Submitted:
09 September 2023
Posted:
12 September 2023
You are already at the latest version
Abstract
Keywords:
Introduction
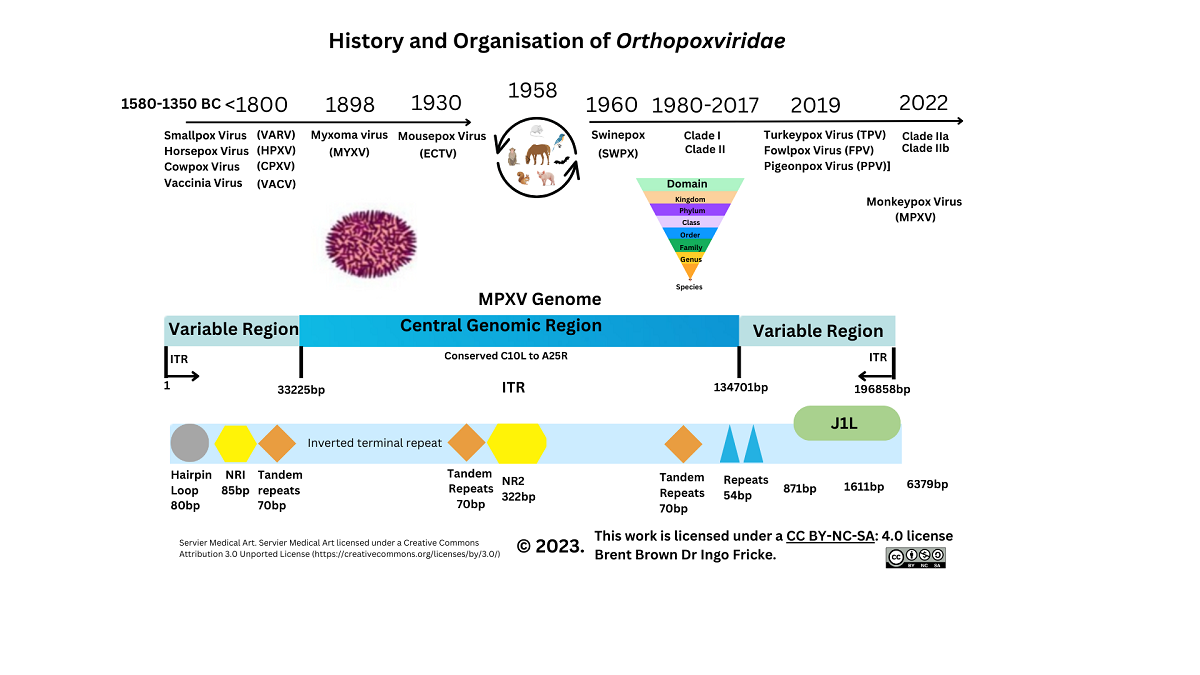
Historical and Epidemiology Overview of Monkeypox Virus
Pathogenesis
Clinical manifestations and Diagnosis
3.2. Cellular Monkeypox and Orthopoxvirus Historical Evolution on Viral Entry
Differences between Smallpox and Monkeypox virus
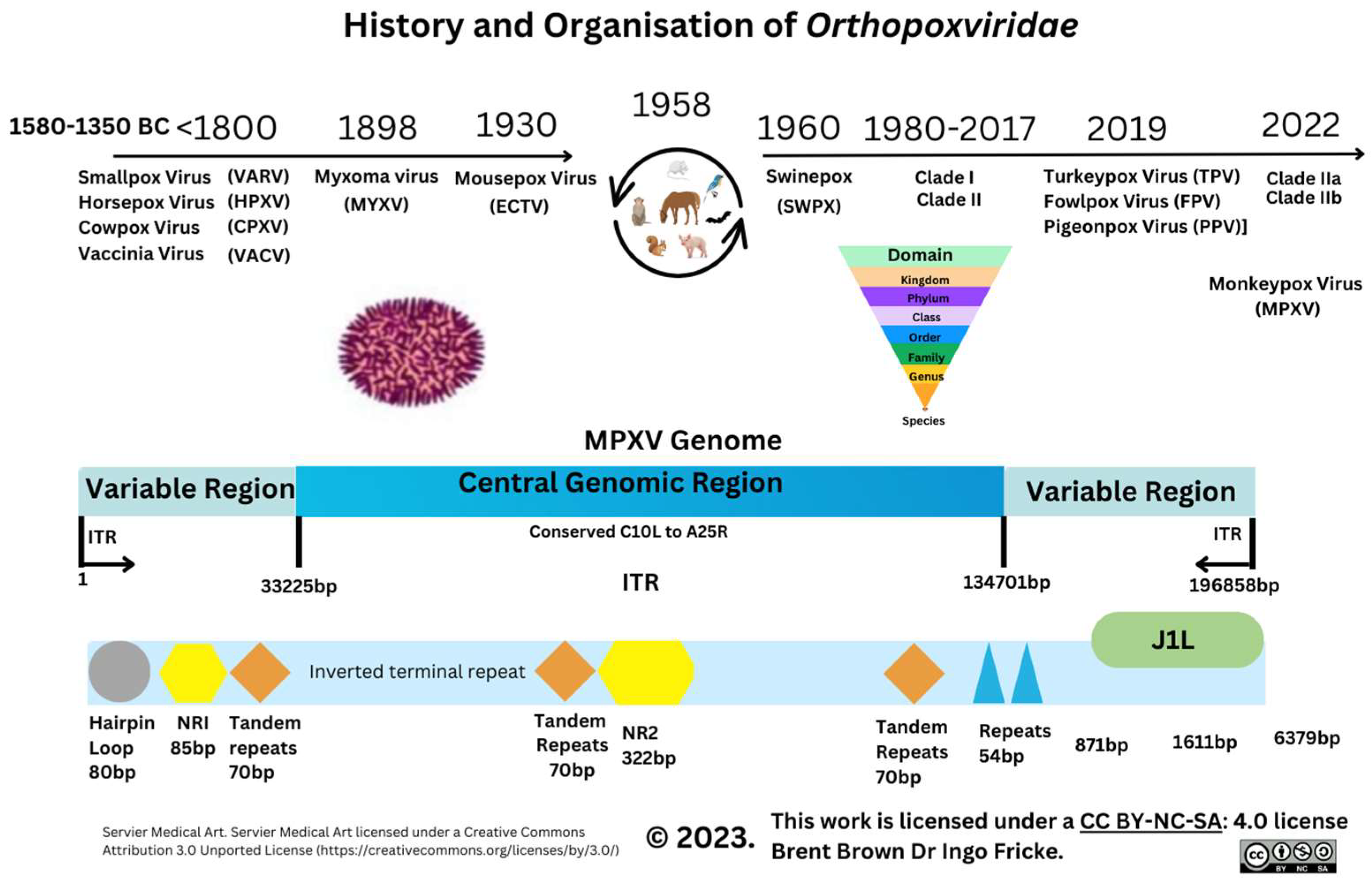
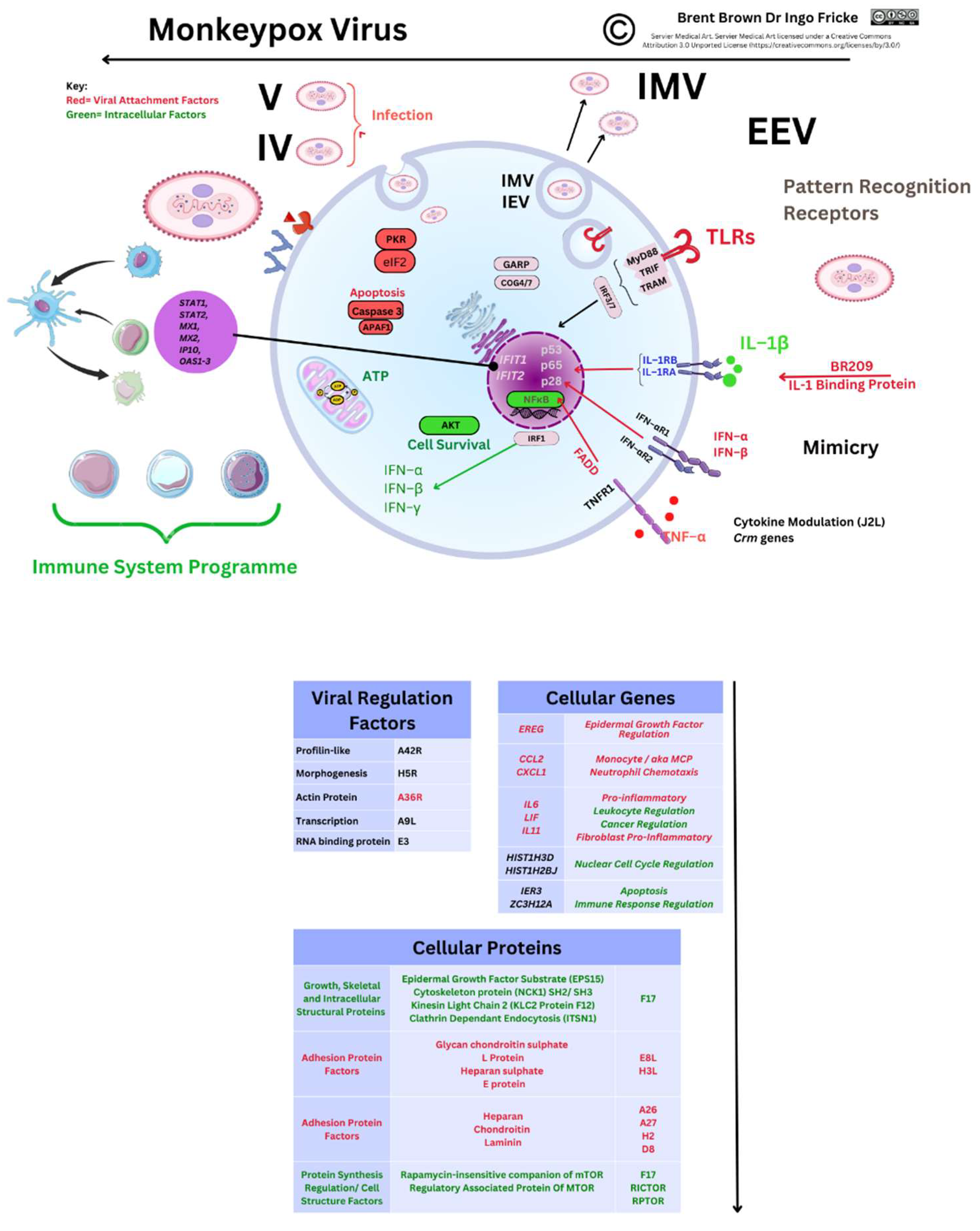
Gene Transcripts and Proteins during Monkeypox Virus Infection
3.5. Recent Monkeypox Virus Protein Characterisation and Research
Orthopoxvirus and Monkeypox Virus 21st Century Immunological Research
Background
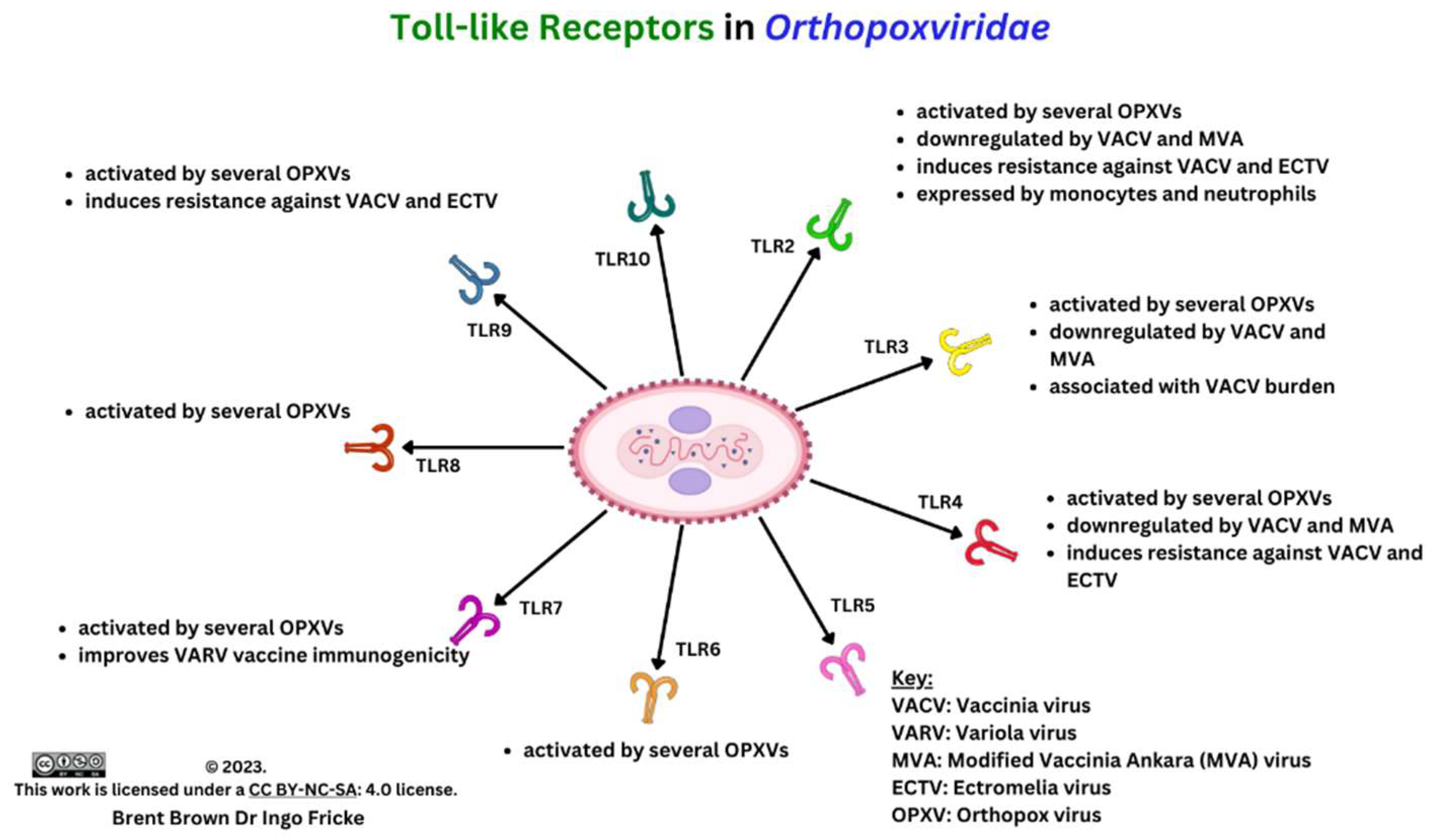
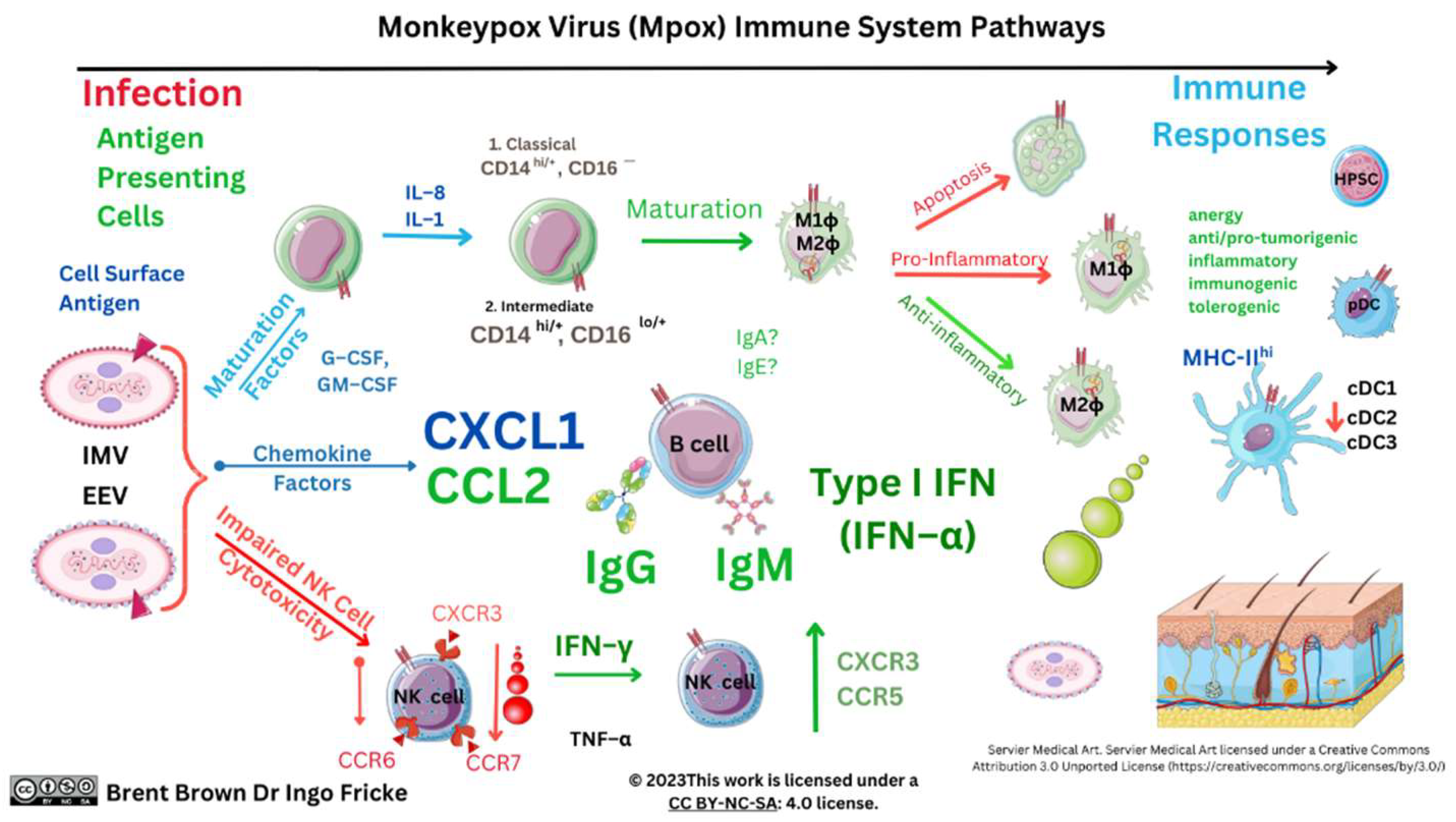
Immunological Response during OPXV Infection
Immunological Responses to Monkeypox Virus and Orthopoxviruses
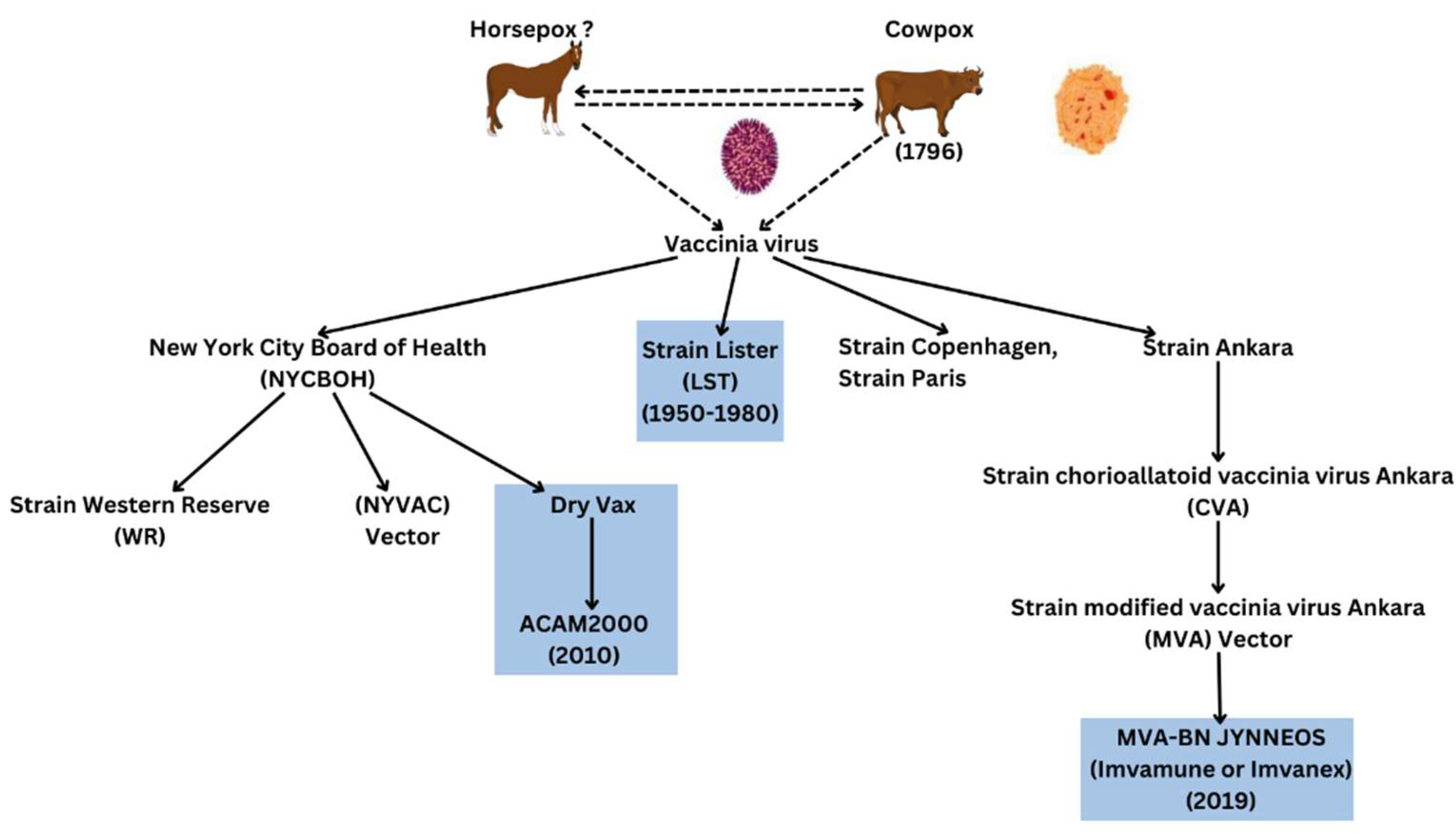
Background to Vaccinia and Orthopoxvirus Role in Cellular Research
Background to Therapeutics, Prevention and Therapy
Discussion
Limitations
Conclusion
Supplementary Materials
Author Contributions
Funding
Availability of data and materials
Conflicts of Interest
Ethical approval
Consent to participate
Consent to publication
Use of Artificial Intelligence (AI) Disclaimer
List of abbreviations
References
- Nuzzo JB, Borio LL, Gostin LO. The WHO Declaration of Monkeypox as a Global Public Health Emergency. JAMA [Internet]. 2022 Aug 16 [cited 2022 Oct 14];328(7):615–6. Available from: https://pubmed.ncbi.nlm.nih.gov/35895041/.
