3.1. Derivation of formulas for Cumulative Number of Infections (CNI) and Daily Number of Infections (DNI)
The potential for infection of a given virus strain is determined by the basic reproduction number known as
R0. For the virus to spread, it must continually re-infect the host population. However, countermeasures by humans must aim to limit these subsequent infections. Essentially, this implies that the interaction and compromise between the virus and host (humans) necessitates the
j-th reproduction number
Rj to be smaller than the (
j-1)-th reproduction
Rj-1, as expressed in the equation
The attenuation constant '
k' in this scenario is influenced by social contact rates and policy measures. After
m-1 rounds of infection spread, the total number of infections in a specific area can be represented as
This function
F(
R0,
k;
m) describes the Cumulative Number of Infections (CNI), which is an increasing function of
m. Assuming the total infected individuals as
N, solving the equation
F(
R0,
k;
m)=
N will produce '
m', the transmission number of the virus strain. The parameter m grows with time
t. Assuming each transmission takes place every
q days, we have
where
t0 is a time shift parameter linked to the start of infection (the time of
m=1). Inserting (2) into (1) we obtain a formula of CNI vs
t represented by four parameters
R0,
k,
q and
t0 that can be used to simulate the change of a cumulative number of infections with time in a region. As seen in
Figure 1, our simulations of the UK-alpha strain (November 2020 to April 2021) and the Hong Kong-delta/omicron strain (February 2022 to April 2022) spreading are well fitted to the data obtained from COVID-19 pandemic updates.
Applying Equations (1) and (2), the Daily Number of Infections (DNI) is derived:
From Eq (3), it is easy to understand why DNI first rises then falls, considering that
R0>1 (for most infectious viruses) and
k<1. Zero DNI is arrived when
dF/
dm =0, which requires
m to be large enough such that
This is a condition for the end of an infection wave.
3.2. Insights from typical figures of CNI vs m
Utilizing Eq (1), we plotted the change of CNIs with variables
k and
m for the given
R0 (
Figure 2). Figures 2A-E show typical cases that help in understanding the development and termination of an infection wave.
From
Figure 2, one can observe that the curve of
F(
R0,
k;
m) increases with m and approaches a stable value when
m>
mst for each
k<
kth. We denote
F(
R0,
kth;
mst) as
Nth. Calculating for
mst =15 in
Figure 2A to 2E, we obtained the threshold
kth and the corresponding CNI,
Nth, for each given
R0 (each virus strain). On the other hand, CNI attaining a stable value means DNI approaching zero. By defining
Eq (4) can be written as
R0<
Ekm. The threshold
kth, the corresponding CNI
Nth and the parameter
Ekm are listed in
Table 1. We found the relation
R0<
Ekm to hold well across all data.
The daily increasing number
dF/dm=0 means the virus infection dies out. The above result shows the higher the basic reproduction number
R0, the more strict public health measure is required to increase
Ekm to satisfy Eq (4) and terminate the wave of virus infection effectively. From
Table 1 we found the cumulative infection numbers
Nth of many virus strains (except Omicron with
R0=18.6) are lower than 10
5 if appropriate
k (lower than
kth) is introduced.
For the virus strain of given
R0, if
F(
R0,
k;
m) (for all
m) exceeds a threshold
Nmax, then the spread of this strain would lead to a wave of COVID-19 pandemic in a region with population larger than
Nmax. In order to end this spread, the necessary condition would be
Equation (5) provides a constraint on
k for strains with
R0. The critical value of
k is denoted as
kcr. Taking
Nmax= 10
7 the values of
kcr are also listed in
Table 1. Note that the parameter
kth is the threshold value of
k required for ending the spread in 15
th generation, but the parameter
kcr is that value for ending the spread in arbitrary generation. The latter constraint is looser than the former. Therefore,
kcr is larger than
kth.
