Submitted:
10 July 2023
Posted:
11 July 2023
You are already at the latest version
Abstract
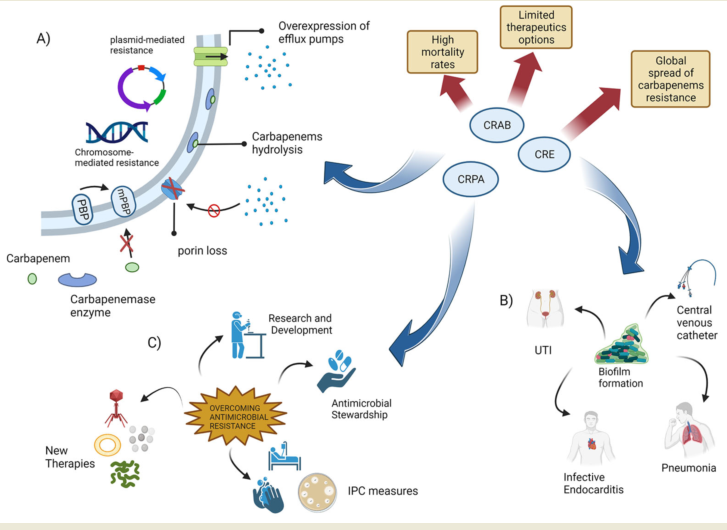
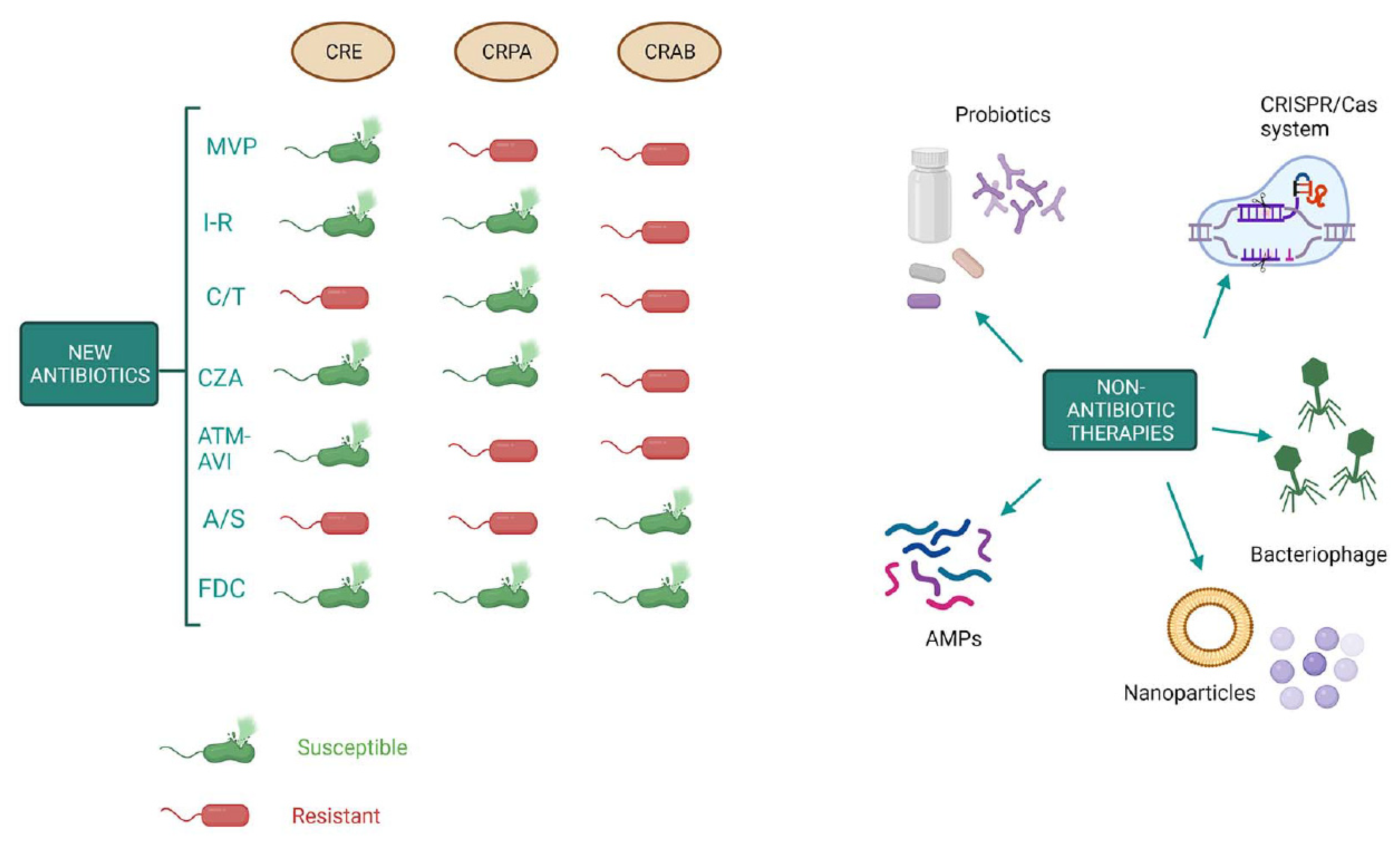
Keywords:
1. Introduction
1.1. Causes and Effects of Antibiotic Resistance
- (1)
- The development of new antibiotics is an extremely expensive endeavour, with a lengthy regulatory process and minimal revenues. This is because antibiotics are used for relatively short periods of time and are often curative, unlike drugs used to treat chronic diseases such as diabetes, asthma or gastrointestinal disorders.
- (2)
- The relatively low cost of antibiotics compared to drugs used to treat neuromuscular diseases or cancer chemotherapy.
- (3)
- Lack of know-how: the research on antibiotics carried out in academia has been scaled back as a result of a lack of financial incentives due to the economic crisis.
- (4)
1.2. Carbapenem Resistance in Gram-negative bacteria
1.3. The emergence of carbapenenases producing Enterobacteriaceae
1.4. The emergence of carbapenem-resistant Acinetobacter baumannii (CRAB) infections
1.5. Emergence of Pseudomonas aeruginosa with difficult-to-treat resistance
1.6. New weapons in the war against “superbugs”
2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods 2022, 11, 2966. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Bronzwaer, S.L.; Cars, O.; Buchholz, U.; Mölstad, S.; Goettsch, W.; Veldhuijzen, I.K.; Kool, J.L.; Sprenger, M.J.; Degener, J.E.; System, P.I.T.E.A.R.S. The Relationship between Antimicrobial Use and Antimicrobial Resistance in Europe. Emerg. Infect. Dis. 2002, 8, 278–282. [Google Scholar] [CrossRef]
- Ogyu, A.; Chan, O.; Littmann, J.; Pang, H.H.; Lining, X.; Liu, P.; Matsunaga, N.; Ohmagari, N.; Fukuda, K.; Wernli, D. National action to combat AMR: a One-Health approach to assess policy priorities in action plans. BMJ Glob. Heal. 2020, 5, e002427. [Google Scholar] [CrossRef]
- Essack, S. Water, sanitation and hygiene in national action plans for antimicrobial resistance. Bull. World Heal. Organ. 2021, 99, 606–608. [Google Scholar] [CrossRef]
- Willemsen, A.; Reid, S.; Assefa, Y. A review of national action plans on antimicrobial resistance: strengths and weaknesses. Antimicrob. Resist. Infect. Control. 2022, 11, 90. [Google Scholar] [CrossRef]
- McCarthy, M. Woman dies after infection with bacteria resistant to all antibiotics available in US. BMJ 2017, 356, j254. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Biondo, C. New Insights into Bacterial Pathogenesis. Pathogens 2022, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2023, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shang, Q.; Li, W.; Guo, W.; Stojadinovic, A.; Mannion, C.; Man, Y.-G.; Chen, T. Antibiotics for cancer treatment: A double-edged sword. J. Cancer 2020, 11, 5135–5149. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- A Odeyemi, O.; Sani, N.A.; Tishchenko, I.; Riveros, C.; Moscato, P.; Robinson, A.M.; Medlicott, N.J.; E Ussher, J.; A Bamidele, F.; Kohane, D.S. Antibiotic resistance and burden of foodborne diseases in developing countries. Futur. Sci. OA 2016, 2, FSO139. [Google Scholar] [CrossRef]