- Schrick L, Tausch SH, Dabrowski PW, Damaso CR, Esparza J, Nitsche A. An Early American Smallpox Vaccine Based on Horsepox. New England Journal of Medicine. 2017, 377, 1491–2. [Google Scholar] [CrossRef]
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, et al. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2016). Arch Virol. 2016, 161, 2921–49. [Google Scholar] [CrossRef]
- Cho CT, Wenner HA. Monkeypox virus. Bacteriol Rev. 1973, 37, 1–18. [Google Scholar] [CrossRef]
- Gessain A, Nakoune E, Yazdanpanah Y. Monkeypox. New England Journal of Medicine. 2022, 387, 1783–93. [Google Scholar] [CrossRef] [PubMed]
- Beer EM, Rao VB. A systematic review of the epidemiology of human monkeypox outbreaks and implications for outbreak strategy. PLoS Negl Trop Dis. 2019, 13, e0007791. [Google Scholar]
- Sereewit J, Lieberman NAP, Xie H, Bakhash SAKM, Nunley BE, Chung B, et al. ORF-Interrupting Mutations in Monkeypox Virus Genomes from Washington and Ohio, 2022. Viruses. 2022, 14, 2393. [Google Scholar] [CrossRef]
- Shchelkunov SN, Totmenin AV, Safronov PF, Mikheev MV, Gutorov VV, Ryazankina OI, et al. Analysis of the Monkeypox Virus Genome. Virology. 2002, 297, 172–94. [Google Scholar] [CrossRef] [PubMed]
- Americo JL, Earl PL, Moss B. Virulence Differences of Monkeypox Virus Clades 1, 2a and 2b.1 in a Small Animal Model. Available from. [CrossRef]
- Woolley SD, Lester R, Devine K, Warrell CE, Groves N, Beadsworth MBJ. Clade IIb A.3 monkeypox virus: an imported lineage during a large global outbreak. Lancet Infect Dis. 2023 Feb.
