1. Introduction
M. rosenbergii, a major freshwater aquaculture prawn species, has become the primarily-consumed freshwater prawn in the world due to the rapid increase in annual yields (Hossain et al. 2017). Although the M. rosenbergii industry has expanded dramatically in the past decades (Ahmed et al. 2010), there is still a big difference in the average yield per unit compared with other shrimp species (Siddiqui et al. 1996; Sui et al. 2019), such as Penaeus vannamei (Martinez-Cordova et al. 1997). There are also significant gender differences in the growth rate and other growth traits between males and females in M. rosenbergii (Dan and Ziva, 1983; Kumar et al. 2000; Dinh and Nguyen, 2014).Therefore, the factors influencing the growth of M. rosenbergii still need to be further explored to provide the theoretical basis for improving molecular breeding rapidly (Sui et al. 2019).As a sexually dimorphic organism, the growth rate of male prawns were significantly higher than those of female prawns (Dinh and Nguyen, 2014). The growth rate of prawns heavily depend on the secretion capacity of digestive enzymes (Dong et al. 2001; Jun-jie et al. 2006; Silva et al. 2018) and the diversity of intestinal microorganisms (Moss et al. 2000; Landsman et al. 2019). Recent studies on the digestive glands and gut microbes have shown that the coordination between the secretion capacity of enzymes (Habte-Tsion et al. 2013; Belsare et al.2017) and the activity of microbes is associated with feed conversion ratio and growth in multiple animals, and it might provide novel methods and strategies for the selection of high growth ratio populations in aquaculture animals. Concurrently, studies have shown that the intestinal microbes of shrimps assist food digestion and improve disease resistance, respectively (Cornejo-Granados et al. 2017). Therefore, further research to find out the coordination of the digestive glands, intestinal microbiome, and sex glands of different sexes in M. rosenbergii, which may cause the differences in the growth performance of different sexes of M. rosenbergii, is very important for improving the efficiency of feed conversion and for improving the growth rate of M. rosenbergii.
Sexual dimorphism of growth traits is quite common in aquatic animals and has been found in fish, shrimps and crabs (Davey and Jellyman, 2005; Tang et al. 2019). However, how the growth advantage is gained by males varies among aquatic animals. Previously, quantitative PCR and microbial taxonomic studies have shown that gut microbes are related to the growth traits of organisms (Carani, 2014; Huang et al. 2016). The potential role of gut microbes in the growth of aquatic animals is still under investigation. Gender differences in growth traits and digestive function in shrimps have been observed, and such gender differences in M. rosenbergii are more significant, especially after adulthood (Tang et al. 2019). Transcriptome analyses of intestinal and digestive glands have shown that there are significant differences in the expression of digestive enzymes between male and female digestive glands, and there is a significant association between the expression of digestive enzymes and the growth traits of male and female shrimps (Martínez-Alarcón et al. 2019). In addition, gut microbes are also a major cause of gender phenotypic differences in aquatic animals (Landsman et al. 2019). However, due to the poor reproducibility of gut microbe analysis, few microbes were reported to be significantly associated with gender phenotype differences. Although there are conservative differences in the types of gut microbes, previous studies have shown that the metabolomics characteristics of gut microbes are significantly conserved, and they also have a great contribution to the phenotypic characteristics of organisms (Xue et al. 2022). Whether there is an association between the digestive gland transcriptome and gut microbiome of prawns, which would affect the growth traits of different sexes, still remains unclear.
At present, whether there is a significant functional coordination between the differentially expressed genes of the digestive gland and gut microbial diversity, which would impact on the growth traits of male and female prawns, still remains unclear. Therefore, it is necessary to integrate phenotypic differences, transcriptome data, and gut microbiome diversity to evaluate the mechanisms underlying the differences in growth traits between male and female in M. rosenbergii. The revelation of these mechanisms will help us to further understand the causes for the growth differences between males and females, and to select prawns with significant advantages in growth rate in M. rosenbergii.
2. Methods and materials
2.1. Animals and samples
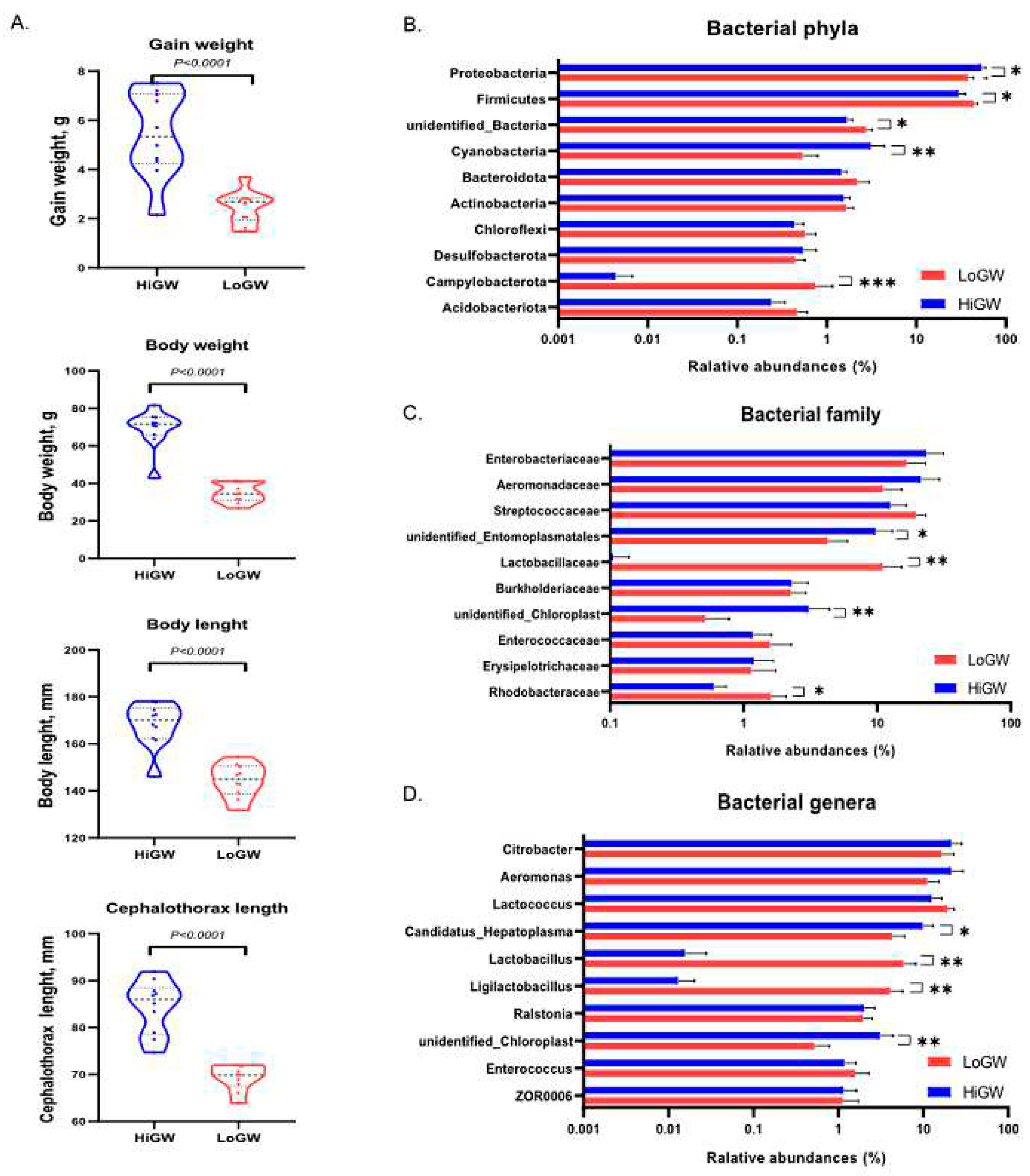
