResearch, West Grove, PA, USA); mouse anti-b-actin. For analyzing NF, FO, and MZ B cell populations, single-cell suspensions from spleens were stained with anti-B220-Pacific Blue, anti-CD21-FITC, and anti-CD23-PE-Cy7 mAbs and sorted using FACSAria (BD Biosciences, San Jose, CA). Splenic T1, T2, and T3 cells were characterized by gating for B220+CD93+ B cells and analyzing with IgM/CD23 positivity.
B cell separation and cell sorting
Resting B cells were purified using B cell isolation kit (Miltenyi Biotech Inc., Auburn CA). Total B cells were also sorted using markers including B220 and CD19. T1 B cells, Follicular B or Marginal zone B cells were isolated with gating strategies involving B220/CD93/IgM/CD23, B220/CD21/CD23 panels as indicated in the results section. Separated cells were stained with anti-CD19 and anti-CD3ε mAbs, and their purity was more than 95%, as confirmed by flow cytometry. Viability of the sorted cells were tested using trphan blue stain exclusion.
Clonotyping
Isotyping pre or post-immune sera and quantifying k or l chains in the sera were done using SBA Clonotyping System-HRP kit, a sandwich ELISA-based assay (SouthernBiotech, Birmingham, AL, USA).
Immunohistochemistry staining
Spleens were embedded in Tissue-Tek (Sakura Finetek, Zoeterwoude, The Netherlands), frozen with the dry-ice-Isopropanol mixture, and kept at -80oC until sectioning. 10 µm cryostat sections were generated, air-dried for 1h at room temperature (RT), and fixed in acetone for 10 min. The sections were blocked with 0.1% BSA/PBS containing anti-CD16/CD32 mAb for 45 min, incubated for 1h at RT in the dark with the rat anti-mouse CD3ε-PE, anti-B220-Cy5, MOMA-1-FITC, anti-mouse IgM-PE or anti-mouse IgD-FITC mAbs. Sections were mounted in an aqueous mounting medium and visualized on Olympus FV1000 Two Photon Confocal microscope. Pictures were taken at 40 × magnification.
Bone marrow transplantation (BMT)
BM cells from tibias and femurs of WT and Iqgap1−/− mice were prepared and resuspended in 2% FBS/RPMI1640 (Invitrogen Life Technologies, Carlsbad, CA). 5 × 106 BM cells (200 µl) were injected intravenously to reconstitute 6-8 weeks old, lethally (1100 rads) irradiated WT or Iqgap1−/− mice 4h after the irradiation. The recipient mice were sacrificed 8 weeks later, and the spleens were analyzed for reconstitution of B cells by flow cytometry.
Western blotting
B cells were purified from spleens and resuspended in HEPES buffer (10mM HEPES/0.5% FCS/RPMI 1640). 15×106 B cells were used for each lane. Cells were stimulated with 50 µg/ml goat-anti-mouse IgM F(ab′)2 for 2 min at 37°C or left unstimulated. Stimulation was terminated by adding one volume of 2× ice-cold lysis buffer (100 mM Tris [pH 7.4], 150 mM NaCl, 2% NP-40, 1% deoxycholic acid, 0.2% SDS, 2 mM sodium orthovanadate, with phosphatase inhibitor cocktail, PhosSTOP (Roche Diagnostics GmbH, Mannheim, Germany) and proteinase inhibitor cocktail (Sigma, St Louis, MO). Lysates were incubated for 30 min on ice, and centrifuged at 15,000g for 15 min at 4°C. For Western blotting, cell lysate aliquot (corresponding to 1×106 cells) were separated by SDS-PAGE, transferred to PVDF membrane, and probed with primary and secondary Abs conjugated with horseradish peroxidase. The membranes were probed with the indicated antibody, and the signal was detected using either SuperSignal West Pico Chemiluminescent substrate or Dura (Thermo Scientific, Waltham, MA). We used the following antibodies: phospho-C-Raf (Ser338) (MilliporeSigma, Burlington MA), -Actin (ACTBD11b7) (Santa Cruz, Dallas, TX). We also used the following antibodies from Cell Signaling Technology (Danvers, MA): phospho-MEK1/2(Ser221, 166F8), MEK1/2 (47E6), phosphor-Erk1/2 (Thr202/Tyr204, D13.14.4E) , Erk1/2 (137F5), phosphor-Jnk1/2 (Thr183/Tyr185, 98F2) , Jnk1/2 (9252) phospho-p38 (Thr180/Tyr182, 3D7), p38, phospho-B-Raf (Ser445), anti-E2A (D2B1), anti-Stat3 (124H6), anti-Stat5A (4H1), anti-Stat5B (34662), anti-Ezh2 (D2C9), anti-IRF4 (15106). The fold change in phosphorylation following 5, 20, or 60 min of activation was calculated using this normalized value by comparing it with that value in the unstimulated lane (normalized phospho-protein value after 5/20/60 min of activation/normalized phospho-protein value in the unstimulated lane).