- Lum FM, Torres-Ruesta A, Tay MZ, Lin RTP, Lye DC, Rénia L, et al. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat Rev Immunol. 2022, 22, 597–613. [Google Scholar] [CrossRef]
- Jacobs BL, Langland JO, Kibler K v. , Denzler KL, White SD, Holechek SA, et al. Vaccinia virus vaccines: Past, present and future. Antiviral Res. 2009, 84, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Knöpfel N, Noguera-Morel L, Latour I, Torrelo A. Viral exanthems in children: A great imitator. Clin Dermatol. 2019, 37, 213–26. [Google Scholar] [CrossRef]
- Patel AB, Pacha O. Skin Reactions to Immune Checkpoint Inhibitors. In 2021. p. 319–30.
- Drago F, Ciccarese G, Gasparini G, Cogorno L, Javor S, Toniolo A, et al. Contemporary infectious exanthems: An update. Vol. 12, Future Microbiology. Future Medicine Ltd.; 2017. p. 171–93.
- Soman, L. Fever with Rashes. The Indian Journal of Pediatrics. 2018, 85, 528–34. [Google Scholar] [CrossRef]
- Kumar N, Acharya A, Gendelman HE, Byrareddy SN. The 2022 outbreak and the pathobiology of the monkeypox virus. J Autoimmun. 2022 Jul;102855.
- Yeh TY, Hsieh ZY, Feehley MC, Feehley PJ, Contreras GP, Su YC, et al. Recombination shapes the 2022 monkeypox (mpox) outbreak. Med. 2022, 3, 824–6. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov SN. Emergence and reemergence of smallpox: The need for development of a new generation smallpox vaccine. Vaccine. 2011 Dec 30;29(SUPPL. 4):D49–53.