The experimental animal protocol was approved by the Laboratory Animal Welfare and Ethics Committee of Zhejiang Academy of Agricultural Sciences (Hangzhou, China). The analyses of the gut microbe diversity, digestive gland and sex glands transcriptomes of M. rosenbergii were conducted in our study. Ten male and 10 female prawns, which were sexually mature, but had not brooded, were selected from a brood stock of 20,000 prawns reared for 182 days in a breeding farm (Xiangshan, China). The 20 animals were transferred to an indoor culture environment with a controlled-temperature of 28 °C, and were fed with 5% of the total weight as described previously (Claudio et al. 1997). The male prawns were fed 33 g per day as the high weight gain group (HiWG), and the female prawns were fed 17 g per day as the low weight gain group (LoWG). After two weeks of feeding, all animals were measured for body weight (BW), body length (BL), carapace length (CL), and absolute weight gain(WG), and 10 animals were selected as the experimental subjects of HiWG and LoWG group respectively. At-test of absolute weight gain showed the sample size enabled 99.6% power and a type 1 error of 1% (effect size = 0.761 and Cohen's d = 2.35).
2.2. 16S rDNA sequencing and data processing
The animal’s gut content was collected from the dissected intestines of 20 prawns, and sent to Novgene Bioinformatics Technology Co., Ltd (Tianjing, China). Total bacterial DNA extraction and sequencing were performed according to standard protocols. Briefly, genomic DNA from each sample was extracted using CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gels. According to the concentration, DNA was diluted to 1ng/μL using sterile water. 16S rRNA gene fragments were amplified using a 341f/806r primer set targeting the V3 and V4 regions of bacterial 16S rDNA (341f: 5'-CCTAYGGGRBGCASCAG -3', 806r: 5'-GGACTACNNGGGTATCTAAT-3') (Giuseppe et al. 2003; Qiong et al. 2007). All PCR reactions were carried out within 15 μL of Phusion® High-Fidelity PCR Master Mix (New EnglandBiolabs). Sequencing libraries were constructed using a TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA). The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and the Agilent Bioanalyzer 2100 system. The library was sequenced on Illumina MiSeq 2×250 platforms according to the published protocol. Sequencing reads were assembled using FLASH (V1.2.7) (Magoč and Salzberg, 2011). The quality filtering of the raw reads were performed on QIIME(V1.9.1) as described (Gregory et al. 2010). All reads were compared using UCHIME algorithm to detect chimera sequences, and then the chimera sequences were removed. After the valid reads were finally obtained, operational taxonomic units (OTUs) were picked using the de novo OTU picking protocol with a similarity threshold of 97% by Uparse software (Uparse v7.0.1001) (Edgar, 2013).
2.3. Digestive gland transcriptome sequencing and data processing
Five male and five female prawns were firstly anesthetized by putting on ice for 1 min. After anesthetizing, the samples of eye-stalk, heart, muscle, hepatopancreas, stomach, testis, ovary, big claw, gill, androgenic glands of each shrimp were collected separately and dissected, and pooled by sex. Total RNA was extracted from each type of samples using an RNeasy Plus Micro Kit (QIGEN Group, Germany). Five micrograms of total RNA extracted from each type of pooled samples were used for the construction of Iso-Seq libraries for 3rd generation sequencing, respectively. The Iso-Seq libraries were prepared according to the Isoform Sequencing protocol (Iso-Seq) as described by Pacific Biosciences (PN 100-092-800-03). Sequencing was performed on a PacBio Sequel platform. The raw read data were generated by PacBio Sequel, and were processed using the SMRTlink 7.0 software. The circular consensus sequences (CCSs) were generated from the sub-reads and then the output reads were classified into full-length and non-full length reads. Full-length reads were corrected using the Illumina RNAseq data with the software LoRDEC to obtain final transcripts. The function of each transcript was annotated with NR, NT, Pfam, KOG/COG, Swiss-Prot, KO and GO, and analyzed by BLAST. The e-value ‘1e-10’was set to construct a unique library with gene identifications for the expression level of each analyzed functional gene.
A total quantity of 3 μg RNA per sample was used as the initial material for RNA sample preparation. RNA was used to generate sequencing libraries with various index labels using a NEBNextUltra™ Directional RNA Library Prep Kit. The clustering of index-coded samples was carried out on a cBot cluster generation system. After cluster generation, the libraries were sequenced on an Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA) and 150 bp paired-end reads were generated. All data generated by this study have been deposited in Gene Expression Omnibus (GEO) with the accession code: GSE212385. Additional nucleotide errors in consensus reads were corrected using the Illumina RNAseq data with the software LoRDEC. After correction, the expression levels of each gene were analyzed, and read counts from each sample presented by FPKM values were obtained and used for following analysis.