T-dependent and T-independent humoral responses
For TI immune response, 6-8 weeks-old mice were immunized intraperitoneally with 100 μg of TNP-Ficoll (Biosearch Technology, Novato, CA) in PBS. 50 µl of blood was collected from the retro-orbital venous plexus with capillary tubes on days 0 and 7 after immunization. For TD response, mice were intraperitoneally injected with 100 µg TNP-KLH in 200 µl of aluminum hydroxide adjuvant, Imject Alum (Pierce, Rockford, IL) and boosted with10 µg of TNP-KLH without adjuvant on day 21. Sera were collected on days 0, 7, and 28. Sera were analyzed by ELISA for basal and TNP-specific immunoglobulin isotypes using SBA Clonotyping system (SouthernBiotech, Birmingham, AL).
Heavy (CDR3) and Light chain (VK-RS) chain rearrangements
Semiquantitative PCR with genomic DNA was done as previously described 29, 40. The following primers were used to define the heavy chain rearrangements. Degenerate Vκ and Igk intron primers and five-fold template dilutions were used for PCR. Spectrotyping (Fragment analysis) following DNA sequencing (3500 Genetic Analyzer, Applied Biosystems) was done using Peak Scanner Software. Electropherogram records of amplified fragments were collected and analyzed. JH region (5′-CTTACCTGAGGAGACGGTGA-3′; Forward primers were the following: MIGHV1, 5′-TCCAGCACAGCCTACATGCAGCTC-3′; MIGHV2, 5′-CAGGTGCAGCTGAAGGAGTCAGG-3′; MIGHV3, 5′-AGGTGCAGCTTCAGGAGTCAGG-3′; MIGHV5, 5′-CAGCTGGTGGAGTCTGGGGGA-3′; MIGHV6, 5′-AAGTGAAGCTTGAGGAGTCTGG-3′; and MIGHV7, 5′-AGGTGAAGCTGGTGGAGTCTGG-3′. The following primers were used to define the light chain rearrangements. Forward primer: 5′-AGCTTCAGTGGCAGTGGRTCWGGRAC-3’; Reverse primer: 5′-ACATGGAAGTTTTCCTGGGAGAATATG-3′
PCR Array analyses
We performed the RT-qPCR using custom-made amplicons for the indicated genes 57. Assays were done using BioMark™ Real-Time PCR System and the Digital Array Chip from Fluidigm Inc (San Francisco, CA, USA) as described earlier 58. This multiplexed digital array uses a microfluidic dynamic array platform in conjunction with multiplexed RT reactions for mRNA profiling. cDNAs were diluted 1:5 with ddH2O, and the assay was performed in a 48×48 dynamic array. Briefly, a 5 μl sample mixture was prepared for each sample containing 1 × TaqMan Universal Master Mix (No UNG), 1 × GE Sample Loading Reagent (Fluidigm PN 85000746) and each of diluted pre-amplified cDNA. 5 μl of Assay mix was prepared with 1 × each of TaqMan miRNA assay and 1 × Assay Loading Reagent (Fluidigm PN 85000736). An IFC controller was used to prime the fluidics array (chip) with control line fluid and then with samples and assay mixes in the appropriate inlets. After loading, the chip was placed in the BioMark for PCR at 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for 1 min. Five house-keeping genes (beta-glucuronidase, Mm.3317; Hprt, Mm.299381; Hsp70ab1, Mm.2180; GAPDH, Mm.343110;, and beta-actin, Mm.328431) were used to normalize the data. Mouse genomic DNA, positive PCR controls, and RT controls were used as suggested by Fluidgm Inc. The data was analyzed with Real-Time PCR Analysis Software in the BioMark instrument (Fluidigm Inc, San Francisco, CA, USA).