- Firth C, Kitchen A, Shapiro B, Suchard MA, Holmes EC, Rambaut A. Using Time-Structured Data to Estimate Evolutionary Rates of Double-Stranded DNA Viruses. Mol Biol Evol. 2010, 27, 2038–51. [Google Scholar] [CrossRef]
- Kerr PJ, Ghedin E, DePasse J V. , Fitch A, Cattadori IM, Hudson PJ, et al. Evolutionary History and Attenuation of Myxoma Virus on Two Continents. PLoS Pathog. 2012, 8, e1002950. [Google Scholar]
- Alakunle E, Moens U, Nchinda G, Okeke MI. Monkeypox Virus in Nigeria: Infection Biology, Epidemiology, and Evolution. Viruses. 2020, 12, 1257. [Google Scholar] [CrossRef]
- Khodakevich L, Jezek Z, Kinzanzka K. ISOLATION OF MONKEYPOX VIRUS FROM WILD SQUIRREL INFECTED IN NATURE. The Lancet. 1986, 327, 98–9. [Google Scholar] [CrossRef] [PubMed]
- Reynolds MG, Yorita KL, Kuehnert MJ, Davidson WB, Huhn GD, Holman RC, et al. Clinical Manifestations of Human Monkeypox Influenced by Route of Infection. J Infect Dis. 2006, 194, 773–80. [Google Scholar] [CrossRef]
- Parker S, Buller RM. A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol. 2013, 8, 129–57. [CrossRef]
- Tu SL, Nakazawa Y, Gao J, Wilkins K, Gallardo-Romero N, Li Y, et al. Characterization of Eptesipoxvirus, a novel poxvirus from a microchiropteran bat. Virus Genes. 2017, 53, 856–67. [Google Scholar] [CrossRef]
- Yang Z, Gray M, Winter L. Why do poxviruses still matter? Cell Biosci. 2021, 11, 96. [Google Scholar]
- Reynolds MG, Guagliardo SAJ, Nakazawa YJ, Doty JB, Mauldin MR. Understanding orthopoxvirus host range and evolution: from the enigmatic to the usual suspects. Curr Opin Virol. 2018 Feb;28:108–15.
- Cohen JM, Bamford A, Eisen S, Emonts M, Ho D, Kadambari S, et al. Care of children exposed to monkeypox. https://eprints.ncl.ac.uk [Internet]. 2022 Oct 1 [cited 2022 Nov 21];21:100514. Available from. [CrossRef]
- Kisalu NK, Mokili JL. Toward Understanding the Outcomes of Monkeypox Infection in Human Pregnancy. J Infect Dis. 2017, 216, 795–7. [Google Scholar] [CrossRef]
- Mbala PK, Huggins JW, Riu-Rovira T, Ahuka SM, Mulembakani P, Rimoin AW, et al. Maternal and Fetal Outcomes Among Pregnant Women With Human Monkeypox Infection in the Democratic Republic of Congo. J Infect Dis. 2017, 216, 824–8. [Google Scholar] [CrossRef]
- O’toole Á, Neher RA, Ndodo N, Borges V, Gannon B, Gomes JP, et al. Putative APOBEC3 deaminase editing in MPXV as evidence for sustained human transmission since at least 2016. Chikwe Ihekweazu 3. Available from. [CrossRef]
- Milewska A, Kindler E, Vkovski P, Zeglen S, Ochman M, Thiel V, et al. APOBEC3-mediated restriction of RNA virus replication. Sci Rep. 2018, 8, 5960. [Google Scholar] [CrossRef]
- Sah R, Mohanty A, Abdelaal A, Reda A, Rodriguez-Morales AJ, Henao-Martinez AF. First Monkeypox deaths outside Africa: no room for complacency. Ther Adv Infect Dis. 2022 Jan 26;9:204993612211240.
- Tomori O, Ogoina D. Monkeypox: The consequences of neglecting a disease, anywhere. Science (1979). 2022, 377, 1261–3. [Google Scholar]
- Du Z, Shao Z, Bai Y, Wang L, Herrera-Diestra JL, Fox SJ, et al. Reproduction number of monkeypox in the early stage of the 2022 multi-country outbreak. J Travel Med. 2022 Dec 27;29(8).
- Li H, Zhang H, Ding K, Wang XH, Sun GY, Liu ZX, et al. The evolving epidemiology of monkeypox virus. Cytokine Growth Factor Rev [Internet]. 2022 Oct 8 [cited 2022 Nov 19]; Available from: https://linkinghub.elsevier.com/retrieve/pii/S1359610122000776. 1359.
- Delamater PL, Street EJ, Leslie TF, Yang YT, Jacobsen KH. Complexity of the Basic Reproduction Number (R0). Emerg Infect Dis. 2019, 25, 1–4. [CrossRef]
- Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, Castelli F, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020, 20, e238–44. [Google Scholar] [CrossRef]
- Leung NHL. Transmissibility and transmission of respiratory viruses. Nat Rev Microbiol. 2021, 19, 528–45. [Google Scholar] [CrossRef]
- Guerra FM, Bolotin S, Lim G, Heffernan J, Deeks SL, Li Y, et al. The basic reproduction number (R 0 ) of measles: a systematic review. Lancet Infect Dis. 2017, 17, e420–8. [Google Scholar] [CrossRef]
- Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, et al. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proceedings of the National Academy of Sciences. 2010, 107, 16262–7. [Google Scholar] [CrossRef]
- Abreu FVS de, Lorene Soares Rocha K, Silva-Oliveira R, Macedo MV, Silva TGM, Gonçalves-dos-Santos ME, et al. Serological Evidence of Orthopoxvirus Infection in Neotropical Primates in Brazil. Pathogens. 2022, 11, 1167. [Google Scholar] [CrossRef] [PubMed]
- Taha TY, Townsend MB, Pohl J, Karem KL, Damon IK, Mbala Kingebeni P, et al. Design and Optimization of a Monkeypox virus Specific Serological Assay. Pathogens. 2023, 12, 396. [Google Scholar] [CrossRef] [PubMed]
- Karem KL, Reynolds M, Braden Z, Lou G, Bernard N, Patton J, et al. Characterization of Acute-Phase Humoral Immunity to Monkeypox: Use of Immunoglobulin M Enzyme-Linked Immunosorbent Assay for Detection of Monkeypox Infection during the 2003 North American Outbreak. Clinical and Vaccine Immunology. 2005, 12, 867–72. [Google Scholar] [CrossRef]
- Hughes LJ, Goldstein J, Pohl J, Hooper JW, Lee Pitts R, Townsend MB, et al. A highly specific monoclonal antibody against monkeypox virus detects the heparin binding domain of A27. Virology. 2014 Sep;464–465:264–73.
- Prkno A, Hoffmann D, Goerigk D, Kaiser M, van Maanen A, Jeske K, et al. Epidemiological Investigations of Four Cowpox Virus Outbreaks in Alpaca Herds, Germany. Viruses. 2017, 9, 344. [Google Scholar] [CrossRef] [PubMed]
- Franke A, Pfaff F, Jenckel M, Hoffmann B, Höper D, Antwerpen M, et al. Classification of Cowpox Viruses into Several Distinct Clades and Identification of a Novel Lineage. Viruses. 2017, 9, 142. [Google Scholar] [CrossRef]
- Atkinson B, Burton C, Pottage T, Thompson K, Ngabo D, Crook A, et al. Infection-competent monkeypox virus contamination identified in domestic settings following an imported case of monkeypox into the <scp>UK</scp>. Environ Microbiol. 2022, 24, 4561–9. [Google Scholar]
- Morgan CN, Whitehill F, Doty JB, Schulte J, Matheny A, Stringer J, et al. Environmental Persistence of Monkeypox Virus on Surfaces in Household of Person with Travel-Associated Infection, Dallas, Texas, USA, 2021. Emerg Infect Dis. 2022, 28, 1982–9.
- Nörz D, Pfefferle S, Brehm TT, Franke G, Grewe I, Knobling B, et al. Evidence of surface contamination in hospital rooms occupied by patients infected with monkeypox, Germany, June 2022. Eurosurveillance. 2022 Jun 30;27(26). 20 June.
- Vaughan A, Aarons E, Astbury J, Brooks T, Chand M, Flegg P, et al. Human-to-Human Transmission of Monkeypox Virus, United Kingdom, October 2018. Emerg Infect Dis. 2020, 26, 782–5.
- Gould S, Atkinson B, Onianwa O, Spencer A, Furneaux J, Grieves J, et al. Air and surface sampling for monkeypox virus in a UK hospital: an observational study. Lancet Microbe. 2022, 3, e904–11. [Google Scholar] [CrossRef]
- McCollum A, diseases IDC infectious, 2014 undefined. Human monkeypox. academic.oup.com [Internet]. [cited 2022 Nov 19]; Available from: https://academic.oup.com/cid/article-abstract/58/2/260/335791. 3357.
- Li H, Zhang H, Ding K, Wang XH, Sun GY, Liu ZX, et al. The evolving epidemiology of monkeypox virus. Cytokine Growth Factor Rev [Internet]. 2022 Oct 8 [cited 2022 Nov 14]; Available from: https://linkinghub.elsevier.com/retrieve/pii/S1359610122000776.
- Bryer J, Freeman EE, Rosenbach M. Monkeypox emerges on a global scale: A historical review and dermatologic primer. J Am Acad Dermatol. 2022, 87, 1069–74. [Google Scholar] [CrossRef]
- Titanji BK, Tegomoh B, Nematollahi S, Konomos M, Kulkarni PA. Monkeypox: A Contemporary Review for Healthcare Professionals. Open Forum Infect Dis [Internet]. 2022 Jul 4 [cited 2022 Nov 14];9(7). Available from: https://academic.oup.com/ofid/article/9/7/ofac310/6615388.
- Zachariou M. Monkeypox: Symptoms seen in London sexual health clinics differ from previous outbreaks, study finds. BMJ [Internet]. 2022 Jul 5 [cited 2022 Nov 14];378:o1659. Available from: https://www.bmj.com/content/378/bmj.o1659. 1659.