2.4. Bioinformatics and statistical analysis
For each representative 16S sequence, the Silva Database (
http://www.arb-silva.de/) (Christian et al. 2019) was used to annotate taxonomic information based on Mothur algorithm. In order to study the phylogenetic relationship of different OTUs and the difference of the dominant genera in different samples, multiple sequence alignments were conducted using the MUSCLE software (Version 3.8.31,
http://www.drive5.com/muscle/) (Edgar, 2013). OTUs abundance was normalized using a standard of sequence number corresponding to the sample with the least sequences. Subsequent analyses of alpha diversity and beta diversity were performed based on this normalized output data. Alpha diversity was applied to analyze the complexity of species diversity through6 indices, including Observed-species, Chao1, Shannon, Simpson, ACE, Good-coverage. All these indices in our samples were calculated with QIIME (Version 1.9.1) and presented with R (Version 2.15.3). Beta diversity analysis was used to evaluate the differences of samples in species complexity. Beta diversities assessed by both weighted and unweighted UniFrac were calculated by QIIME software (Version 1.9.1).
Differential expression analysis of each gene between the two groups was performed using the R DESeq package (1.10.1). The resulted P values were adjusted using the Benjamini and Hochberg’s approach to control the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq were assigned as differentially expressed genes (DEGs).Quantitative reverse transcription PCR (qRT-PCR) was performed in triplicate for each sample using a Power SYBRGreen RT-PCR Reagents Kit (Applied Biosystems, Carlsbad, CA, USA). Actin was used as a reference gene. Quantification of all chosen mRNA expressions was conducted using the comparative CT method.
In order to find the differences between the groups under each classification level (Phylum, Class, Order, Family, Genus, Species), we did the Wilcoxon rank-sum test between the two groups (P value < 0.05). Growth traits were compared between the two groups using a one-way t test (P value <0.05). We selected the top 100 microbial genera to calculate the correlation coefficients with Pearson method, and then selected those with the significant (P-value<0.05) correlation coefficients greater than 0.6 or less than -0.6. According to the results of the correlation analysis, the networks were visualized using Cytoscape to reveal the relationship between the gut microbes and digestive gland DEGs or growth traits, in which the genus-level bacteria taxa with a relative abundance > 0.01% were used in the correlation analysis, and only those with a correlation coefficient> 0.3 or < -0.3 and a P value< 0.05 were used in co-occurrence network analysis.
The third-generation transcriptome sequencing generated a total of 82.8 Gb and 148.5 Gb of raw data for male and female animals, respectively, which were used to establish the reference gene sequences and annotations of male and female adult prawn whole transcriptome. Taking the third-generation transcriptome sequencing results as the reference, the second-generation sequencing analysis of the digestive glands and gonad transcriptomes of 10 shrimps (5 males and 5 males) was performed, and the average raw data of each type of sample was 1.51 ± 0.28 Gb. After assembly, a total of 30,260 unique transcribed genes were obtained, and 26,401 functional annotations were completed.
3. Results
3.1. Animal growth traits, 16S characteristics of gut microbes and the statistics of transcriptome data
The experimental animals were fed with the same amount of feed every day to ensure there were no surpluses of feed for the animals, so that there were no differences in relative feeding amount (
P > 0.05).However, after the feeding experiment, there were significant differences between HiWG (male animals) and LoWG (female animals) groups in 4 growth traits: absolute gain weight (GW), body weight (BW), body length (BL) and carapace length (CL). The HiWG prawns had a better performance than the LoWG prawns in the 4 traits (
P < 0.01) (
Figure 1A). The two groups of
M. rosenbergii did not differ in the relative growth ratio (GW/BW prior to the experiment) (
P > 0.05).
A total of 37.2 Gb of raw sequencing data were obtained from 16S gut microbiome sequencing, with an average of 1.7 ± 0.3 Gb of raw data and 83,371 reads per sample (
Table 1). A total of 36.1 Gb of clean data were retained after quality filtering and removing redundant reads. After trimming the reads, 82,549 reads were obtained on average. After quality control, an average of 67, 106 valid reads were obtained. All the valid reads were clustered into OTUs (Operational Taxonomic Units) with 97% identity, and a total of 3,207 OTUs were obtained, among which 1,429 OTUs were annotated to the genera level.
3.2. Gut microbiome diversity determined by 16Ssequencing
The whole microbial NMDS analysis, which based on 16Ssequencing, revealed 3 clusters (Stress = 0.123) between the two groups. Analyses of 16S sequencing results from the gut microbiome identified a total of (83371±8756 sequences per sample) 57 phyla, 683 genera, and 3207 species of bacteria (data not shown), of which top 10 phyla, 10 family, 29 genera were used for the main bacterial analysis in this study (each taxa with a relative abundance >0.5% in at least one sample) (
Table 2). Two of these major bacterial phyla significantly differed in the relative abundance between the HiWG and LoWG groups (P < 0.05) (
Figure 1B). At the family level, the abundance of
Latobacillaceae and
Rhodobacteraceae were more abundant in LoWG than in the HiWG group (P < 0.05) (
Figure 1C). At the genera level, unclassified
Chloroplast and
Candidatus Hepatoplasma were more abundant in the HiWG animals than the LoWG (P < 0.05) (
Figure 1D).