- Nwobodo, D.C.; Ugwu, M.C.; Anie, C.O.; Al-Ouqaili, M.T.S.; Ikem, J.C.; Chigozie, U.V.; Saki, M. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J. Clin. Lab. Anal. 2022, 36, e24655. [Google Scholar] [CrossRef]
- Lobanovska, M.; Pilla, G. Penicillin's Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Biol. Med. 2017, 90, 135–145. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: part 1: causes and threats. P T: A Peer-Rev. J. Formul. Manag. 2015, 40, 277–283. [Google Scholar]
- Valli, S.; Suvathi, S.S.; Aysha, O.; Nirmala, P.; Vinoth, K.P.; Reena, A. Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian Pac. J. Trop. Biomed. 2012, 2, 469–473. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Llor, C.; Bjerrum, L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Bin Emran, T.; Dhama, K.; Ripon, K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Heal. 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Xu, C.; Kong, L.; Gao, H.; Cheng, X.; Wang, X. A Review of Current Bacterial Resistance to Antibiotics in Food Animals. Front. Microbiol. 2022, 13, 822689. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Can. J. Infect. Dis. Med Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Burmeister, A.R. Horizontal Gene Transfer: Figure 1. Evol. Med. Public Heal. 2015, 2015, 193–194. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Evans, D.R.; Griffith, M.P.; Sundermann, A.J.; A Shutt, K.; I Saul, M.; Mustapha, M.M.; Marsh, J.W.; Cooper, V.S.; Harrison, L.H.; Van Tyne, D. Systematic detection of horizontal gene transfer across genera among multidrug-resistant bacteria in a single hospital. Elife 2020, 9. [Google Scholar] [CrossRef]
- Fiore, D.C.; Fettic, L.P.; Wright, S.D.; Ferrara, B.R. Antibiotic overprescribing: Still a major concern. J. Fam. Pr. 2017, 66, 730–736. [Google Scholar]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; Salamat, M.K.F.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Broek, A.K.v.D.; Beishuizen, B.H.H.; Haak, E.A.F.; Duyvendak, M.; Oever, J.T.; Sytsma, C.; van Triest, M.; Wielders, C.C.H.; Prins, J.M. A mandatory indication-registration tool in hospital electronic medical records enabling systematic evaluation and benchmarking of the quality of antimicrobial use: a feasibility study. Antimicrob. Resist. Infect. Control. 2021, 10, 103. [Google Scholar] [CrossRef]
- Broek, A.K.v.D.; van Hest, R.M.; Lettinga, K.D.; Jimmink, A.; Lauw, F.N.; Visser, C.E.; Prins, J.M. The appropriateness of antimicrobial use in the outpatient clinics of three hospitals in the Netherlands. Antimicrob. Resist. Infect. Control. 2020, 9, 40. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, W.; Zhu, Y.; Gong, Q.; Yu, W.; Lu, X. Antibiotics at subinhibitory concentrations improve the quorum sensing behavior ofChromobacterium violaceum. FEMS Microbiol. Lett. 2013, 341, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Anderson, A.; Macchiarelli, G.; Hellwig, E.; Cieplik, F.; Vach, K.; Al-Ahmad, A. Subinhibitory Antibiotic Concentrations Enhance Biofilm Formation of Clinical Enterococcus faecalis Isolates. Antibiotics 2021, 10, 874. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Tello, A.; Austin, B.; Telfer, T. Selective Pressure of Antibiotic Pollution on Bacteria of Importance to Public Health. Environ. Health Perspect. 2012, 120, 1100–1106. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Genet. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ștefan, M.G.; Loghin, F. Antibiotics in the environment: causes and consequences. Med. Pharm. Rep. 2020, 93, 231–240. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Liu, K.; Guo, Y.; Ding, C.; Han, J.; Li, P. Global review of macrolide antibiotics in the aquatic environment: Sources, occurrence, fate, ecotoxicity, and risk assessment. J. Hazard. Mater. 2022, 439, 129628. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Ghimpețeanu, O.M.; Pogurschi, E.N.; Popa, D.C.; Dragomir, N.; Drăgotoiu, T.; Mihai, O.D.; Petcu, C.D. Antibiotic Use in Livestock and Residues in Food—A Public Health Threat: A Review. Foods 2022, 11, 1430. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.; Kasimanickam, M.; Kasimanickam, R. Antibiotics Use in Food Animal Production: Escalation of Antimicrobial Resistance: Where Are We Now in Combating AMR? Med Sci. 2021, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial use in food animals and human health: time to implement ‘One Health’ approach. Antimicrob. Resist. Infect. Control. 2020, 9, 181. [Google Scholar] [CrossRef]
- Reardon, S. Antibiotic use in farming set to soar despite drug-resistance fears. Nature 2023, 614, 397. [Google Scholar] [CrossRef]
- Iskandar, K.; Murugaiyan, J.; Halat, D.H.; El Hage, S.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef]
- A Dutescu, I.; A Hillier, S. Encouraging the Development of New Antibiotics: Are Financial Incentives the Right Way Forward? A Systematic Review and Case Study. Infect. Drug Resist. 2021, ume 14, 415–434. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Biondo, C.; Ponzo, E.; Midiri, A.; Ostone, G.B.; Mancuso, G. The Dark Side of Nosocomial Infections in Critically Ill COVID-19 Patients. Life 2023, 13, 1408. [Google Scholar] [CrossRef]
- Principi, N.; Silvestri, E.; Esposito, S. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front. Pharmacol. 2019, 10, 513. [Google Scholar] [CrossRef] [PubMed]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.-S.; Harnod, D.; Hsueh, P.-R. Global Threat of Carbapenem-Resistant Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2022, 12, 823684. [Google Scholar] [CrossRef]
- A Elshamy, A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: epidemiology, detection and treatment options. Futur. Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensiv. Care 2020, 8, 13. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Tehrani, K.H.M.E.; Martin, N.I. β-lactam/β-lactamase inhibitor combinations: an update. MedChemComm 2018, 9, 1439–1456. [Google Scholar] [CrossRef]
- Paterson, D.L.; Doi, Y. Carbapenemase-Producing Enterobacteriaceae. Semin. Respir. Crit. Care Med. 2015, 36, 74–84. [Google Scholar] [CrossRef]
- Amin, M.; Navidifar, T.; Shooshtari, F.S.; Goodarzi, H. Association of the genes encoding Metallo-β-Lactamase with the presence of integrons among multidrug-resistant clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2019, ume 12, 1171–1180. [Google Scholar] [CrossRef]