- Chen X, Yuan W, Yang X, Shi Y, Zeng X, Huang J, et al. Ultrasensitive and Specific Identification of Monkeypox Virus Congo Basin and West African Strains Using a CRISPR/Cas12b-Based Platform. Microbiol Spectr. 2023 Feb 22. .
- Zheng Y, Song X, Fredj Z, Bian S, Sawan M. Challenges and perspectives of multi-virus biosensing techniques: A review. Anal Chim Acta. 2023 Mar;1244:340860.
- Sui Y, Xu Q, Liu M, Zuo K, Liu X, Liu J. CRISPR-Cas12a-based detection of monkeypox virus. Journal of Infection. 2022, 85, 702–69. [Google Scholar]
- Mistry DA, Wang JY, Moeser ME, Starkey T, Lee LYW. A systematic review of the sensitivity and specificity of lateral flow devices in the detection of SARS-CoV-2. BMC Infect Dis. 2021, 21, 828. [Google Scholar]
- Kwon S, Shin HY. Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19. Life. 2021, 11, 1356. [CrossRef]
- Colavita F, Mazzotta V, Rozera G, Abbate I, Carletti F, Pinnetti C, et al. Kinetics of viral DNA in body fluids and antibody response in patients with acute Monkeypox virus infection. iScience. 2023, 26, 106102. [Google Scholar] [CrossRef]
- Li Y, Olson VA, Laue T, Laker MT, Damon IK. Detection of monkeypox virus with real-time PCR assays. Journal of Clinical Virology. 2006, 36, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Shchelkunov SN, Shcherbakov DN, Maksyutov RA, Gavrilova E v. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. J Virol Methods. 2011, 175, 163–9. [Google Scholar] [CrossRef] [PubMed]
- Peiró-Mestres A, Fuertes I, Camprubí-Ferrer D, Marcos MÁ, Vilella A, Navarro M, et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Eurosurveillance. 2022 Jul 14;27(28). 20 June.
- Orba Y, Sasaki M, Yamaguchi H, Ishii A, Thomas Y, Ogawa H, et al. Orthopoxvirus infection among wildlife in Zambia. Journal of General Virology. 2015, 96, 390–4. [Google Scholar] [CrossRef]
- Davi SD, Kissenkötter J, Faye M, Böhlken-Fascher S, Stahl-Hennig C, Faye O, et al. Recombinase polymerase amplification assay for rapid detection of Monkeypox virus. Diagn Microbiol Infect Dis. 2019, 95, 41–5. [Google Scholar] [CrossRef]
- Minasov G, Inniss NL, Shuvalova L, Anderson WF, Satchell KJF. Structure of the Monkeypox virus profilin-like protein A42R reveals potential functional differences from cellular profilins. Acta Crystallogr F Struct Biol Commun. 2022, 78, 371–7. [Google Scholar] [CrossRef]
- Murk K, Ornaghi M, Schiweck J. Profilin Isoforms in Health and Disease – All the Same but Different. Front Cell Dev Biol. 2021 Aug 12;9.
- Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O’Neill LAJ. A46R and A52R from vaccinia virus are antagonists of host IL-1 and toll-like receptor signaling. Proceedings of the National Academy of Sciences. 2000, 97, 10162–7. [Google Scholar] [CrossRef] [PubMed]
- Talbot-Cooper C, Pantelejevs T, Shannon JP, Cherry CR, Au MT, Hyvönen M, et al. Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling. Cell Host Microbe. 2022, 30, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Rubins KH, Hensley LE, Relman DA, Brown PO. Stunned Silence: Gene Expression Programs in Human Cells Infected with Monkeypox or Vaccinia Virus. PLoS One. 2011, 6, e15615. [Google Scholar]
- Goel RR, Kotenko S V. , Kaplan MJ. Interferon lambda in inflammation and autoimmune rheumatic diseases. Nat Rev Rheumatol. 2021, 17, 349–62. [Google Scholar] [CrossRef] [PubMed]
- Chen SN, Gan Z, Hou J, Yang YC, Huang L, Huang B, et al. Identification and establishment of type IV interferon and the characterization of interferon-υ including its class II cytokine receptors IFN-υR1 and IL-10R2. Nat Commun. 2022, 13, 999. [Google Scholar] [CrossRef]
- Moss, B. Poxvirus Cell Entry: How Many Proteins Does it Take? Viruses. 2012, 4, 688–707. [Google Scholar] [CrossRef] [PubMed]
- Moss B. Membrane fusion during poxvirus entry. Semin Cell Dev Biol. 2016 Dec;60:89–96.
- Schin AM, Diesterbeck US, Moss B. Insights into the Organization of the Poxvirus Multicomponent Entry-Fusion Complex from Proximity Analyses in Living Infected Cells. J Virol. 2021 Jul 26;95(16).
- Kaler J, Hussain A, Flores G, Kheiri S, Desrosiers D. Monkeypox: A Comprehensive Review of Transmission, Pathogenesis, and Manifestation. Cureus. 2022 Jul 3;
- Gong Q, Wang C, Chuai X, Chiu S. Monkeypox virus: a re-emergent threat to humans. Virol Sin [Internet]. 2022 Jul 9; Available from: http://www.ncbi.nlm.nih.gov/pubmed/35820590. /: from: http.
- Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. Poxvirus multiprotein entry-fusion complex. Proc Natl Acad Sci U S A [Internet]. 2005 Dec 20 [cited 2022 Sep 24];102(51):18572–7. Available from: https://www.pnas.org/doi/abs/10.1073/pnas.0509239102.
- Huang Y, Mu L, Wang W. Monkeypox: epidemiology, pathogenesis, treatment and prevention. Signal Transduct Target Ther. 2022, 7, 373. [Google Scholar] [CrossRef]
- Hendrickson RC, Wang C, Hatcher EL, Lefkowitz EJ. Orthopoxvirus Genome Evolution: The Role of Gene Loss. Viruses. 2010, 2, 1933–67. [Google Scholar] [CrossRef]
- Weaver JR, Isaacs SN. Monkeypox virus and insights into its immunomodulatory proteins. Immunol Rev. 2008, 225, 96–113. [Google Scholar] [CrossRef]
- Lin CL, Chung CS, Heine HG, Chang W. Vaccinia Virus Envelope H3L Protein Binds to Cell Surface Heparan Sulfate and Is Important for Intracellular Mature Virion Morphogenesis and Virus Infection In Vitro and In Vivo. J Virol. 2000, 74, 3353–65. [Google Scholar] [CrossRef]
- Kaever T, Matho MH, Meng X, Crickard L, Schlossman A, Xiang Y, et al. Linear Epitopes in Vaccinia Virus A27 Are Targets of Protective Antibodies Induced by Vaccination against Smallpox. J Virol. 2016, 90, 4334–45. [Google Scholar] [CrossRef]
- Estep RD, Messaoudi I, O’Connor MA, Li H, Sprague J, Barron A, et al. Deletion of the Monkeypox Virus Inhibitor of Complement Enzymes Locus Impacts the Adaptive Immune Response to Monkeypox Virus in a Nonhuman Primate Model of Infection. J Virol. 2011, 85, 9527–42. [Google Scholar] [CrossRef]
- Arndt WD, Cotsmire S, Trainor K, Harrington H, Hauns K, Kibler K v. , et al. Evasion of the Innate Immune Type I Interferon System by Monkeypox Virus. J Virol. 2015, 89, 10489–99. [Google Scholar] [CrossRef]
- Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020, 20, 537–51. [Google Scholar] [CrossRef] [PubMed]
- Brown E, Senkevich TG, Moss B. Vaccinia Virus F9 Virion Membrane Protein Is Required for Entry but Not Virus Assembly, in Contrast to the Related L1 Protein. J Virol [Internet]. 2006 Oct [cited 2022 Sep 24];80(19):9455–64. Available from: https://journals.asm.org/doi/10.1128/JVI.01149-06.
- Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol [Internet]. 2013 Sep [cited 2022 Sep 26];5(9). Available from: https://pubmed.ncbi.nlm.nih.gov/23838441/.
- Rubins KH, Hensley LE, Bell GW, Wang C, Lefkowitz EJ, Brown PO, et al. Comparative analysis of viral gene expression programs during poxvirus infection: A transcriptional map of the vaccinia and monkeypox genomes. PLoS One. 2008, 3, 1–12. [Google Scholar]
- Rampogu S, Kim Y, Kim SW, Lee KW. An overview on monkeypox virus: Pathogenesis, transmission, host interaction and therapeutics. Front Cell Infect Microbiol. 2023 Feb 10;13.
- Realegeno S, Priyamvada L, Kumar A, Blackburn JB, Hartloge C, Puschnik AS, et al. Conserved Oligomeric Golgi (COG) Complex Proteins Facilitate Orthopoxvirus Entry, Fusion and Spread. Viruses. 2020, 12, 707. [Google Scholar] [CrossRef]
- Maluquer de Motes, C. Poxvirus cGAMP nucleases: Clues and mysteries from a stolen gene. PLoS Pathog. 2021, 17, e1009372. [Google Scholar] [CrossRef] [PubMed]
- Eaglesham JB, Pan Y, Kupper TS, Kranzusch PJ. Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS–STING signalling. Nature. 2019, 566, 259–63. [Google Scholar] [CrossRef] [PubMed]
- Realegeno S, Puschnik AS, Kumar A, Goldsmith C, Burgado J, Sambhara S, et al. Monkeypox Virus Host Factor Screen Using Haploid Cells Identifies Essential Role of GARP Complex in Extracellular Virus Formation. J Virol. 2017 Jun;91(11). J Virol.