RT-qPCR
For Irf4, Irf4, b-catenin, Bcl-xL, Ccnd3, Ccnd1, and Bim mRNA quantification, sorted BM or splenic B cells from WT and Iqgap1−/− mice were processed for RNA extraction (RNeasy Mini Kit; QIAGEN). Next, cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. Real-time PCR was performed by using a previously published SYBR green protocol with an ABI7900 HT thermal cycler 59. Transcript in each sample was assayed in triplicate, and the mean cycle threshold was used to calculate the x-fold change and control changes for each gene using ΔΔCt. Each experiment used the control (housekeeping) gene actb for global normalization.
Graphs
DeltaGraph 5 (Red Rock Software Inc, Salt Lake City, UT) or PRISM GraphPad 9.0 (San Diego, CA) were used for statistical analysis of data and generation of figure graphs
Statistical methods
Number of animals used to generate each panel are shown within the figures or main text. Data presented in each figure were from a minimum of three independent experiments. Raw data were analyzed with the unpaired Student’s t-test and analysis of variance, followed by the least-significant difference test for comparisons within or between groups. Significant P values (<0.05) are presented in each one of the figures.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary Figure S1. Dysregulated early B cell development in the BM of Iqgap1−/− mice is B cell-intrinsic. Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host BM were harvested and analyzed. A, B) Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells. Staining with anti-B220 antibody indicated the presence of both B220High and B220Low B cells in both the WT and Iqgap1−/− mice. Total cell numbers in each subset were enumerated from six reconstituted mice and shown on right-side panels. C) Absolute numbers of reconstituted B220+IgM− pro/pre-B, B220+IgM+ immature B, and B220Hi recirculating B cells were defined using anti-B220 and anti-IgM antibodies, and their numbers were calculated from live lymphocyte gates. Data shown on the left are representative panels. Data shown are from two independent experiments with three mice each. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. Supplementary Figure S2. Impairment in the splenic B cell development in Iqgap1−/− mice is cell-intrinsic. Indicated host mice were sub-lethally irradiated and reconstituted with donor-derived 5 × 105 BM cells. Eight weeks later, cells from the host spleens were harvested and analyzed. A) Absolute total splenic B cells were comparable between the WT and Iqgap1−/− mice. Viable cells within the lymphocyte gate were analyzed for the absolute number of total B cells using an anti-B220 antibody. Data shown represent the B220+ B cells per spleen from both strains. B) Absolute numbers of immature and mature B cells do not differ between the WT and Iqgap1−/− mice. Total splenic B cells were stained with anti-B220, anti-IgM, and anti-IgD antibodies. Data shown represent the B220+IgM+ immature B and B220+IgM+IgD+ or B220+IgM−IgD+ mature B cells. Data represent absolute numbers of immature or mature B cells per spleen from both strains. Data are shown with the mean ± SEM. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph. Supplementary Figure S3. Expression levels Ccnd3 is moderately increased, and β-catenin is significantly decreased in T1 B cells. Total mRNA was isolated from sorted splenic T1 B cells. RTqPCR was performed to quantify the expression levels of β-catenin, Bcl-xl, Ccnd3, Ccnd1, and Bim. Statistical significance was calculated using Student’s t-test, and p-values are shown below each graph.