- de Barsy, M.; Mercuri, P.S.; Oueslati, S.; Elisée, E.; Huang, T.-D.; Sacré, P.; Iorga, B.I.; Naas, T.; Galleni, M.; Bogaerts, P. Detection and Characterization of VIM-52, a New Variant of VIM-1 from a Klebsiella pneumoniae Clinical Isolate. Antimicrob. Agents Chemother. 2021, 65, e0266020. [Google Scholar] [CrossRef]
- Fraenkel, C.-J.; Starlander, G.; Tano, E.; Sütterlin, S.; Melhus. The First Swedish Outbreak with VIM-2-Producing Pseudomonas aeruginosa, Occurring between 2006 and 2007, Was Probably Due to Contaminated Hospital Sinks. Microorganisms 2023, 11, 974. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.P.; Woodford, N. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med Microbiol. 2013, 62, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.-A.; Matsumura, Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019, 33. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-Like β-Lactamases: Global Epidemiology, Treatment Options, and Development Pipeline. Antimicrob. Agents Chemother. 2022, 66, e0021622. [Google Scholar] [CrossRef]
- Alemayehu, E.; Fiseha, T.; Gedefie, A.; Tesfaye, N.A.; Ebrahim, H.; Ebrahim, E.; Fiseha, M.; Bisetegn, H.; Mohammed, O.; Tilahun, M.; et al. Prevalence of carbapenemase-producing Enterobacteriaceae from human clinical samples in Ethiopia: a systematic review and meta-analysis. BMC Infect. Dis. 2023, 23, 277. [Google Scholar] [CrossRef]
- O'Connell, N.; Gasior, S.; Slevin, B.; Power, L.; Barrett, S.; Bhutta, S.; Minihan, B.; Powell, J.; Dunne, C. Microbial epidemiology and clinical risk factors of carbapenemase-producing Enterobacterales amongst Irish patients from first detection in 2009 until 2020. Infect. Prev. Pr. 2022, 4, 100230. [Google Scholar] [CrossRef]
- Tilahun, M.; Kassa, Y.; Gedefie, A.; Belete, M.A. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021, ume 14, 4363–4374. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Marra, M.; Zummo, S.; Biondo, C. Urinary Tract Infections: The Current Scenario and Future Prospects. Pathogens 2023, 12, 623. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Cruz, C.S.D.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef] [PubMed]
- Aranega-Bou, P.; Verlander, N.; Paton, S.; Bennett, A.; Aiken, Z.; Akinremi, O.; Ali, A.; Cawthorne, J.; Cleary, P.; Crook, D.W.; et al. Carbapenem-resistant Enterobacteriaceae dispersal from sinks is linked to drain position and drainage rates in a laboratory model system. J. Hosp. Infect. 2019, 102, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Chia, P.Y.; Sengupta, S.; Kukreja, A.; Ponnampalavanar, S.S.; Ng, O.T.; Marimuthu, K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob. Resist. Infect. Control. 2020, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Suay-García, B.; Pérez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef]
- Sugawara, E.; Kojima, S.; Nikaido, H. Klebsiella pneumoniae Major Porins OmpK35 and OmpK36 Allow More Efficient Diffusion of β-Lactams than Their Escherichia coli Homologs OmpF and OmpC. J. Bacteriol. 2016, 198, 3200–3208. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wu, C.; Gao, H.; Xu, C.; Dai, M.; Huang, L.; Hao, H.; Wang, X.; Cheng, G. Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 2020, 11. [Google Scholar] [CrossRef]