- Cui W, Huang H, Duan Y, Luo Z, Wang H, Zhang T, et al. Crystal structure of monkeypox H1 phosphatase, an antiviral drug target. Protein Cell. 2022 Nov 5;
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009, 458, 1185–90. [Google Scholar] [CrossRef]
- Tang Z, Mao Y, Meng Y, Qiu X, Bajinka O, Wu G, et al. A bioinformatics approach to systematically analyze the molecular patterns of monkeypox virus-host cell interactions. Available from: https://doi.org/10.1101/2022.10.12.511850 (bioRxiv, pre-print). [CrossRef]
- Arlt A, Schäfer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol. 2011 Jun;90(6–7):545–52.
- Fung KY, Louis C, Metcalfe RD, Kosasih CC, Wicks IP, Griffin MDW, et al. Emerging roles for IL-11 in inflammatory diseases. Cytokine. 2022 Jan;149:155750.
- Widjaja AA, Chothani S, Viswanathan S, Goh JWT, Lim WW, Cook SA. IL11 Stimulates IL33 Expression and Proinflammatory Fibroblast Activation across Tissues. Int J Mol Sci. 2022, 23, 8900. [Google Scholar] [CrossRef]
- Thuong NTT, Hawn TR, Chau TTH, Bang ND, Yen NTB, Thwaites GE, et al. Epiregulin (EREG) variation is associated with susceptibility to tuberculosis. Genes Immun. 2012, 13, 275–81. [Google Scholar] [CrossRef]
- Odell ID, Steach H, Gauld SB, Reinke-Breen L, Karman J, Carr TL, et al. Epiregulin is a dendritic cell–derived EGFR ligand that maintains skin and lung fibrosis. Sci Immunol. 2022 Dec 16;7(78).
- Pradhan SA, Rather MI, Tiwari A, Bhat VK, Kumar A. Evidence that TSC2 acts as a transcription factor and binds to and represses the promoter of Epiregulin. Nucleic Acids Res. 2014, 42, 6243–55. [Google Scholar] [CrossRef]
- Cao W, Luo L lin, Chen W wei, Liang L, Zhang R ran, Zhao Y lin, et al. Polymorphism in the EREG gene confers susceptibility to tuberculosis. BMC Med Genet. 2019, 20, 7. [Google Scholar]
- Hop PJ, Luijk R, Daxinger L, van Iterson M, Dekkers KF, Jansen R, et al. Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference. Genome Biol. 2020, 21, 220. [Google Scholar]
- Berasain C, Avila MA. Amphiregulin. Semin Cell Dev Biol. 2014 Apr;28:31–41.
- Yousaf M, Ismail S, Ullah A, Bibi S. Immuno-informatics profiling of monkeypox virus cell surface binding protein for designing a next generation multi-valent peptide-based vaccine. Front Immunol. 2022 Nov 2;13.
- Korbecki J, Barczak K, Gutowska I, Chlubek D, Baranowska-Bosiacka I. CXCL1: Gene, Promoter, Regulation of Expression, mRNA Stability, Regulation of Activity in the Intercellular Space. Int J Mol Sci. 2022, 23, 792. [Google Scholar] [CrossRef] [PubMed]
- Zaucha GM, Jahrling PB, Geisbert TW, Swearengen JR, Hensley L. The Pathology of Experimental Aerosolized Monkeypox Virus Infection in Cynomolgus Monkeys (Macaca fascicularis). Laboratory Investigation. 2001, 81, 1581–600. [Google Scholar] [CrossRef]
- Brown B, Ojha V, Fricke I, Al-Sheboul SA, Imarogbe C, Gravier T, et al. Innate and Adaptive Immunity during SARS-CoV-2 Infection: Biomolecular Cellular Markers and Mechanisms. Vaccines (Basel). 2023, 11, 408. [Google Scholar] [CrossRef]
- Davies ML, Parekh NJ, Kaminsky LW, Soni C, Reider IE, Krouse TE, et al. A systemic macrophage response is required to contain a peripheral poxvirus infection. PLoS Pathog. 2017, 13, e1006435. [Google Scholar]
- Byrd D, Shepherd N, Lan J, Hu N, Amet T, Yang K, et al. Primary Human Macrophages Serve as Vehicles for Vaccinia Virus Replication and Dissemination. J Virol. 2014, 88, 6819–31. [Google Scholar] [CrossRef]
- Bourquain D, Schrick L, Tischer BK, Osterrieder K, Schaade L, Nitsche A. Replication of cowpox virus in macrophages is dependent on the host range factor p28/N1R. Virol J. 2021, 18, 173. [Google Scholar] [CrossRef]
- Quigley M, Martinez J, Huang X, Yang Y. A critical role for direct TLR2-MyD88 signaling in CD8 T-cell clonal expansion and memory formation following vaccinia viral infection. Blood. 2009, 113, 2256–64. [Google Scholar] [CrossRef]
- Bortolotti D, Gentili V, Rizzo S, Schiuma G, Beltrami S, Strazzabosco G, et al. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms [Internet]. 2021 Aug;9(9):1820. Available from. [CrossRef]
- TLR2 on CD4+ and CD8+ T cells promotes late control of Mycobacterium tuberculosis infection. Available from (bioRxiv, pre-print). [CrossRef]
- Schattner, M. Platelet TLR4 at the crossroads of thrombosis and the innate immune response. J Leukoc Biol [Internet]. 2018, 105, 873–80. [Google Scholar] [CrossRef]
- Dai R, Huang X, Yang Y. γδT Cells Are Required for CD8+ T Cell Response to Vaccinia Viral Infection. Front Immunol. 2021 Oct 8;12.
- Serrano R, Wesch D, Kabelitz D. Correction: Serrano, R.; Wesch, D.; Kabelitz, D. Activation of Human γδ T Cells: Modulation by Toll-Like Receptor 8 Ligands and Role of Monocytes. Cells 2020, 9, 1977. [Google Scholar] [CrossRef] [PubMed]
- Akhtar N, Kaushik V, Grewal RK, Wani AK, Suwattanasophon C, Choowongkomon K, et al. Immunoinformatics-Aided Design of a Peptide Based Multiepitope Vaccine Targeting Glycoproteins and Membrane Proteins against Monkeypox Virus. Viruses. 2022, 14, 2374. [Google Scholar] [CrossRef]
- Yaghoubi A, Khazaei M, Avan A, Hasanian SM, Cho WC, Soleimanpour S. p28 Bacterial Peptide, as an Anticancer Agent. Front Oncol. 2020 Aug 6;10.
- van Vliet K, Mohamed MR, Zhang L, Villa NY, Werden SJ, Liu J, et al. Poxvirus Proteomics and Virus-Host Protein Interactions. Microbiology and Molecular Biology Reviews. 2009, 73, 730–49. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Navarro A, González-Soria I, Caldiño-Bohn R, Bobadilla NA. Integrative view of serpins in health and disease: the contribution of serpinA3. American Journal of Physiology-Cell Physiology. 2020 Oct 28;ajpcell.00366.2020.
- Nathaniel R, MacNeill AL, Wang YX, Turner PC, Moyer RW. Cowpox virus CrmA, Myxoma virus SERP2 and baculovirus P35 are not functionally interchangeable caspase inhibitors in poxvirus infections. Journal of General Virology. 2004, 85, 1267–78. [Google Scholar] [CrossRef] [PubMed]
- Graham SC, Bahar MW, Abrescia NGA, Smith GL, Stuart DI, Grimes JM. Structure of CrmE, a Virus-encoded Tumour Necrosis Factor Receptor. J Mol Biol. 2007, 372, 660–71. [Google Scholar] [CrossRef]
- Gallwitz S, Schutzbank T, Heberling RL, Kalter SS, Galpin JE. Smallpox: Residual Antibody after Vaccination. J Clin Microbiol. 2003, 41, 4068–70. [Google Scholar] [CrossRef]
- Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting Edge: Long-Term B Cell Memory in Humans after Smallpox Vaccination. The Journal of Immunology. 2003, 171, 4969–73. [Google Scholar] [CrossRef]
- Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol Rev. 2006, 211, 320–37. [Google Scholar] [CrossRef]
- Hammarlund E, Dasgupta A, Pinilla C, Norori P, Früh K, Slifka MK. Monkeypox virus evades antiviral CD4 + and CD8 + T cell responses by suppressing cognate T cell activation. Proceedings of the National Academy of Sciences. 2008, 105, 14567–72. [Google Scholar] [CrossRef]
- Song H, Josleyn N, Janosko K, Skinner J, Reeves RK, Cohen M, et al. Monkeypox Virus Infection of Rhesus Macaques Induces Massive Expansion of Natural Killer Cells but Suppresses Natural Killer Cell Functions. PLoS One. 2013, 8, e77804. [Google Scholar]
- Agrati C, Cossarizza A, Mazzotta V, Grassi G, Casetti R, de Biasi S, et al. Immunological Signature in Human Cases of Monkeypox Infection in 2022 Outbreak. SSRN Electronic Journal. 2022.
