Submitted:
06 July 2023
Posted:
10 July 2023
You are already at the latest version
Abstract
Keywords:
INTRODUCTION
MATERIALS AND METHODS
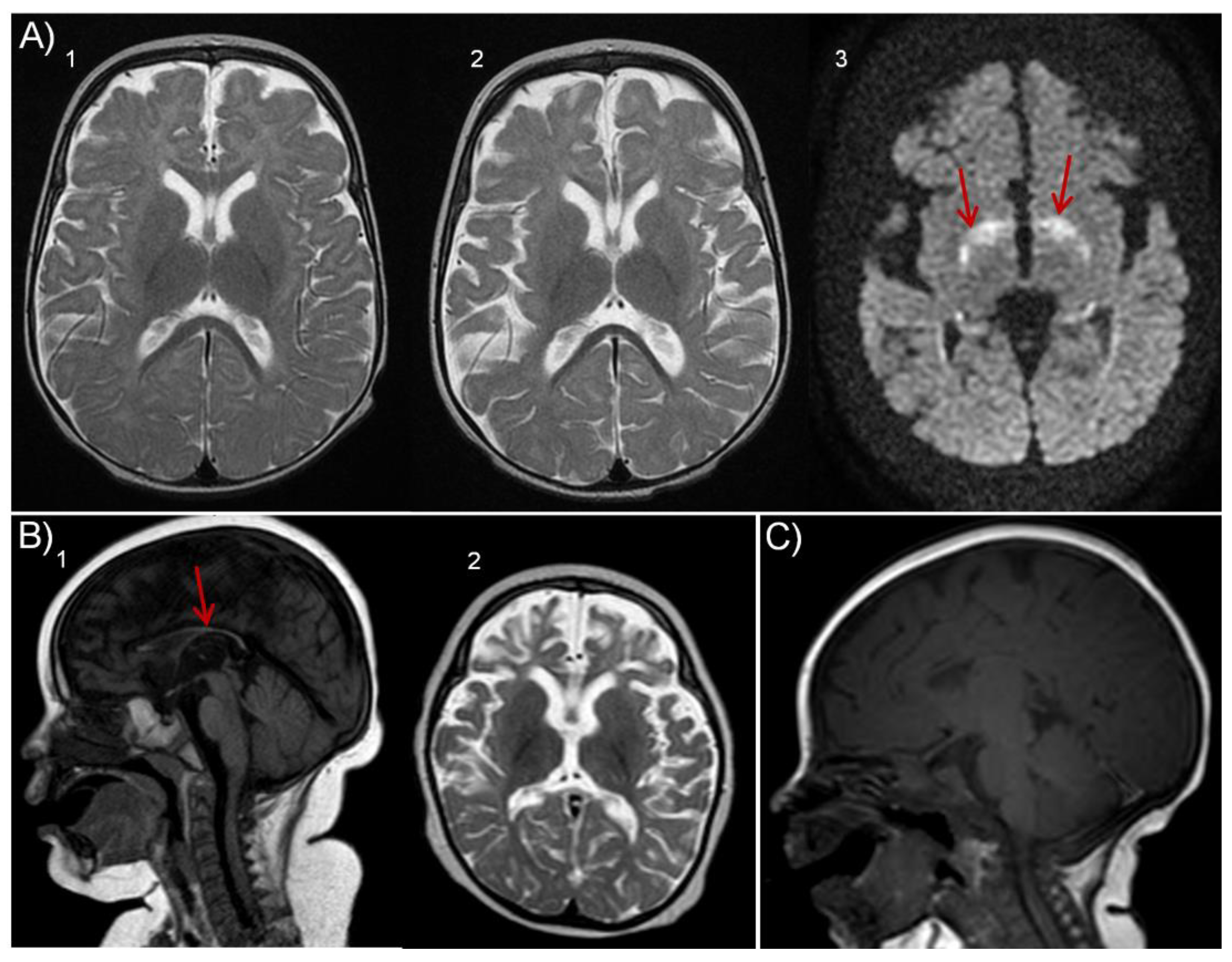
Cases Report
Whole Exome Sequencing
Protein Expression Analysis
RNA Sequencing
Functional Annotation of Differentially Expressed Genes
RESULTS
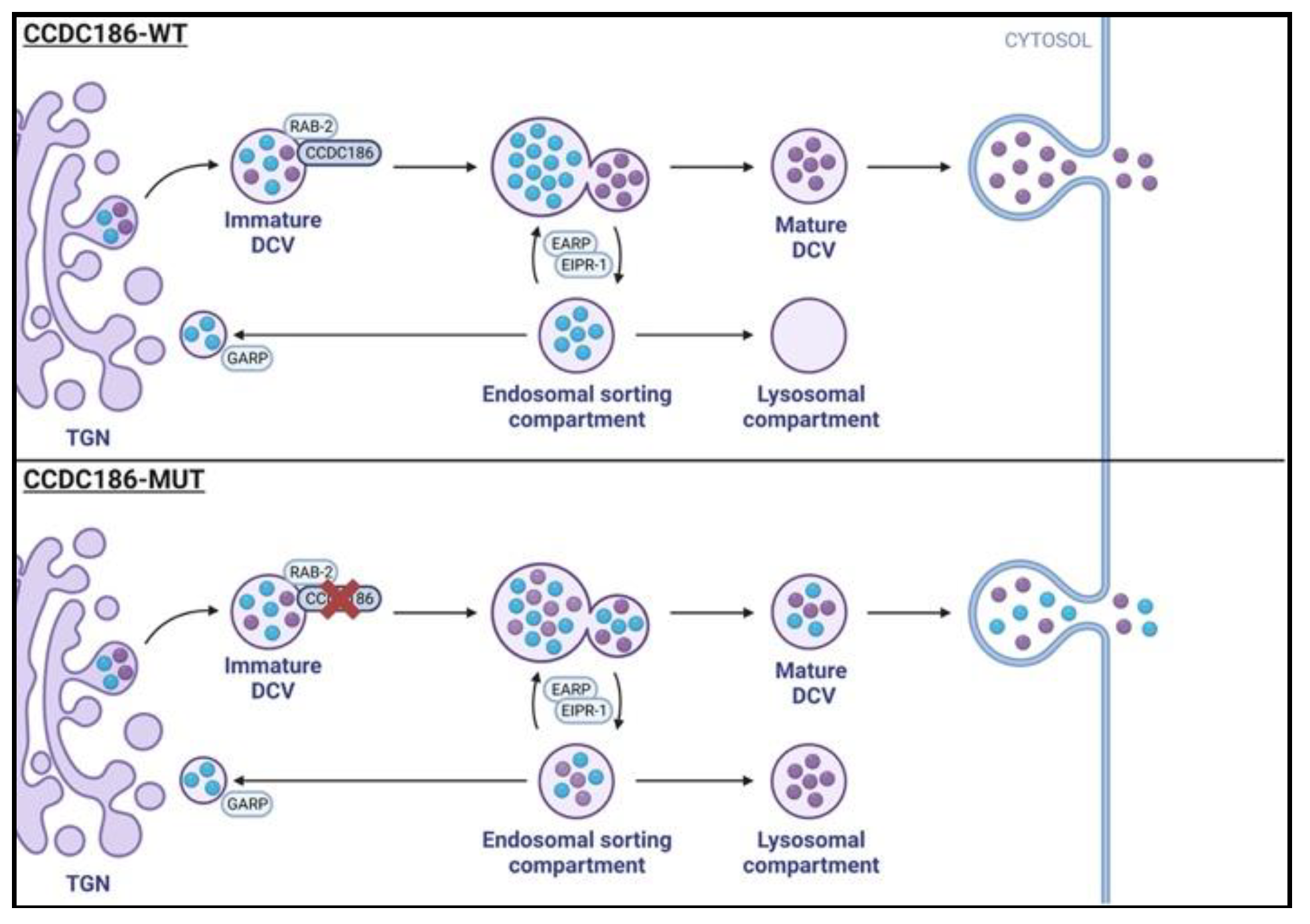
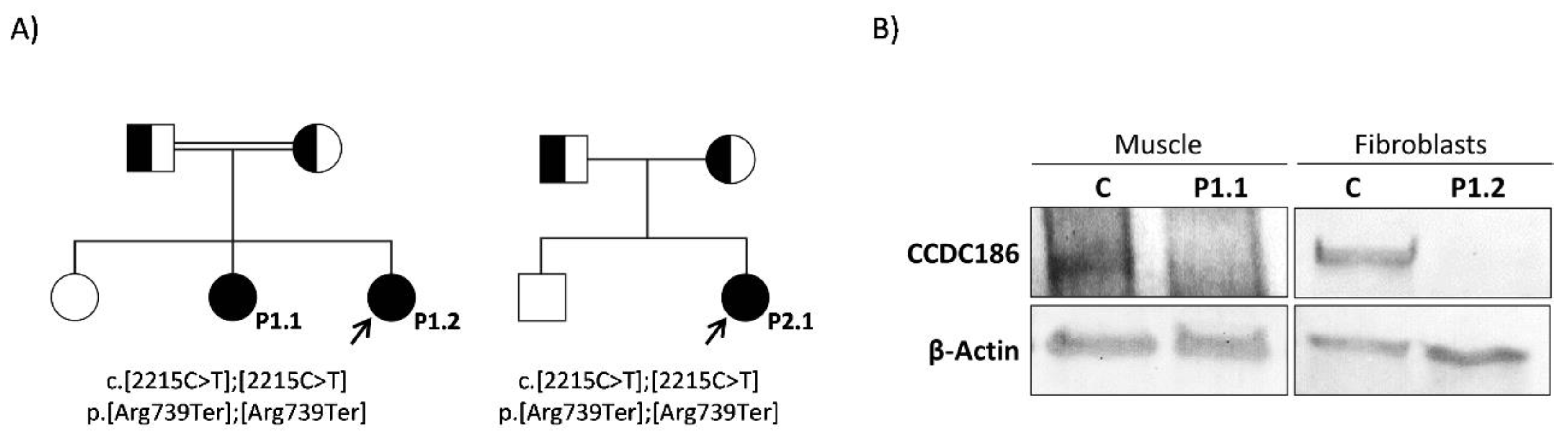
Identification of CCDC186 Variant
Analysis of CCDC186 Protein Expression
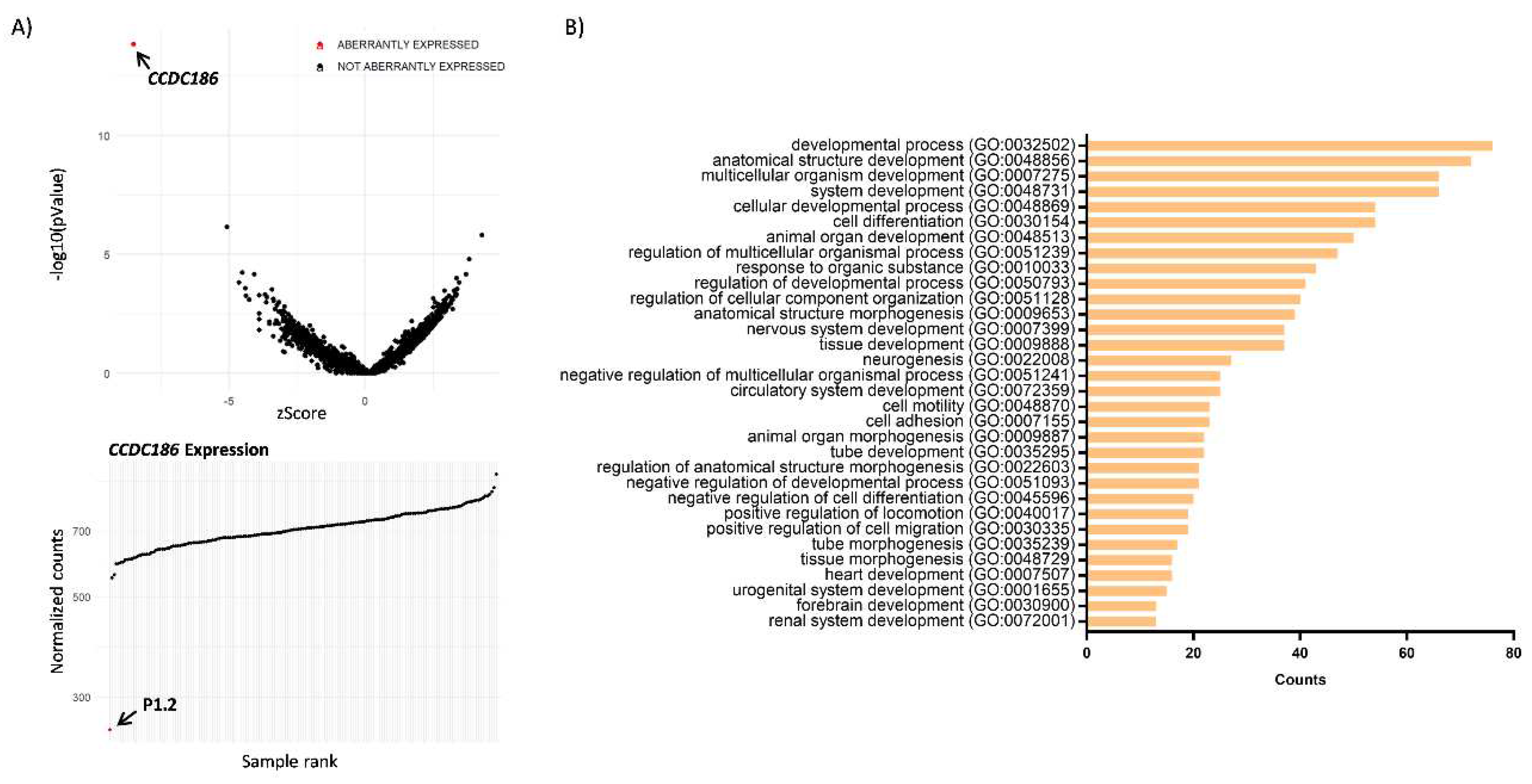