Author Contributions
RKL designed the study and performed experiments, data collection, analysis, and drafted the original manuscript. SM conceived of the study, participated in its design, and performed data analysis and writing of the manuscript. All authors have read and approved the final draft of the manuscript.
Funding
NIH R01 AI102893 (S.M.) and NCI R01 CA179363 (S.M.); Alex Lemonade Stand Foundation (S.M.); HRHM Program of MACC Fund/Children’s Hospital of Wisconsin (S.M.), Nicholas Family Foundation (S.M.); Gardetto Family (S.M.); MCW-Cancer Center-Large Seed Grant (S.M.); and Advancing Healthier Wisconsin (S.M.).
Acknowledgments
We thank Dr. Andre Bernards, MGH Cancer Center, Massachusetts General Hospital, MA, for providing the original IQGAP1 knockout mice. We thank Fluidgm corporation for their help with the Digital Array Chip analyses. We also thank the former and current lab members who helped in performing experiments, genotyping mice, and discussions.
Conflicts of Interest
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics Statement
All mice used in this study were utilized responsibly, and all protocols were approved by the institutional IACUC committee at the Medical College of Wisconsin (MCW), Milwaukee, WI.
References
- Sacks, D.B. The role of scaffold proteins in MEK/ERK signalling. Biochem. Soc. Trans 34, 833-836 (2006). [CrossRef]
- Ren, J.G., Li, Z. & Sacks, D.B. IQGAP1 modulates activation of B-Raf. Proc Natl Acad Sci U S A 104, 10465-10469 (2007). [CrossRef]
- McNulty, D.E., Li, Z., White, C.D., Sacks, D.B. & Annan, R.S. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J Biol. Chem 286, 15010-15021 (2011). [CrossRef]
- Sbroggio, M. et al. ERK1/2 activation in heart is controlled by melusin, focal adhesion kinase and the scaffold protein IQGAP1. J Cell Sci 124, 3515-3524 (2011). [CrossRef]
- Liu, X.Y. et al. IQGAP1/ERK regulates fear memory formation via histone posttranslational modifications induced by HDAC2. Neurobiol Learn Mem 171, 107210 (2020). [CrossRef]
- Briggs, M.W. & Sacks, D.B. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep 4, 571-574 (2003). [CrossRef]
- Weissbach, L. et al. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J. Biol. Chem 269, 20517-20521 (1994). [CrossRef]
- Wang, S. et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J. Cell Sci 120, 567-577 (2007). [CrossRef]
- Schmidt, V.A. et al. IQGAP2 functions as a GTP-dependent effector protein in thrombin-induced platelet cytoskeletal reorganization. Blood 101, 3021-3028 (2003). [CrossRef]
- Cupit, L.D., Schmidt, V.A., Miller, F. & Bahou, W.F. Distinct PAR/IQGAP expression patterns during murine development: implications for thrombin-associated cytoskeletal reorganization. Mamm. Genome 15, 618-629 (2004). [CrossRef]
- Brill, S. et al. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell Biol 16, 4869-4878 (1996). [CrossRef]
- Nojima, H. et al. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nat. Cell Biol 10, 971-978 (2008). [CrossRef]
- Abel, A.M. et al. IQGAP1: insights into the function of a molecular puppeteer. Mol Immunol 65, 336-349 (2015). [CrossRef]
- Brown, M.D. & Sacks, D.B. Protein scaffolds in MAP kinase signalling. Cell Signal 21, 462-469 (2009). [CrossRef]
- Awasthi, A. et al. Rap1b facilitates NK cell functions via IQGAP1-mediated signalosomes. J Exp Med 207, 1923-1938 (2010). [CrossRef]