Virus strains with low R0 values have higher kcr and kth values. As a result, they can spread in regions with smaller populations under looser public health measures. This theoretical prediction can explain why strains like SARS-CoV in 2003 only spread in restricted regions and soon disappeared globally. On the contrary, virus strains with high R0 values, such as Omicron, have lower kcr and kth values. To satisfy Equations (4) and (5), the parameter k needs to meet very strict constraints. In cases where social management measures are not stringent enough, the number of infected individuals will quickly surpass the Nmax limit, triggering a global pandemic wave, possibly culminating in a type of coexistence between viruses and humans.
In summary, the termination of an infection wave is determined by the condition
dF/dm =0 on the CNI diagram (
Figure 2), which requires a sufficiently large
m and the condition in Equation (4) satisfied, i.e.
R0<
Ekm=(1/
kth)
7. Additionally, the necessary condition for a pandemic to die out in a population of
Nmax is expressed by Equation (5), imposing a limitation on
k, namely
k<
kcr.
3.3. Prediction on the Second Wave of pandemics
Early models based on previous pandemics such as SARS, MERS, and the 2009 H1N1 outbreak can effectively predict the occurrence of the first wave of a disease. However, their predictive power decreases when it comes to anticipating the possibility of a second wave [
9]. This raises the question: Why does the second wave of COVID-19 infections often follow the first wave? The present model aims to address this issue.
As shown in
Figure 2, there are multiple curves (
F versus
m) for a given
R0 value. These curves differ from each other by the parameter
k. When the number of daily new infections (DNI) approaches zero, any fluctuation in
k significantly influences the spread of the virus. Therefore, changes in public health measures can induce variations in virus spread along different curves. Generally, as public health measures are relaxed, the parameter
k increases, causing the virus to transition from one curve of
F to another steeper curve. This signifies the onset of a new wave of virus spread. For instance, in
Figure 2A, the spread of the Omicron variant (with an
R0 of 18.6) is plotted. Assuming the initial spread is along the curve with
k equal to 0.613, a stable state is reached at
m=15. At this point, the
k value increases to 0.722. In response to this change, the virus begins spreading along a new curve with
k=0.722, starting from
m=4, as the value of
F(18.6, 0.722;
m=4) is equal to the original CNI value,
F(18.6, 0.613;
m=15). This example explains how the second wave of virus infection occurs. However, in cases where the parameter
k decreases when DNI approaches zero, the curve of
F will transition to a flatter one. This indicates that the first wave of viral infection will end soon and no second wave will occur.
By examining
Figure 2A to
Figure 2E, we can analyze how the occurrence of a second wave depends on the
R0 value of the virus. For instance, as
k changes from 0.85 to 0.9, the CNI (at
m=15) for high
R0 viruses increases hundreds of times, whereas it only increases tens of times for low
R0 viruses. Therefore, our model predicts that multi-wave infections are more likely to occur with viruses that have high
R0 values.
In the aforementioned discussions, we have assumed that no virus mutation occurs and that only one type of virus is spreading. In reality, changes in public health measures may be accompanied by virus mutations. In this case, the change in public health measures would cause the jump of F not only between different curves with a given R0 but also between different R0 values, providing more opportunities for the occurrence of a second wave during virus spread.
Another crucial point to consider is that the change in k can simultaneously induce a change in q, as the eigen-time (inherent time) m depends on q (Equation (2)). In the case of a single wave, the parameter m can be used to represent time dependence, and q can be simply set to 1 (known as normalization). However, when studying two continuous waves, the dependence on q should be clearly indicated due to the different eigen-times m. We have mentioned that the dependence of F on the change in k results in a jump from one curve to another. Meanwhile, the dependence of F on the change in q only affects the lengthening or shortening of the abscissa of the graph without altering the shape of the curve. When the public health measures change and a jump between curves occurs, the abscissa of the graph of the second wave simultaneously lengthens or shortens.