- Song H, Sidney J, Wiseman RW, Josleyn N, Cohen M, Blaney JE, et al. Characterizing monkeypox virus specific CD8+ T cell epitopes in rhesus macaques. Virology. 2013 Dec;447(1–2):181–6.
- Lazear E, Sun MM, Wang X, Geurs TL, Nelson CA, Campbell JA, et al. Structural basis of cowpox evasion of NKG2D immunosurveillance 1 2. Available from. [CrossRef]
- Depierreux DM, Smith GL, Ferguson BJ. Transcriptional reprogramming of natural killer cells by vaccinia virus shows both distinct and conserved features with mCMV. Front Immunol. 2023 Feb 23;14.
- Buller CW, Mathew PA, Mathew SO. Roles of NK Cell Receptors 2B4 (CD244), CS1 (CD319), and LLT1 (CLEC2D) in Cancer. Cancers (Basel). 2020, 12, 1755. [Google Scholar] [CrossRef] [PubMed]
- Sun Z, Li Y, Zhang Z, Fu Y, Han X, Hu Q, et al. CD160 Promotes NK Cell Functions by Upregulating Glucose Metabolism and Negatively Correlates With HIV Disease Progression. Front Immunol. 2022 Aug 19;13.
- Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, Bray M, et al. Smallpox vaccine–induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005, 11, 740–7. [Google Scholar] [CrossRef] [PubMed]
- Karem KL, Reynolds M, Hughes C, Braden Z, Nigam P, Crotty S, et al. Monkeypox-Induced Immunity and Failure of Childhood Smallpox Vaccination To Provide Complete Protection. Clinical and Vaccine Immunology. 2007, 14, 1318–27. [Google Scholar] [CrossRef] [PubMed]
- Johnson RF, Dyall J, Ragland DR, Huzella L, Byrum R, Jett C, et al. Comparative Analysis of Monkeypox Virus Infection of Cynomolgus Macaques by the Intravenous or Intrabronchial Inoculation Route. J Virol. 2011, 85, 2112–25. [Google Scholar] [CrossRef]
- Xuan DTM, Yeh IJ, Wu CC, Su CY, Liu HL, Chiao CC, et al. Comparison of Transcriptomic Signatures between Monkeypox-Infected Monkey and Human Cell Lines. J Immunol Res. 2022 Sep 1;2022:1–17.
- Bourquain D, Nitsche A. Cowpox virus but not Vaccinia virus induces secretion of CXCL1, IL-8 and IL-6 and chemotaxis of monocytes in vitro. Virus Res. 2013, 171, 161–7. [Google Scholar] [CrossRef]
- Kindrachuk J, Arsenault R, Kusalik A, Kindrachuk KN, Trost B, Napper S, et al. Systems Kinomics Demonstrates Congo Basin Monkeypox Virus Infection Selectively Modulates Host Cell Signaling Responses as Compared to West African Monkeypox Virus. Molecular & Cellular Proteomics. 2012, 11, M111–015701. [Google Scholar]
- Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science (1979). 2017 Apr 21;356(6335).
- Hijdra D, Vorselaars ADM, Grutters JC, Claessen AME, Rijkers GT. Phenotypic Characterization of Human Intermediate Monocytes. Front Immunol [Internet]. 2013;4. Available from. [CrossRef]
- Xuan DTM, Yeh IJ, Wu CC, Su CY, Liu HL, Chiao CC, et al. Comparison of Transcriptomic Signatures between Monkeypox-Infected Monkey and Human Cell Lines. J Immunol Res. 2022 Sep 1;2022:1–17.
- Brown B, Gravier T, Fricke I, Al-Sheboul SA, Carp TN, Leow CY, et al. Immunopathogenesis of Nipah Virus Infection and Associated Immune Responses. Immuno. 2023, 3, 160–81. [Google Scholar]
- Moutaftsi M, Tscharke DC, Vaughan K, Koelle DM, Stern L, Calvo-Calle M, et al. Uncovering the interplay between CD8, CD4 and antibody responses to complex pathogens. Future Microbiol. 2010, 5, 221–39. [Google Scholar] [CrossRef]
- Grifoni A, Zhang Y, Tarke A, Sidney J, Rubiro P, Reina-Campos M, et al. Defining antigen targets to dissect vaccinia virus and monkeypox virus-specific T cell responses in humans. Cell Host Microbe. 2022, 30, 1662–1670. [Google Scholar] [CrossRef]
- Yefet R, Friedel N, Tamir H, Polonsky K, Mor M, Hagin D, et al. A35R and H3L are Serological and B Cell Markers for Monkeypox Infection. Available from. [CrossRef]
- Rowley DA, Fitch FW. The road to the discovery of dendritic cells, a tribute to Ralph Steinman. Cell Immunol. 2012, 273, 95–8. [Google Scholar] [CrossRef] [PubMed]
- Lechmann M, Berchtold S, Steinkasserer A, Hauber J. CD83 on dendritic cells: more than just a marker for maturation. Trends Immunol. 2002, 23, 273–5. [Google Scholar] [CrossRef] [PubMed]
- Kelly B, ONeill LAJ. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res [Internet]. 2015, 25, 771–84. [Google Scholar] [CrossRef]
- Marongiu L, Protti G, Facchini FA, Valache M, Mingozzi F, Ranzani V, et al. Maturation signatures of conventional dendritic cell subtypes in COVID-19 suggest direct viral sensing. Eur J Immunol. 2022, 52, 109–22. [Google Scholar] [CrossRef] [PubMed]
- Dudek AM, Martin S, Garg AD, Agostinis P. Immature, Semi-Mature, and Fully Mature Dendritic Cells: Toward a DC-Cancer Cells Interface That Augments Anticancer Immunity. Front Immunol. 2013;4.
- Flechsig C, Suezer Y, Kapp M, Tan SM, Löffler J, Sutter G, et al. Uptake of antigens from modified vaccinia Ankara virus-infected leukocytes enhances the immunostimulatory capacity of dendritic cells. Cytotherapy. 2011, 13, 739–52. [Google Scholar] [CrossRef]
- Zielinski CE. T helper cell subsets: diversification of the field. Eur J Immunol. 2023 Feb 15;2250218.
- Matic S, Popovic S, Djurdjevic P, Todorovic D, Djordjevic N, Mijailovic Z, et al. SARS-CoV-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PLoS One. 2020, 15, e0241097. [Google Scholar]
- Al-Sheboul SA, Brown B, Shboul Y, Fricke I, Imarogbe C, Alzoubi KH. An Immunological Review of SARS-CoV-2 Infection and Vaccine Serology: Innate and Adaptive Responses to mRNA, Adenovirus, Inactivated and Protein Subunit Vaccines. Vaccines (Basel) [Internet]. 2022, 11, 51. [Google Scholar] [CrossRef]
- Hansen SJ, Rushton J, Dekonenko A, Chand HS, Olson GK, Hutt JA, et al. Cowpox virus inhibits human dendritic cell immune function by nonlethal, nonproductive infection. Virology. 2011, 412, 411–25. [Google Scholar] [CrossRef]
- Spel L, Luteijn RD, Drijfhout JW, Nierkens S, Boes M, Wiertz EJH. Endocytosed soluble cowpox virus protein CPXV012 inhibits antigen cross-presentation in human monocyte-derived dendritic cells. Immunol Cell Biol. 2018, 96, 137–48. [Google Scholar] [CrossRef]
- Leite Pereira A, Jouhault Q, Marcos Lopez E, Cosma A, Lambotte O, le Grand R, et al. Modulation of Cell Surface Receptor Expression by Modified Vaccinia Virus Ankara in Leukocytes of Healthy and HIV-Infected Individuals. Front Immunol. 2020 Sep 8;11.
- Thiele F, Tao S, Zhang Y, Muschaweckh A, Zollmann T, Protzer U, et al. Modified Vaccinia Virus Ankara-Infected Dendritic Cells Present CD4 + T-Cell Epitopes by Endogenous Major Histocompatibility Complex Class II Presentation Pathways. J Virol. 2015, 89, 2698–709. [Google Scholar] [CrossRef]
- Chahroudi A, Garber DA, Reeves P, Liu L, Kalman D, Feinberg MB. Differences and Similarities in Viral Life Cycle Progression and Host Cell Physiology after Infection of Human Dendritic Cells with Modified Vaccinia Virus Ankara and Vaccinia Virus. J Virol. 2006, 80, 8469–81. [Google Scholar] [CrossRef] [PubMed]
- Breloer M, Fleischer B. CD83 regulates lymphocyte maturation, activation and homeostasis. Trends Immunol. 2008, 29, 186–94. [Google Scholar] [CrossRef]
- Riaz B, Islam SMS, Ryu HM, Sohn S. CD83 Regulates the Immune Responses in Inflammatory Disorders. Int J Mol Sci. 2023, 24, 2831. [Google Scholar] [CrossRef]
- Chen L, Zhu Y, Zhang G, Gao C, Zhong W, Zhang X. CD83-stimulated monocytes suppress T-cell immune responses through production of prostaglandin E2. Proceedings of the National Academy of Sciences. 2011, 108, 18778–83. [CrossRef]
- Wu YJ, Song YN, Geng XR, Ma F, Mo LH, Zhang XW, et al. Soluble CD83 alleviates experimental allergic rhinitis through modulating antigen-specific Th2 cell property. Int J Biol Sci. 2020;16(2):216–27.