- Abel, A.M. et al. IQ Domain-Containing GTPase-Activating Protein 1 Regulates Cytoskeletal Reorganization and Facilitates NKG2D-Mediated Mechanistic Target of Rapamycin Complex 1 Activation and Cytokine Gene Translation in Natural Killer Cells. Front Immunol 9, 1168 (2018). [CrossRef]
- Wang, J.C. et al. The Rap1-cofilin-1 pathway coordinates actin reorganization and MTOC polarization at the B cell immune synapse. J Cell Sci 130, 1094-1109 (2017). [CrossRef]
- Fukata, M. et al. Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J Biol Chem 272, 29579-29583 (1997). [CrossRef]
- Tanos, B.E., Yeaman, C. & Rodriguez-Boulan, E. An emerging role for IQGAP1 in tight junction control. Small GTPases 9, 375-383 (2018). [CrossRef]
- Li, S., Wang, Q., Chakladar, A., Bronson, R.T. & Bernards, A. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Mol Cell Biol 20, 697-701 (2000). [CrossRef]
- Gorman JA, G.T., Billadeau DA. IQGAP1 mediates lymphocyte cytoskeletal polarization and peripheral effector functions. The Journal of Immunology 182 (2009). [CrossRef]
- Pillai, S., Cariappa, A. & Moran, S.T. Marginal zone B cells. Annu. Rev. Immunol 23, 161-196 (2005). [CrossRef]
- Carey, J.B., Moffatt-Blue, C.S., Watson, L.C., Gavin, A.L. & Feeney, A.J. Repertoire-based selection into the marginal zone compartment during B cell development. J Exp Med 205, 2043-2052 (2008). [CrossRef]
- Novobrantseva, T.I. et al. Rearrangement and expression of immunoglobulin light chain genes can precede heavy chain expression during normal B cell development in mice. J Exp Med 189, 75-88 (1999). [CrossRef]
- Mandal, M. et al. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol 10, 1110-1117 (2009). [CrossRef]
- Khan, S.N. et al. Editing and escape from editing in anti-DNA B cells. Proc Natl Acad Sci U S A 105, 3861-3866 (2008). [CrossRef]
- Luning Prak, E.T., Monestier, M. & Eisenberg, R.A. B cell receptor editing in tolerance and autoimmunity. Ann N Y Acad Sci 1217, 96-121 (2011). [CrossRef]
- Walter, J.E. et al. Expansion of immunoglobulin-secreting cells and defects in B cell tolerance in Rag-dependent immunodeficiency. J Exp Med 207, 1541-1554 (2010). [CrossRef]
- Panigrahi, A.K. et al. RS rearrangement frequency as a marker of receptor editing in lupus and type 1 diabetes. J Exp Med 205, 2985-2994 (2008). [CrossRef]
- Roy, M., Li, Z. & Sacks, D.B. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol 25, 7940-7952 (2005). [CrossRef]
- Jeong, H.W., Li, Z., Brown, M.D. & Sacks, D.B. IQGAP1 binds Rap1 and modulates its activity. J Biol. Chem 282, 20752-20762 (2007). [CrossRef]
- Ren, J.G., Li, Z. & Sacks, D.B. IQGAP1 modulates activation of B-Raf. Proc. Natl. Acad. Sci. U. S. A 104, 10465-10469 (2007). [CrossRef]
- Mason, C.S. et al. Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J 18, 2137-2148 (1999). [CrossRef]
- Yin, X.L., Chen, S., Yan, J., Hu, Y. & Gu, J.X. Identification of interaction between MEK2 and A-Raf-1. Biochim. Biophys. Acta 1589, 71-76 (2002). [CrossRef]
- Macias, M.J., Wiesner, S. & Sudol, M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 513, 30-37 (2002). [CrossRef]
- Roy, M., Li, Z. & Sacks, D.B. IQGAP1 binds ERK2 and modulates its activity. J Biol. Chem 279, 17329-17337 (2004). [CrossRef]
- Fleming, H.E. & Paige, C.J. Pre-B cell receptor signaling mediates selective response to IL-7 at the pro-B to pre-B cell transition via an ERK/MAP kinase-dependent pathway. Immunity 15, 521-531 (2001). [CrossRef]