- Alenazy, R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology 2022, 11, 1328. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F.; et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Principe, L.; Mauri, C.; Conte, V.; Pini, B.; Giani, T.; Rossolini, G.M.; Luzzaro, F. First report of NDM-1-producing Klebsiella pneumoniae imported from Africa to Italy: Evidence of the need for continuous surveillance. J. Glob. Antimicrob. Resist. 2017, 8, 23–27. [Google Scholar] [CrossRef]
- Mendes, G.; Ramalho, J.F.; Duarte, A.; Pedrosa, A.; Silva, A.C.; Méndez, L.; Caneiras, C. First Outbreak of NDM-1-Producing Klebsiella pneumoniae ST11 in a Portuguese Hospital Centre during the COVID-19 Pandemic. Microorganisms 2022, 10, 251. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Chen, L.; Ai, W.; Zhou, Y.; Wu, C.; Guo, Y.; Wu, X.; Wang, B.; Rao, L.; Xu, Y.; Zhang, J.; et al. Outbreak of IncX8 Plasmid–Mediated KPC-3–Producing Enterobacterales Infection, China. Emerg. Infect. Dis. 2022, 28, 1421–1430. [Google Scholar] [CrossRef]
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.-P.; Touati, A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 587–604. [Google Scholar] [CrossRef]
- Sivaramakrishnan, A.; Mack, D.; El-Mugamar, H.; Jacques, J.; Paget, S.; Phee, L.; Carter, Y. Epidemiology and control measures of an OXA-48-producing Enterobacteriaceae hospital outbreak. Infect. Prev. Pr. 2020, 2, 100021. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Galera, S.; Bravo-Ferrer, J.M.; Paniagua, M.; Kostyanev, T.; de Kraker, M.E.; Feifel, J.; Sojo-Dorado, J.; Schotsman, J.; Cantón, R.; Daikos, G.L.; et al. Risk factors for infections caused by carbapenem-resistant Enterobacterales: an international matched case-control-control study (EURECA). eClinicalMedicine 2023, 57, 101871. [Google Scholar] [CrossRef]
- Trecarichi, E.M.; Tumbarello, M. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence 2017, 8, 470–484. [Google Scholar] [CrossRef]
- Nasomsong, W.; Nulsopapon, P.; Changpradub, D.; Pongchaidecha, M.; Pungcharoenkijkul, S.; Juntanawiwat, P.; Simsiriporn, W.; Santimaleeworagun, W. The Potential Use of Ceftazidime-Avibactam Against Carbapenem Resistant Klebsiella pneumoniae Clinical Isolates Harboring Different Carbapenemase Types in a Thai University Hospital. Drug Des. Dev. Ther. 2021, ume 15, 3095–3104. [Google Scholar] [CrossRef]
- Cruz-López, F.; Martínez-Meléndez, A.; Morfin-Otero, R.; Rodriguez-Noriega, E.; Maldonado-Garza, H.J.; Garza-González, E. Efficacy and In Vitro Activity of Novel Antibiotics for Infections With Carbapenem-Resistant Gram-Negative Pathogens. Front. Cell. Infect. Microbiol. 2022, 12, 884365. [Google Scholar] [CrossRef]
- Asif, M.; Alvi, I.A.; Rehman, S.U. Insight into Acinetobacter baumannii: pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect. Drug Resist. 2018, ume 11, 1249–1260. [Google Scholar] [CrossRef]
- Jiang, Y.; Ding, Y.; Wei, Y.; Jian, C.; Liu, J.; Zeng, Z. Carbapenem-resistant Acinetobacter baumannii: A challenge in the intensive care unit. Front. Microbiol. 2022, 13, 1045206. [Google Scholar] [CrossRef] [PubMed]
- Gedefie, A.; Demsiss, W.; Belete, M.A.; Kassa, Y.; Tesfaye, M.; Tilahun, M.; Bisetegn, H.; Sahle, Z. Acinetobacter baumannii Biofilm Formation and Its Role in Disease Pathogenesis: A Review. Infect. Drug Resist. 2021, ume 14, 3711–3719. [Google Scholar] [CrossRef]