3.3. Gut microbial functions determined by 16S sequencing
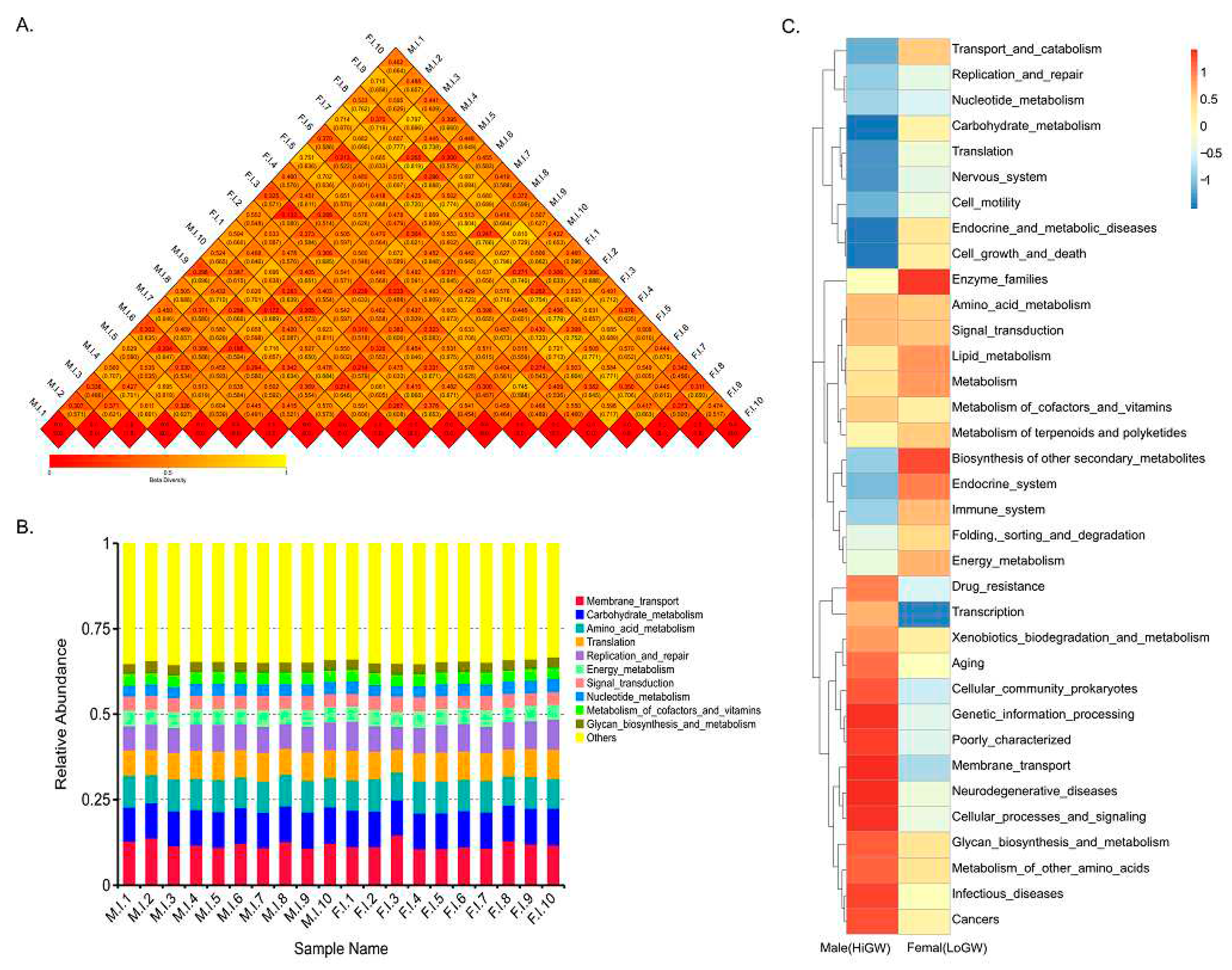
The correlation analysis of the beta diversity index of each sample showed that different groups were significantly clustered together (
Figure 2A). The functional analysis of gut microbes showed that the main functions of gut microbes in the two groups were enriched in the transmembrane transport pathway, followed by carbohydrate metabolism and amino acid metabolism pathways (
Figure 2B). The analysis of intestinal hygiene function between the HiWG and LoWG groups found that the terms of amino acid metabolism, membrane transport, glycan synthesis and metabolic pathways were significantly higher in the HiWG group with a high growth rate than the LoWG group with a low growth rate, while the lipid metabolism and endocrine metabolism were significantly higher in the HiWG group than that of LoGW group with a low growth rate. An opposite trend in the term of energy metabolism was observed (
Figure 2C).
3.4. Digestive glandular and gonadal DEGs and functional analyses
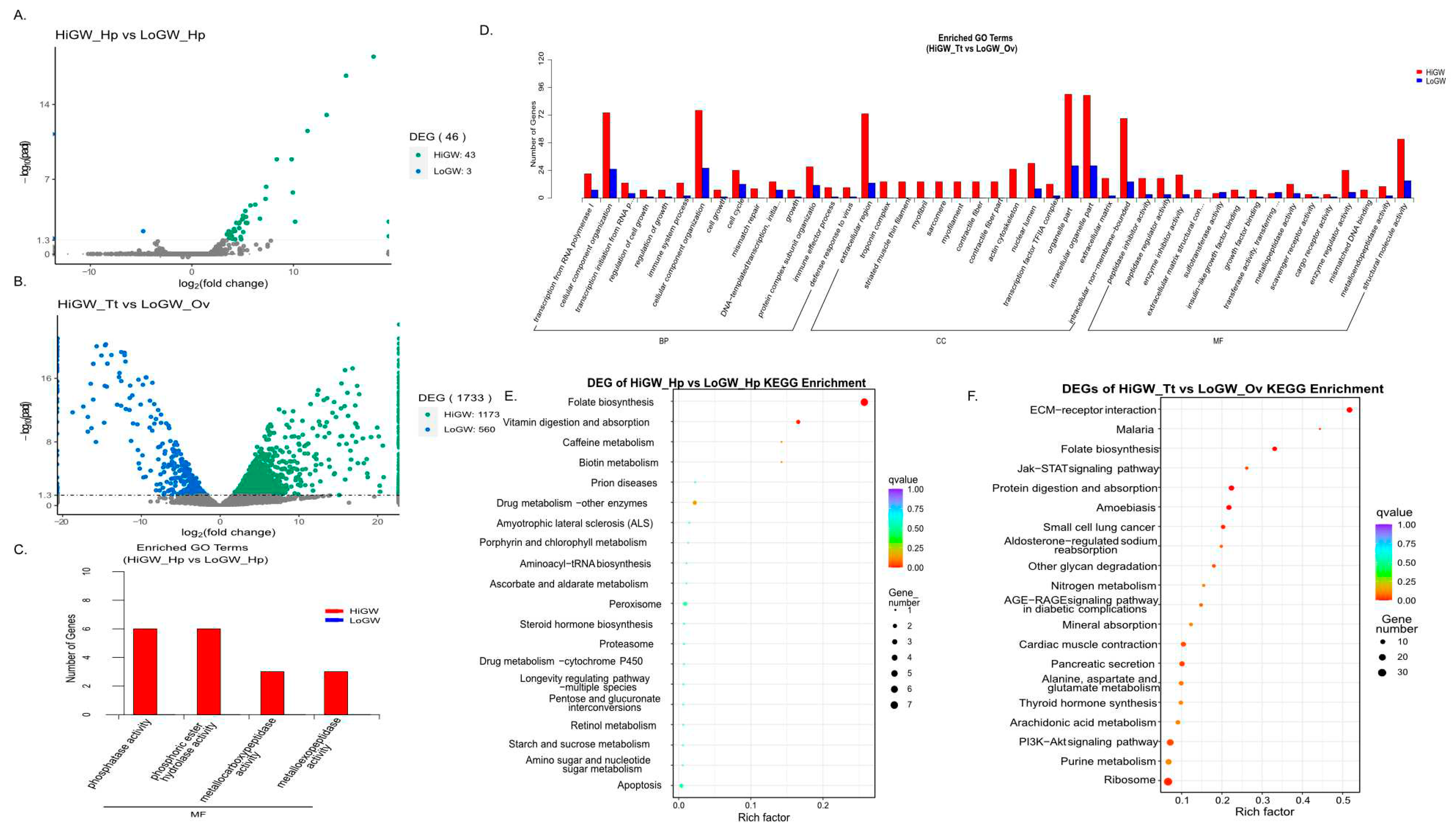
A total of 1779 DEGs were identified between the hepatopancreas and gonad of the two groups of
M. rosenbergii (
Supplementary Table S1 and
Figure 3A). According to the DEG analysis results, in the hepatopancreas, the expression levels of 43 genes were found to be significantly higher in the HiWG group than in the LoWG group, and only 3 genes were highly expressed in the hepatopancreas of the LoWG group (
Figure 3A). In the gonads of
M. rosenbergii, 1173 genes were identified to be significantly highly expressed in the testis of the HiWG group than the gonad of the LoWG group. 560 differential genes were expressed at a higher level in the LoWG group (
Figure 3B). To further investigate the functions of DEGs in the digestive glands and gonads, GO and KEGG enrichment analyses of the identified hepatopancreatic DEGs and gonadal DEGs were performed, respectively. The hepatopancreatic DEGs were significantly enriched in four GO terms, including phosphatase activity, phosphoric ester hydrolase activity, metallocarboxy peptidase activity, and metalloexo peptidase activity, which were related to digestion, absorption and molecule transport processes (
Figure 3C). The KEGG pathway analysis of these DEGs revealed that they were significantly enriched in the pathways of folate biosynthesis, vitamin digestion and absorption, and biotin metabolism (
Figure 3E). The analysis of gonadal DEGs showed that the testis DEGs functioned in the terms of cellular component organization, cell cycle, and organelle composition as shown by the GO analysis (