- Yasuda, T. et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity 28, 499-508 (2008). [CrossRef]
- Lu, R., Medina, K.L., Lancki, D.W. & Singh, H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev 17, 1703-1708 (2003). [CrossRef]
- Johnson, K. et al. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity 28, 335-345 (2008). [CrossRef]
- Rowland, S.L., DePersis, C.L., Torres, R.M. & Pelanda, R. Ras activation of Erk restores impaired tonic BCR signaling and rescues immature B cell differentiation. J Exp Med 207, 607-621 (2010). [CrossRef]
- Morgan, C.J., Hedman, A.C., Li, Z. & Sacks, D.B. Endogenous IQGAP1 and IQGAP3 do not functionally interact with Ras. Sci Rep 9, 11057 (2019). [CrossRef]
- Brown, M.D. & Sacks, D.B. IQGAP1 in cellular signaling: bridging the GAP. Trends Cell Biol 16, 242-249 (2006). [CrossRef]
- Kurella, V.B. et al. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J. Biol. Chem 284, 14857-14865 (2009). [CrossRef]
- Ren, J.G., Li, Z. & Sacks, D.B. IQGAP1 integrates Ca2+/calmodulin and B-Raf signaling. J. Biol. Chem 283, 22972-22982 (2008). [CrossRef]
- Sbroggio, M. et al. IQGAP1 regulates ERK1/2 and AKT signalling in the heart and sustains functional remodelling upon pressure overload. Cardiovasc. Res 91, 456-464 (2011). [CrossRef]
- King, A.J., Wireman, R.S., Hamilton, M. & Marshall, M.S. Phosphorylation site specificity of the Pak-mediated regulation of Raf-1 and cooperativity with Src. FEBS Lett 497, 6-14 (2001). [CrossRef]
- Zang, M., Hayne, C. & Luo, Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J. Biol. Chem 277, 4395-4405 (2002). [CrossRef]
- Joyal, J.L. et al. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry. J. Biol. Chem 272, 15419-15425 (1997). [CrossRef]
- Marais, R., Light, Y., Paterson, H.F. & Marshall, C.J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J 14, 3136-3145 (1995). [CrossRef]
- Vetterkind, S., Poythress, R.H., Lin, Q.Q. & Morgan, K.G. Hierarchical scaffolding of an ERK1/2 activation pathway. Cell Commun. Signal 11, 65 (2013). [CrossRef]
- Hardy, R.R. & Hayakawa, K. A developmental switch in B lymphopoiesis. Proc. Natl. Acad. Sci. U. S. A 88, 11550-11554 (1991). [CrossRef]
- Loder, F. et al. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp. Med 190, 75-89 (1999). [CrossRef]
- Li, Y.S., Hayakawa, K. & Hardy, R.R. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med 178, 951-960 (1993). [CrossRef]
- Shaffer, A.L. & Schlissel, M.S. A truncated heavy chain protein relieves the requirement for surrogate light chains in early B cell development. J Immunol 159, 1265-1275 (1997). [CrossRef]
- van Zelm, M.C. et al. Ig gene rearrangement steps are initiated in early human precursor B cell subsets and correlate with specific transcription factor expression. J Immunol 175, 5912-5922 (2005). [CrossRef]
- Spurgeon, S.L., Jones, R.C. & Ramakrishnan, R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One 3, e1662 (2008). [CrossRef]
- Kolbert, C.P. et al. Multi-platform analysis of microRNA expression measurements in RNA from fresh frozen and FFPE tissues. PLoS One 8, e52517 (2013). [CrossRef]
- Yu, S. et al. Quantitative real-time polymerase chain reaction for the verification of genomic imbalances detected by microarray-based comparative genomic hybridization. Genet Test Mol Biomarkers 13, 751-760 (2009). [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).