RNA Sequencing
Functional Annotation of Differentially Expressed Genes
DISCUSSION
Funding
Institutional Review Board Statement
Acknowledgments
References
- Cattin-Ortola J, Topalidou I, Dosey A, et al. The dense core vesicle maturation protein CCCP-1 binds RAB-2 and membranes through its C-terminal domain. Traffic 2017;18:720–732. [CrossRef]
- Gondre-Lewis MC, Park JJ, Loh YP. Cellular mechanisms for the biogenesis and transport of synaptic and dense core vesicles. Int Rev Cell Mol Biol 2012;299:27–115. [CrossRef]
- Ailion, M., Hannemann, M., Dalton, S. et al.Two Rab2 interactors regulate dense-core vesicle maturation. Neuron 2014; 82: 167-180. [CrossRef]
- Wang G, Li R, Yang Y, et al. Disruption of the Golgi protein Otg1 gene causes defective hormone secretion and aberrant glucose homeostasis in mice. Cell Biosci2016;6:41. [CrossRef]
- Monies D, Abouelhoda M, AlSayed M et al. The landscape of genetic diseases in Saudi Arabia based on the first 1000 diagnostic panels and exomes. Hum Genet. 2017;136: 921-939. [CrossRef]
- Brugger, M., Becker-Dettling, F., Brunet, T., Strom, T., Meitinger, T., Lurz, E., Borggraefe, I., Wagner, M. A homozygous truncating variant in CCDC186 in an individual with epileptic encephalopathy. Ann. Clin. Transl. Neurol. 8: 278-283, 2021. [CrossRef]
- Laurie S, Fernandez-Callejo M, Marco-Sola S, Trotta JR, Camps J, Chacón A, Espinosa A, Gut M, Gut I, Heath S, et al. From Wet-Lab to Variations: Concordance and Speed of Bioinformatics Pipelines for Whole Genome and Whole Exome Sequencing. Hum. Mutat. 2016, 37:1263–1271.