Figure 3D), while KEGG Pathway analysis showed significant enrichment in ECM-receptor interaction, folate biosynthesis, protein digestion and absorption and other pathways. In addition, the upregulated genes in the LoWG animals’ ovaries were significantly enriched in Ubiquinone and other terpenoid-quinone biosynthesis and Jak- STAT signaling pathway, which are related to lipid metabolism (
Figure 3F).
3.5. Gut microbial interactions and their associations with growth traits
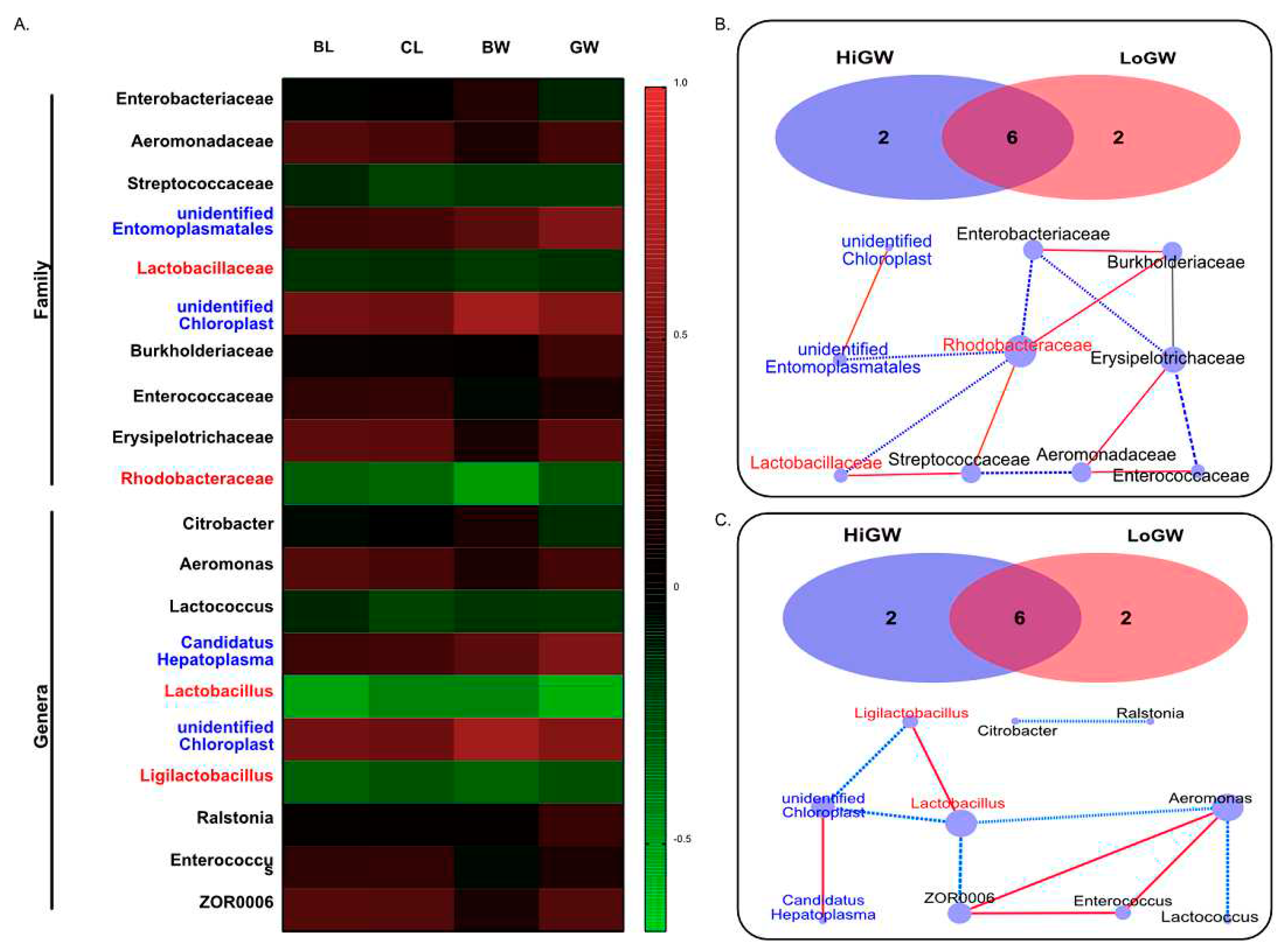
Spearman analyses were conducted on the correlations of the four growth traits with the top 10 abundant gut microbial in family and genera level, and the results showed that unclassified
Entomoplasmatales and
Chloroplast in family level, and unclassified
Chloroplast and
Candidatus Hepatoplasma in genera level were highly enriched in the HiWG group were significant positive correlations (
P<0.05) with the body weight (BW) and gain weight (GW) were observed, and the correlation coefficient between unclassified
Chloroplast and
Candidatus Hepatoplasma with GW reached 0.525, 0.545 respectively (
Figure 4A). The correlation network analysis was carried out with 10 selected gut microbial in family and genera level. Correlation network analysis revealed a total of 25 significant associations were found. Among them, 10 gut microbial family and genera presented tight associations. Two gut microbial in family and genera level were found tightly association with other microbial in HiWG group, and two gut microbial in family and genera level also found tightly association with other microbial in LoWG respectively (
Figure 4B,C).
The results of correlation network analysis in family level showed that there were 6 co-occurrences with a negative spearman coefficient, and the minimum spearman coefficient reached -0.582. The correlation network analysis in genera level showed that there were 4 co-occurrences with a positive spearman coefficient, and the maximum spearman coefficient reached 0.848 between
ZOR0006 and
Enterococcus. The top four key gut microbial genera are unclassfied
Chloroplast,
Lactobacillus,
Aeromonas and
Ligilactobacillus (
Figure 4B). Among the positive co-occurrences, the smallest significant spearman coefficient was 0.463 and was detected between
Candidatus Hepatoplasma and unclassified
Chloroplast (
Figure 4C).
Candidatus Hepatoplasma and unclassified
Chloroplast exhibited positive correlations in the HiWG group, and exhibited negative correlations in the LoWG group.