The occurrence of the second wave of infection is more likely when public health measures are relaxed. This change in the second wave is accompanied by a change in the
q value. These predictions align with experimental data. For example, in the UK from May to September 2021, public health measures were relaxed, and a second wave of infection followed the first wave (
Figure 1A). Similarly, in Hong Kong several months after May 2022, the looser public health measures led to a second wave occurring after the first wave (
Figure 1B). (The data on the change of public health measures can be found at
https://www.bsg.ox.ac.uk/research/covid-19-government-response-tracker)
In summary, the dependence of CNI on k (given a specific R0) is determined by the jump between different curves on the graph F(R0,k;m) versus m, referred to as k-transformation. The dependence of CNI on the duration of m is obtained by stretching the abscissa m on the graph, known as q-transformation. The k-transformation and q-transformation, occurring when the first wave is nearing its end, are the causes of a continuous second wave. The change in k is attributed to the modification of public health measures, while the modification of q is due to the change in physical time within a unit of m. The changes in k and q provide an explanation for the experimental data on continuous multi-wave infections.
3.4. Cross-Spread of Two Viruses: Discriminant Function
Viral infections often involve the simultaneous presence of two or more viruses. To accurately simulate the cross-spread of two viruses, it is necessary to consider the differences in eigen-times (
m1 and
m2) between these viruses and their relationship with the physical time
t. Consequently, additional parameters
q and
t0 (as given in Eq (2)) must be taken into account. By utilizing the four parameters
R0,
k,
q and
t0, the CNI
F(
R0,
k;
m) can be expressed as:
When the CNIs of two virus strains
F1(
t;
a1,
b1,
c1,
d1) and F
2(
t;
a2,
b2,
c2,
d2) intersect at
tcr,
and
This represents a transition in the population of virus strains from
F1 to
F2. As it is challenging to directly solve the intersection equation (Eq (7)), we introduce a function:
which satisfies:
By utilizing Eqs (2)(3)(6) and (8)
D21 can be formulated as a simple quadratic function of time
where
In order to determine if a real root of D21 exists, we define:
The value of Δ determines the existence of the real root in the quadratic form
D21. This form, known as the discriminant function, serves as a tool to identify the occurrence of
tcr and the domain of its existence. The prediction rules can be summarized as follows:
Rule 1: When Δ>0, the quadratic form intersects with
t axis at
ts and
tm (
tm>
ts),
These values partition the time into three distinct domains:
t<
ts in the first domain,
ts<
t<
tm in the second domain and
t>
tm in the third domain.
Rule 2: In the first domain, ta=qm0-t0 is defined as the initial time where m0>>1, indicating m1,2=(t+d1,2)/c1,2>>1. If D21>0, there will be no intersection of F1(t) and F2(t) in the domain between ta and ts when the initial values of F at ta satisfy F1(ta)<F2(ta), but there may be one tcr (the number of tcr is either 1 or 0) in the domain when F1(ta)>F2(ta). If D21<0, there will be no intersection of F1(t) and F2(t) in the domain when the initial values satisfy F1(ta)>F2(ta), but there may be one tcr (the number of tcr is either 1 or 0) when F1(ta)<F2(ta).
Rule 3: In the second domain, if D21>0 there will be no intersection of F1(t) and F2(t) in the domain when the F-values at t=ts satisfy F1(ts)<F2(ts), but there may be one tcr (the number of tcr is either 1 or 0) when F1(ts)>F2(ts). If D21<0 there will be no intersection of F1(t) and F2(t) in the domain when the F-values at t=ts satisfy F1(ts)>F2(ts), but there may be one tcr (i.e. the number of tcr is either 1 or 0) when F1(ts)<F2(ts). The F-values at t=ts are determined by F1(t) and F2(t) in the first domain.
Rule 4: In the third domain, if D21>0 there will be no intersection of F1(t) and F2(t) in the domain when the F-values at t=tm satisfy F1(tm)<F2(tm), but there may be one tcr (the number of tcr is either 1 or 0) when F1(tm)>F2(tm). If D21<0 there will be no intersection of F1(t) and F2(t) in the domain when the F-values at t=tm satisfy F1(tm)>F2(tm), but there may be one tcr (i.e. the number of tcr is either 1 or 0) when F1(tm)<F2(tm). The F-values at t=tm are determined by F1(t) and F2(t) in the second domain.
Rule 5: There can be at most one tcr in a given domain because the symbol of D21 is definite in any domain. The magnitude of the domain is an important factor to predict the occurrence of a tcr. For example, in the second domain the magnitude is tm-ts=Δ1/2/(2|α|), and the necessary condition for the occurrence of tcr is a large enough (tm-ts) or Δ.