- Bartal, C.; Rolston, K.V.I.; Nesher, L. Carbapenem-resistant Acinetobacter baumannii: Colonization, Infection and Current Treatment Options. Infect. Dis. Ther. 2022, 11, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Hillyer, T.; Benin, B.M.; Sun, C.; Aguirre, N.; Willard, B.; Sham, Y.Y.; Shin, W.S. A novel strategy to characterize the pattern of β-lactam antibiotic-induced drug resistance in Acinetobacter baumannii. Sci. Rep. 2023, 13, 9177. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-J.; Xiao, Z.-G.; Lv, X.-J.; Huang, H.-T.; Liao, C.; Hui, C.-Y.; Xu, Y.; Li, H.-F. Drug-resistant Acinetobacter baumannii: From molecular mechanisms to potential therapeutics (Review). Exp. Ther. Med. 2023, 25, 209. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Genet. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Shields, R.K.; Paterson, D.L.; Tamma, P.D. Navigating Available Treatment Options for Carbapenem-Resistant Acinetobacter baumannii-calcoaceticus Complex Infections. Clin. Infect. Dis. 2023, 76, S179–S193. [Google Scholar] [CrossRef]
- Ma, C.; McClean, S. Mapping Global Prevalence of Acinetobacter baumannii and Recent Vaccine Development to Tackle It. Vaccines 2021, 9, 570. [Google Scholar] [CrossRef]
- Castanheira, M.; E Mendes, R.; Gales, A.C. Global Epidemiology and Mechanisms of Resistance of Acinetobacter baumannii-calcoaceticus Complex. Clin. Infect. Dis. 2023, 76, S166–S178. [Google Scholar] [CrossRef]
- Sy, C.L.; Chen, P.-Y.; Cheng, C.-W.; Huang, L.-J.; Wang, C.-H.; Chang, T.-H.; Chang, Y.-C.; Chang, C.-J.; Hii, I.-M.; Hsu, Y.-L.; et al. Recommendations and guidelines for the treatment of infections due to multidrug resistant organisms. J. Microbiol. Immunol. Infect. 2022, 55, 359–386. [Google Scholar] [CrossRef]
- Russo, A.; Bruni, A.; Gullì, S.; Borrazzo, C.; Quirino, A.; Lionello, R.; Serapide, F.; Garofalo, E.; Serraino, R.; Romeo, F.; et al. Efficacy of cefiderocol- vs colistin-containing regimen for treatment of bacteraemic ventilator-associated pneumonia caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19. Int. J. Antimicrob. Agents 2023, 62, 106825. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Ribeiro, V.S.T. Pathogenesis of the Pseudomonas aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef]
- Paprocka, P.; Durnaś, B.; Mańkowska, A.; Król, G.; Wollny, T.; Bucki, R. Pseudomonas aeruginosa Infections in Cancer Patients. Pathogens 2022, 11, 679. [Google Scholar] [CrossRef]
- Losito, A.R.; Raffaelli, F.; Del Giacomo, P.; Tumbarello, M. New Drugs for the Treatment of Pseudomonas aeruginosa Infections with Limited Treatment Options: A Narrative Review. Antibiotics 2022, 11, 579. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031–19. [Google Scholar] [CrossRef]
- Coyne, A.J.K.; El Ghali, A.; Holger, D.; Rebold, N.; Rybak, M.J. Therapeutic Strategies for Emerging Multidrug-Resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 2022, 11, 661–682. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa : Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, A.; Dinh, A.Q.; Hanson, B.M.; Shropshire, W.C.; A Rizvi, S.; Rydell, K.; Tran, T.T.; Wanger, A.; A Arias, C.; Miller, W.R. Evolving landscape of carbapenem-resistant Pseudomonas aeruginosa at a single centre in the USA. JAC-Antimicrobial Resist. 2023, 5, dlad070. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Ji, B.; Zhang, X.; Anbo, M.; Jelsbak, L. Whole-genome sequencing reveals high-risk clones of Pseudomonas aeruginosa in Guangdong, China. Front. Microbiol. 2023, 14, 1117017. [Google Scholar] [CrossRef]