- Kastenmuller W, Drexler I, Ludwig H, Erfle V, Peschel C, Bernhard H, et al. Infection of human dendritic cells with recombinant vaccinia virus MVA reveals general persistence of viral early transcription but distinct maturation-dependent cytopathogenicity. Virology. 2006, 350, 276–88. [Google Scholar] [CrossRef]
- Liu L, Chavan R, Feinberg MB. Dendritic Cells are preferentially targeted among hematolymphocytes by Modified Vaccinia Virus Ankara and play a key role in the induction of virus-specific T cell responses in vivo. BMC Immunol. 2008;9(1):15.
- Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, et al. Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway. PLoS Pathog. 2014, 10, e1003989. [Google Scholar]
- Crabé S, Guay-Giroux A, Tormo AJ, Duluc D, Lissilaa R, Guilhot F, et al. The IL-27 p28 Subunit Binds Cytokine-Like Factor 1 to Form a Cytokine Regulating NK and T Cell Activities Requiring IL-6R for Signaling. The Journal of Immunology. 2009, 183, 7692–702. [Google Scholar] [CrossRef] [PubMed]
- Bauer S, Bathke B, Lauterbach H, Pätzold J, Kassub R, Luber CA, et al. A major role for TLR8 in the recognition of vaccinia viral DNA by murine pDC? Proceedings of the National Academy of Sciences. 2010 Sep 7;107(36).
- Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef]
- Heymann DL, Szczeniowski M, Esteves K. Re-emergence of monkeypox in Africa: A review of the past six years. Br Med Bull. 1998;54(3):693–702.
- Hammarlund E, Lewis MW, Carter S V. , Amanna I, Hansen SG, Strelow LI, et al. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat Med. 2005, 11, 1005–11. [Google Scholar] [CrossRef]
- Rizk JG, Lippi G, Henry BM, Forthal DN, Rizk Y. Prevention and Treatment of Monkeypox. Drugs [Internet]. 2022 Jun 1 [cited 2022 Nov 14];82(9):957. Available from: /pmc/articles/PMC9244487/. /.
- Harrison, C. Monkeypox response relies on three vaccine suppliers. Nat Biotechnol. 2022, 40, 1306–7. [Google Scholar] [CrossRef]
- Pittman PR, Hahn M, Lee HS, Koca C, Samy N, Schmidt D, et al. Phase 3 Efficacy Trial of Modified Vaccinia Ankara as a Vaccine against Smallpox. New England Journal of Medicine. 2019, 381, 1897–908. [Google Scholar] [CrossRef]
- Xu M, Liu C, Du Z, Bai Y, Wang Z, Gao C. Real-world effectiveness of monkeypox vaccines: a systematic review. J Travel Med. 2023 Sep 5;30(5).
- Damon IK, Damaso CR, McFadden G. Are We There Yet? The Smallpox Research Agenda Using Variola Virus. PLoS Pathog. 2014, 10, e1004108. [Google Scholar]
- Wang J, Shahed-AI-Mahmud M, Chen A, Li K, Tan H, Joyce R. An Overview of Antivirals against Monkeypox Virus and Other Orthopoxviruses. J Med Chem. 2023 Mar 24. 2023.
- Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, et al. An Orally Bioavailable Antipoxvirus Compound (ST-246) Inhibits Extracellular Virus Formation and Protects Mice from Lethal Orthopoxvirus Challenge. J Virol. 2005, 79, 13139–49. [Google Scholar] [CrossRef] [PubMed]
- Li P, Du Z, Lamers MM, Incitti R, Tejeda-Mora H, Li S, et al. Mpox virus infects and injures human kidney organoids, but responding to antiviral treatment. Cell Discov. 2023, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Florescu DF, Keck MA. Development of CMX001 (Brincidofovir) for the treatment of serious diseases or conditions caused by dsDNA viruses. Expert Rev Anti Infect Ther. 2014, 12, 1171–8. [CrossRef]
- Quenelle DC, Lampert B, Collins DJ, Rice TL, Painter GR, Kern ER. Efficacy of CMX001 against Herpes Simplex Virus Infections in Mice and Correlations with Drug Distribution Studies. J Infect Dis. 2010, 202, 1492–9.
- Smee DF, Dagley A, Downs B, Hagloch J, Tarbet EB. Enhanced Efficacy of Cidofovir Combined with Vaccinia Immune Globulin in Treating Progressive Cutaneous Vaccinia Virus Infections in Immunosuppressed Hairless Mice. Antimicrob Agents Chemother. 2015, 59, 520–6. [Google Scholar] [CrossRef]
- Tollefson AE, Spencer JF, Ying B, Buller RML, Wold WSM, Toth K. Cidofovir and brincidofovir reduce the pathology caused by systemic infection with human type 5 adenovirus in immunosuppressed Syrian hamsters, while ribavirin is largely ineffective in this model. Antiviral Res. 2014 Dec;112:38–46.
- Tylden GD, Hirsch HH, Rinaldo CH. Brincidofovir (CMX001) Inhibits BK Polyomavirus Replication in Primary Human Urothelial Cells. Antimicrob Agents Chemother. 2015, 59, 3306–16. [CrossRef]
- Farahat RA, Shah R, El-Sakka AA BAKMLFSRAAAB. Human monkeypox disease (MPX). Infezioni in Medicina. 2022 Sep 1;30(3).
- Overton ET, Lawrence SJ, Stapleton JT, Weidenthaler H, Schmidt D, Koenen B, et al. A randomized phase II trial to compare safety and immunogenicity of the MVA-BN smallpox vaccine at various doses in adults with a history of AIDS. Vaccine. 2020, 38, 2600–7. [Google Scholar] [CrossRef]
- Lahariya C, Thakur A, Dudeja N. Monkeypox Disease Outbreak (2022): Epidemiology, Challenges, and the Way Forward. Indian Pediatr. 2022, 59, 636–42. [Google Scholar] [CrossRef]
- Knight C, Andreani J, Garrett N, Winter M, Golubchik T, Breuer J, et al. Absence of detectable monkeypox virus DNA in 11,000 English blood donations during the 2022 outbreak. Transfusion (Paris). 2023 Feb 8.
- Shepherd W, Beard PM, Brookes SM, Frost A, Roberts H, Russell K, et al. The risk of reverse zoonotic transmission to pet animals during the current global monkeypox outbreak, United Kingdom, June to mid-September 2022. Eurosurveillance. 2022 Sep 29;27(39). 20 September.
- Mitjà O, Alemany A, Marks M, Lezama Mora JI, Rodríguez-Aldama JC, Torres Silva MS, et al. Mpox in people with advanced HIV infection: a global case series. The Lancet. 2023 Feb.
- Bloch EM, Sullivan DJ, Shoham S, Tobian AAR, Casadevall A, Gebo KA. The Potential Role of Passive Antibody-Based Therapies as Treatments for Monkeypox. mBio [Internet]. 2022 Oct 31 [cited 2022 Nov 14];e0286222. Available from: https://journals.asm.org/doi/10.1128/mbio.02862-22. 0286.
- Ortiz-Martínez Y, Zambrano-Sanchez G, Rodríguez-Morales AJ. Monkeypox and HIV/AIDS: When the outbreak faces the epidemic. https://doi.org/101177/09564624221114191 [Internet]. 2022 Jul 13 [cited 2022 Nov 21];33(10):949–50. Available from: https://journals.sagepub.com/doi/full/10.1177/09564624221114191. [CrossRef]
- Adnan N, Haq Z ul, Malik A, Mehmood A, Ishaq U, Faraz M, et al. Human monkeypox virus: An updated review. Medicine. 2022, 101, e30406. [Google Scholar] [CrossRef]
- Bunge EM, Hoet B, Chen L, Lienert F, Weidenthaler H, Baer LR, et al. The changing epidemiology of human monkeypox—A potential threat? A systematic review. PLoS Negl Trop Dis. 2022, 16, e0010141. [Google Scholar]
- Kmiec D, Kirchhoff F. Monkeypox: A New Threat? Int J Mol Sci. 2022, 23, 7866. [Google Scholar] [CrossRef] [PubMed]
- Silhan J, Klima M, Otava T, Skvara P, Chalupska D, Chalupsky K, et al. Discovery and structural characterization of monkeypox virus methyltransferase VP39 inhibitors reveal similarities to SARS-CoV-2 nsp14 methyltransferase. Nat Commun. 2023, 14, 2259. [Google Scholar] [CrossRef]
- Lim CK, Roberts J, Moso M, Liew KC, Taouk ML, Williams E, et al. Mpox diagnostics: Review of current and emerging technologies. J Med Virol. 2023 Jan 3;95(1).
- Perdiguero B, Esteban M. The Interferon System and Vaccinia Virus Evasion Mechanisms. Journal of Interferon & Cytokine Research. 2009, 29, 581–98. [Google Scholar]
- Matume ND, Tebit DM, Gray LR, Turner SD, Rekosh D, Bessong PO, et al. Characterization of APOBEC3 variation in a population of HIV-1 infected individuals in northern South Africa. BMC Med Genet. 2019.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