Rule 6: When Δ < 0, the quadratic form D21 does not intersect with t axis. In this case, there is only one domain and the rule is the same as that in the first domain given by Rule 2.
Figure 3 presents examples of cross-spread of two virus strains, 1 and 2. The left panel shows the discriminant function, and the right panel displays the cross-spread of the two strains. The influence of the change in parameters (as an example, we only assume the
R0 value of strain 1 changes) on the intersection of two strains is shown in the figure. In
Figure 3A, there is no intersection. In
Figure 3B and 3C, there are two intersections in the second and third domain, respectively. In
Figure 3D, there is one intersection in the second domain. These intersection occurrences are in agreement with aforementioned prediction rules.
In the first domain we have introduced ta =qm0-t0 as the initial time. To study the cross spread in the time interval between m=1 and ta where dF/dt in D21 is difficult to be defined one should use F(t)-ladder method as follows:
The step size of CNIs of the first few steps in two ladders 1 and 2 are given by
,
,
,
,
,
,
,
,
, etc., and
,
,
,
,
,
,
,
,
, etc., respectively (Eq (1)). Strain 1 spreads from the 1
st step
a1 at
t=
c1-
d1 on ladder 1 and strain 2 spreads from the 1
st step a
2 at
t=
c2-
d2 on ladder 2. The CNI and arrival time t of the two strains on their respective ladders are listed in
Table 2. For example, let’s take
c1=7,
d1=15,
c2=5,
d2=10. The earlier arrival times, in turn, are
t=
c1-
d1 =-8,
c2-
d2 =-5, 2
c1-
d1 =-1, 2
c2-
d2 =0, 3
c2-
d2 =5, 3
c1-
d1 =6, etc. By using
Table 2, one can easily calculate the CNI at each arrival time as the parameters
a1,
b1,
a2,
b2 are given, compare the CNI values on two ladders and obtain the information on the cross spread of two strains.
3.5. Examples of the Cross-Spread of Two Viruses