Lactococcus and
Ligilactobacillus exhibited opposite trend in this study.
3.6. Interaction between gut microbes with DEGs from the hepatopancreas and gonads
The 4 gut microbial genera, which had tight relationships with the 4 phenotypic traits at the 90% confidence level in adult
M. rosenbergii, were selected to conduct the association analysis with the hepatopancreatic and gonadal DEGs. The results showed that 4 gut microbial genera have no directed correlation with all DEGs from the hepatopancreas and gonads. For further exploring the interaction between gut microbes with DEGs, the correlation analysis between gut microbial in species level with DEGs from the hepatopancreas and gonads were conducted. Among all gut microbial species, the 9 gut microbial, which all had tight relationships with the 4 phenotypic traits, were selected. The results showed that the
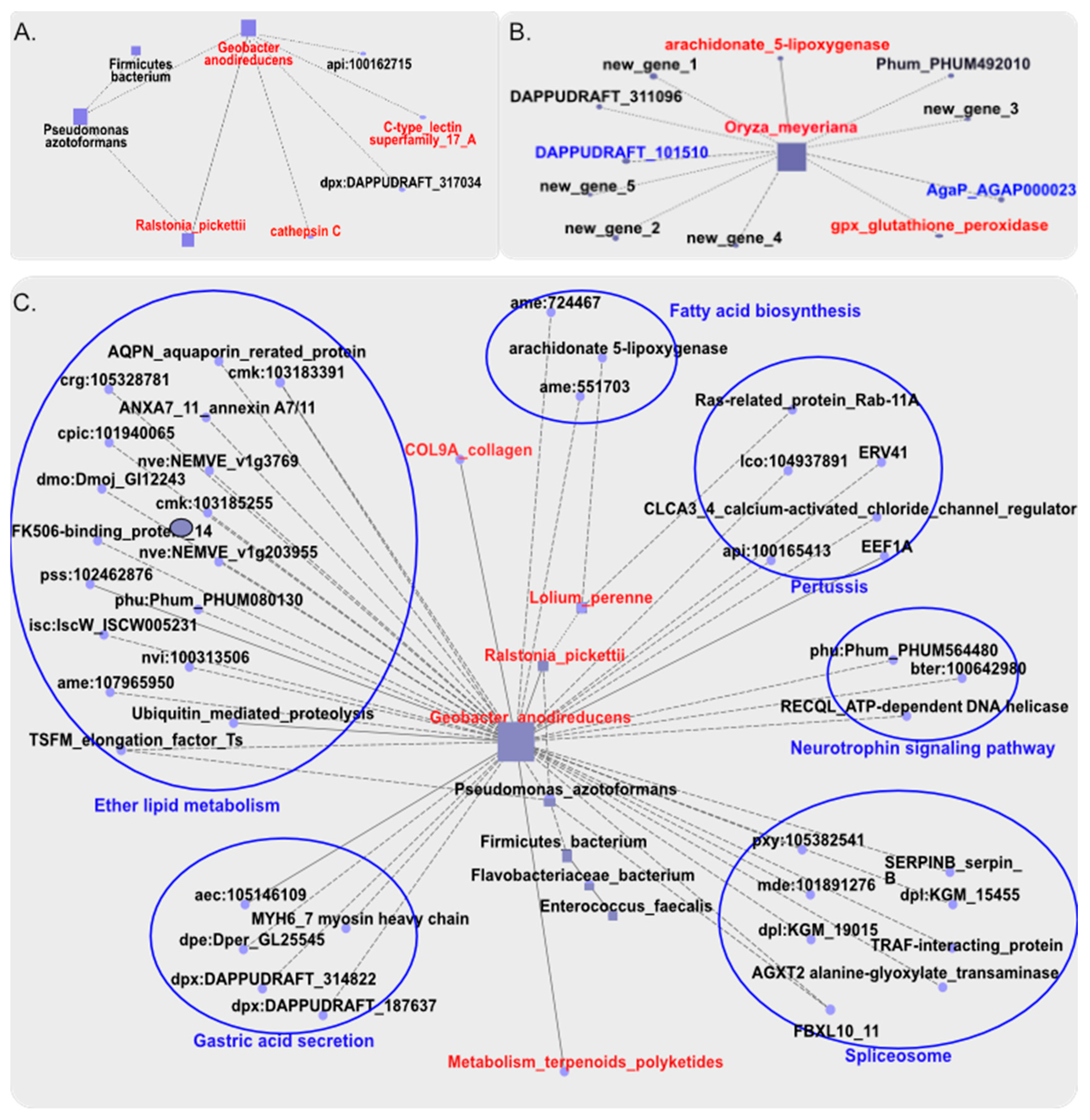
Lolium perenne miacrobial species belong to unclassified
Chloroplast microbial genera which existed a 0.27 relationship coefficient with
Ralstonia microbial genera, might associated with
Ralstonia pickettii, and both associated undirected with
Geobacter anodirecteducens which had a significant correlation with GW in the HiWG group, had a significant negative correlation with the expression level of
cathepsin C in the hepatopancreas. The relative abundance of the gut microbial species,
Geobacter anodirecteducens, presented a negative correlation with
C type lectin super family gene 17Ain the HiWG group (
Figure 5A). A positive correlation between
superoxide dismutase gene and
Lactococcus garvieae abundance
, which was highly enriched in LoWG group and belong to
Lactobacillus genera, was identified. The abundance of
Oryza meyeriana, which was enriched in the HiWG group and belong to unclassified
Chloroplast microbial genera, was associated with
Ras related protein Rab-11A expression, which was highly expressed in the ovary of the LoWG animals. There was a significant positive correlation between the expression level of
arachidonic acid 5-lipoxygenaseand
glutathione peroxidase genes and
Oryza meyeriana abundance (
Figure 5B). The abundance of
Lolium perenne, Ralstonia pickettii, Pseudomonas azotoformans and
Geobacter anodirecteducens, which were highly enriched in the gut of the HiWG group, had a significant correlation with the genes highly expressed in the testis of the HiWG prawns, among which
Geobacter anodirecteducens has a key role in promoting protein digestion and absorption (