- Brechtmann F, Mertes C, Matusevičiūtė A, et al. OUTRIDER: A Statistical Method for Detecting Aberrantly Expressed Genes in RNA Sequencing Data. Am J Hum Genet. 2018;103(6):907-917.
- 9. Mertes C, Scheller IF, Yépez VA, et al. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat Commun. 2021;12(1):529.
- 10. Yépez VA, Mertes C, Müller MF, et al. Detection of aberrant gene expression events in RNA sequencing data. Nat Protoc. 2021;16(2):1276-1296.
- Clinical implementation of RNA sequencing for Mendelian disease diagnostics. Yépez VA, Gusic M, Kopajtich R. et al. Genome Med. 2022;14(1):38. IF: 15.2 (D1). [CrossRef]
| Brugger M et al. (2021) | This report | ||||
|---|---|---|---|---|---|
| P1.1 | P 1.2 | P2.1 | |||
| Age at onset | 4 months | Neonatal | Neonatal | Neonatal | |
| Exitus | Unknown. Age at last follw-up 15 months | 7 months | 4 years | 10 months | |
| Origin | Senegalese | Spanish gypsy | Spanish gypsy | Spanish gypsy | |
| Consanguinity | Yes | Yes | Yes | No | |
| Congenital malformations | Infundibular pulmonary stenosis |
No | Laryngomalacia | No | |
| Growth | IUGR | Yes | Yes | No | No |
| Failure to thrive | Yes | Yes | Yes | Yes | |
| Microcephaly | Yes | Yes | Yes (severe) | Yes (severe) | |
| Development | Developmental delay | Yes (severe) | Yes (severe) | Yes | Yes (severe) |
| Neurological findings | Muscular hypotonia | Yes | Yes | Yes | Yes |
| Refractory seizures | Yes | Yes | Yes | Yes | |
| MRI findings | Yes Frontotemporal atrophy | Yes Thin corpus callosum |
Yes Thin corpus callosum Progressive frontotemporal atrophy |
Yes Thin corpus callosum Progressive atrophy Hypomyelination |
|
| Heart findings | NR | No | Yes, hypertrophic cardiomyopathy | No | |
| Gastrointestinal findings | Yes Vomiting, Swallowing Obstipation Exocrine pancreas insufficiency |
Yes Gastroesofageal reflux |
Yes Intestinal ischemia |
Yes Gastroesofageal reflux Swallowing Exocrine pancreas insufficiency |
|
| Endocrinologic findings | Yes Hypothyroidism, Suspected endocrine pancreas insufficiency |
Yes Low IGF1 |
Yes GH deficiency, non-ketotic hypoglycemia |
Yes Low Insulin, cortisol, IGF1, BP3IGFl. Low Elastase |
|
| Respiratory findings | NR | Yes Apneas |
Yes Apneas |
Yes Apneas |
|
| Ophthalmologic findings | Yes Lack of fixation |
Yes Lack of fixation |
No | Yes Lack of fixation |
|
| Auditory findings | Yes Hyperacusia, left side | No | No | No | |
| Genotype | c.[767C>G]; [767C>G] | c. [2215C>T]; [2215C>T] | c. [2215C>T]; [2215C>T] | c. [2215C>T]; [2215C>T] | |
| Effect on protein | p. [Ser256Ter]; [Ser256Ter] | p.[Arg739Ter]; [Arg739Ter] | p.[Arg739Ter]; [Arg739Ter] | p.[Arg739Ter]; [Arg739Ter] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