The epidemics that occurred in the UK from November 2020 to February 2022 provide a clear demonstration of the cross-spread phenomenon involving multiple viruses. This process can be divided into five stages, namely: (A) the alpha epidemic, (B) the delta invasion and cross-spread of two strains, (C) the delta dominant stage, (D) the omicron invasion and cross-spread of delta and omicron, and (E) the omicron dominant stage. In this section, we will specifically focus on studying the cross-spread of two strains during stage B and D.
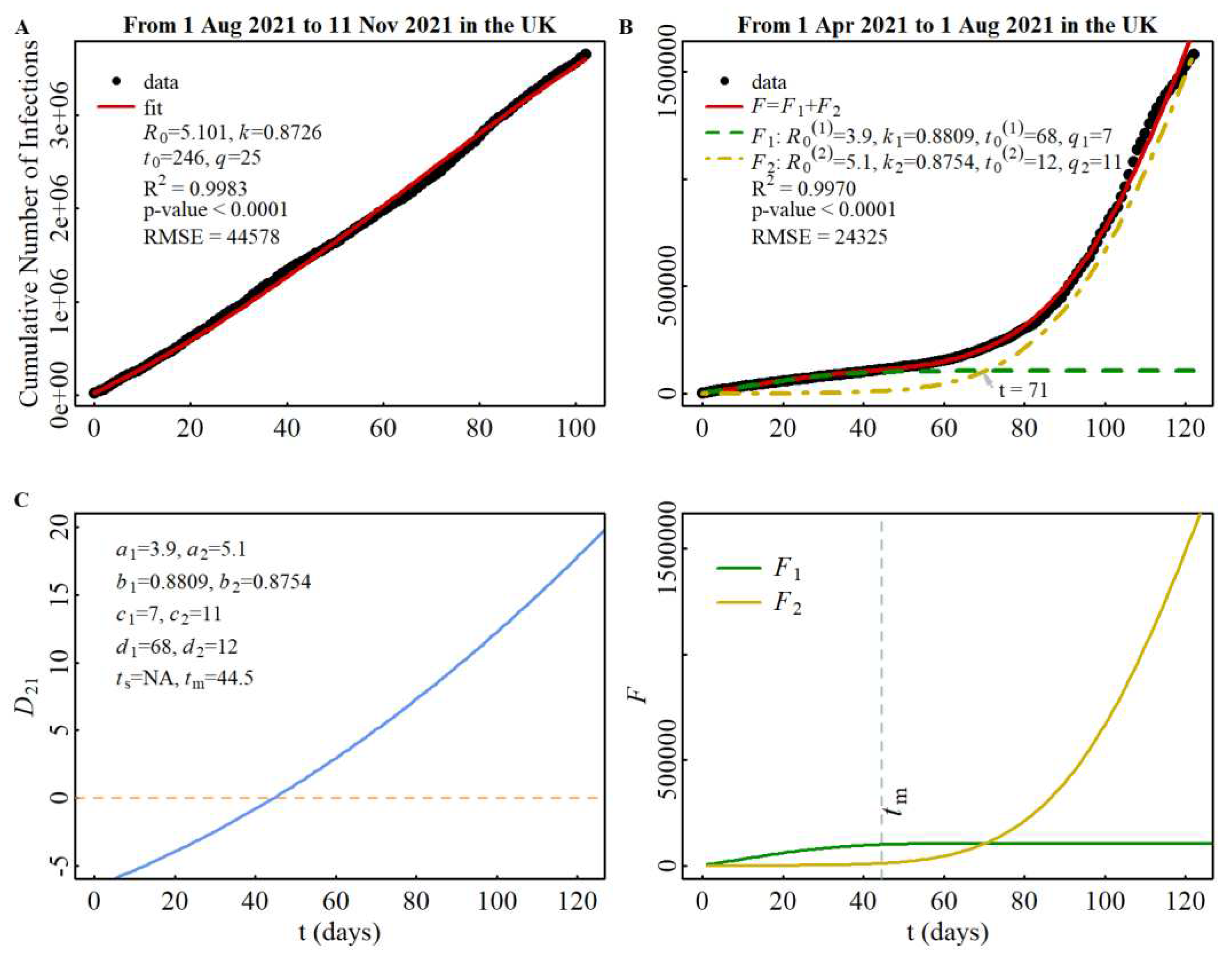
Firstly, let us examine the cross-spread between the alpha strain (designated as strain 1) and the delta strain (designated as strain 2) during stage B. The spread of the alpha strain during stage A has been represented in
Figure 1A, where we obtained the parameter
a1=
R0(1)=3.9. The spread of the delta strain during stage C has been illustrated in
Figure 4A, and we derived the parameter
a2=
R0(2)=5.1 based on this data. By using
R0(1) and
R0(2) as inputs, we simulated the cross-spread of the two strains, as depicted in
Figure 4B. Furthermore, utilizing all the parameters
ai,
bi,
ci,
di (
i=1,2) obtained from Figures 1A, 4A, and 4B, we constructed the discriminant function of the cross-spread and plotted the intersection of the two strains during stage B in
Figure 4C. Interestingly, we discovered that one
tcr occurs at
t=71 in the region where
t>
tm (
tm=44.5), which is consistent with the prediction outlined in
Rule 4.
Similarly, we investigated the cross-spread between the delta strain (strain 1) and the omicron strain (strain 2) during stage D. The results of this analysis are shown in Figures 5A, 5B, and 5C. From
Figure 5C, we observed that a
tcr appears at
t=42 in the region where
t>
tm (
tm=28.5), which is in agreement with the prediction made by
Rule 4.
In both simulations, we assumed April 1 as the initial time for stage B, and November 11 as the initial time for stage D. Due to the uncertainty surrounding the exact timing of the delta invasion in stage B and the omicron invasion in stage D, we shifted the initial times t of stages B and D by several days. Remarkably, we found that the same values of k1, k2, q1, q2, t0(1) were obtained, and only t0(2) varied while still maintaining the invariant t+ t0(2).