Figure 5C) and the COL9A gene expression was positively correlated with its abundance strongly, and have strongly negative correlated with the genes involved in fatty acid biosynthesis and metabolism pathway, indicating that the gut microbiota and specific gene expression in the testis of male prawns could coordinately promote the digestion, absorption and metabolism of amino acids and inhibit fatty acid synthesis, thereby providing a large amount of amino acid as raw materials for protein synthesis needed to increasing the growth for male shrimp.
4. Discussion
M. rosenbergii, the third highest freshwater aquaculture production in China and in the world, is an important source of high-quality animal protein for human, and its global production increases 5% annually owing to its growth rate advantage (Hossain and Chakraborty, 2017). Consistent with the dimorphic characteristics of body size in most aquatic animals, there are also significant gender differences in the body size of adult M. rosenbergii (Carlson and Langkilde, 2017). In the same rearing environment, sexually mature male prawns are significantly larger than female prawns, and their weights can be 2 to 3 times of the female animal (Barki et al. 1997), which is consistent with the results of this study. At the same time, this study found that the absolute weekly-gain weight of sexually mature male M. rosenbergii was also significantly higher than that of females, but there was no significant difference in gain weight rate after sexual maturity. This result indicates that the difference in the growth rate between the sexes primarily happens at the youth developmental stage before sexual maturity. The causes for the differences in the sexually dimorphic growth traits of M. rosenbergii are a hot topic in the field (Chong et al. 2019). Studies on the determination of sexual development are a key direction to explore the phenotypic differences of M.rosenbergii (Wilhelm and Pask, 2018). The different levels of hormones secreted by the gonads of M. rosenbergii result in differences in the differentiation and proliferation of male and female cells (Ren et al. 2021), which ultimately lead to differences in the growth rate and the characteristics of morphological differences. Regarding the impact of gut microbes on growth traits, studies on other aquatic animals have shown that gut microbes have a significant impact on animal growth and development. In addition, gut microbes also affect the immune system of shrimps, which in turn affects their development, leading to phenotypic differences.