- Tran, T.T.; Cabrera, N.L.; Gonzales-Luna, A.J.; Carlson, T.J.; Alnezary, F.; Miller, W.R.; Sakurai, A.; Dinh, A.Q.; Rydell, K.; Rios, R.; et al. Clinical characteristics, microbiology and outcomes of a cohort of patients treated with ceftolozane/tazobactam in acute care inpatient facilities, Houston, Texas, USA. JAC-Antimicrobial Resist. 2023, 5, dlac131. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, D.; Wang, Y.; Ni, W. Cefiderocol for the Treatment of Multidrug-Resistant Gram-Negative Bacteria: A Systematic Review of Currently Available Evidence. Front. Pharmacol. 2022, 13, 896971. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, A.; Almotairy, A.R.Z.; Henidi, H.; Alshehri, O.Y.; Aldughaim, M.S. Nanoparticles as Drug Delivery Systems: A Review of the Implication of Nanoparticles’ Physicochemical Properties on Responses in Biological Systems. Polymers 2023, 15, 1596. [Google Scholar] [CrossRef]
- Hetta, H.F.; Ramadan, Y.N.; Al-Harbi, A.I.; Ahmed, E.A.; Battah, B.; Ellah, N.H.A.; Zanetti, S.; Donadu, M.G. Nanotechnology as a Promising Approach to Combat Multidrug Resistant Bacteria: A Comprehensive Review and Future Perspectives. Biomedicines 2023, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, O.O.; Nontongana, N.; Okoh, J.C.; Okoh, A.I. The Potential of Antibiotics and Nanomaterial Combinations as Therapeutic Strategies in the Management of Multidrug-Resistant Infections: A Review. Int. J. Mol. Sci. 2022, 23, 15038. [Google Scholar] [CrossRef]
- Wang, S.; Zeng, X.; Yang, Q.; Qiao, S. Antimicrobial Peptides as Potential Alternatives to Antibiotics in Food Animal Industry. Int. J. Mol. Sci. 2016, 17, 603. [Google Scholar] [CrossRef]
- Souza, G.d.C.d.; Roque-Borda, C.A.; Pavan, F.R. Beta-lactam resistance and the effectiveness of antimicrobial peptides against KPC-producing bacteria. Drug Dev. Res. 2022, 83, 1534–1554. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell. Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Skurnik, M. Can Bacteriophages Replace Antibiotics? Antibiotics 2022, 11, 575. [Google Scholar] [CrossRef]
- Diallo, K.; Dublanchet, A. A Century of Clinical Use of Phages: A Literature Review. Antibiotics 2023, 12, 751. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Liang, W. The Application of the CRISPR-Cas System in Antibiotic Resistance. Infect. Drug Resist. 2022, ume 15, 4155–4168. [Google Scholar] [CrossRef]
- Walker-Sünderhauf, D.; Klümper, U.; Pursey, E.; Westra, E.R.; Gaze, W.H.; van Houte, S. Removal of AMR plasmids using a mobile, broad host-range CRISPR-Cas9 delivery tool. Microbiology 2023, 169, 001334. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Q.; Hu, X.; Liu, W. Current Status of Probiotics as Supplements in the Prevention and Treatment of Infectious Diseases. Front. Cell. Infect. Microbiol. 2022, 12, 789063. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