Microbes in the animal gut are thought to be important drivers of several metabolic processes in the host, including those related to growth and development. Therefore, the characterization of the microbial flora is a very attractive topic for aquaculture research. The association between the gut microbiome and growth traits has been investigate recently and reported not only in aquatic animals, but also in other vertebrates (Elin et al. 2018). A report on shrimp gut microbes has shown that the polymorphism of gut microbes can not only affect the digestive process of shrimps (Wen-Fang et al. 2018), but also improve the health by influencing the immune system, thereby achieving rapid growth and development (Yilong et al. 2019). Since the survival rate and disease resistance of shrimps are directly related to growth traits, the current research on how shrimp gut microbes promote growth mainly focuses on the regulation of shrimp gut microbes on animal health, and their indirect effects on animal growth and development (Seibert and Pinto, 2012; Chaiyapechara et al. 2012). A previous study shows that at the phylum level, Gamma proteobacteriais the major component of the gut microbes of tiger shrimp and Litopenaeus vannamei (Chaiyapechara et al. 2012; Rungrassamee et al. 2016), while our study found that the intestinal microbes of M. rosenbergii are mainly enriched with two similar contents, Proteobacteria and Firmicutes. Their abundance accounted for more than 87% of the gut microflora of M. rosenbergii, which is similar to the gut microflora of mouse (Guo et al. 2021). An investigation on shrimp gut microbes show that the main shrimp gut microbes primarily regulate growth performance and immune ability by regulating processes such as carbohydrate and protein digestion and metabolism, as well as the susceptibility of the digestive tract to foreign pathogens (Holmes et al. 2007). In this study, we also found that the gut microbes of male prawns were significantly enriched with pathways of protein digestion and absorption, and the gut microbes of the female group had a significant advantage of lipid metabolism. This result further explained the reasons for the different growth and development in different sex shrimp from the perspective of gut microbes.
Further correlation analysis of gut microbes in our study found that Candidatus Hepatoplasma, unclassified Chloroplast were highly enriched in the male group, and Lactobacillus and Ligilactobacillus were highly enriched in the female group. The abundance of these gut microbes was associated with the growth traits. The two bacteria genera highly abundant in the male gut were associated with 6 other bacteria genera and 4 phenotypics, and participated in pathways of protein digestion and amino acid metabolism, while the female group also had two bacteria genera existed negative association between abundant with phtenotypic traits. Further investigation of the relationship between gut microbes with digestive glandular and gonadal DEGs revealed that Lolium perenne and Ralstonia pickettii might association on Geobacter anodirecteducens abundance, a flora enriched for amino acid metabolism and protein metabolism in males, was tightly associated with collagen gene expression, which was differentially expressed in the gonads with a function of protein digestion and absorption. This association indicates that the male gut microbe, Lolium perenne, Ralstonia pickettii, Geobacter anodirecteducens, can coordinate with collagen to regulate protein digestion, absorption and metabolism. The other gene associated with the abundance of Geobacter anodirecteducens is lectin gene, whose protein product promotes the accumulation of proteins, thereby promoting growth. In addition, the higher expression of arachidonic acid 5-lipoxy synthase in the male hepatopancreas also has a tight relationship with gut microbe, Oryza meyeriana, which can inhibit lipids, promote lipid metabolism, and accelerate growth. An opposite correlation was observed in the females in our study.
Candidatus Hepatoplasma belongs to the phyla Desulfobacter of gram-negative bacteria, a group of microorganisms that are widespread in the environment (Röling et al. 2001; Holmes et al. 2007). It has been reported that these microorganisms can enhance protein synthesis (Wilkins et al. 2011). Another gut microbe that is highly enriched in the fast-growing male prawns identified in this study, Oryza meyeriana, is a gram-negative bacterium of phyla Proteobacteria that can promote metabolism and improved disease resistance (Barve, 2013). As the main cultured species of freshwater prawns, M. rosenbergii, like other crustaceans, has a strong demand for protein metabolism at the early stage of growth (Denis et al. 1998), and protein intake determines the size of weight gain (Nasir and Allen, 2002). In addition, studies on Litopenaeus vannamei have also shown that the protein metabolism pathways of shrimps that are beneficial to animal growth and development under stress response conditions are significantly improved (Chang et al, 2017). This study confirmed that the rapid growth and development of M. rosenbergii also depends on key genes in the hepatopancreas and gonad to promote protein digestion and absorption, and these genes can coordinate together with gut microbes to promote protein absorption and metabolism, thereby promoting the rapid growth and development of prawns. Our study found that collagen gene is highly expressed in the higher weight gain group, which is consistent with L. vannamei under stress conditions (Atrux-Tallau et al. 2011). And our study also found that collagen expression had a significant association with the gut microbe, Geobacter anodirecteducens. In addition, the highly expressed gene glutathione peroxidase found in the high weight gain group was also inhibited before female sexual maturity in other organisms, indicating that after sexual maturity of female prawns, the ovary needs to synthesize a large amount of lipids to store energy. And glutathione peroxidase, which is necessary for the lipid metabolism, has a inhibitory effect on growth (Simşek et al. 1998), which also indicates that the rapid growth and development before sexual maturity of M. rosenbergii mainly depend on the accumulation of protein, supplementation of lipids. And the gut microbes may supplement this function.
5. Conclusion
In the absence of a host reference genome, 16s RNA sequencing and transcriptome profiling are currently the best approaches to study the association between gut microbiome or differential genes with the fast-growing trait. The correlation analysis showed that there were significant associations among the gut microbes of M. rosenbergii, and the gut microbiota was also associated with the growth traits of body weight and weight gain, both in the HiWG group and the LoWG groups. The abundance of Geobacter anodirecteducens correlation with the expression of glutathione peroxidase and collagen in HiWG group have synergistic and positive effects with the hepatopancreatic and gonadal on protein digestion and absorption and might promote growth and development. These associations were not detected in the female LoWG group.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, Jindong Ren; methodology, validation, investigation, writing-review and editing, Hongxiang Xu and Jindong Ren; resource, Xiaojun Xu; funding acquisition, Jindong Ren; supervision and project administration, Bao Lou and Demin Zhang.
Funding
This study was financially supported by Zhejiang Provincial Natural Science foundation with grant number LY21C190001.
Institutional Review Board Statement
The study was approved by the Laboratory Animal Welfare and Ethics Committee of Zhejiang Academy of Agricultural Sciences (Hangzhou, China).
Acknowledgments
We would like to express gratitude to Dongmin Zhu at Yonggang Aquatic Seedlings Co., Ltd., Ningbo, Zhejiang province, for his kind help in collecting samples.
Conflicts of Interest
The authors declare no competing interest.
References
- Ahmed N, Allison EH, Muir JF (2010) Rice-fields to prawn farms: a blue revolution in southwest Bangladesh? Aquaculture International 18:555–574. [CrossRef]
- Atrux-Tallau N, Callejon S, Migdal C, Padois K, Bertholle V, Denis A, Chavagnac-Bonneville M, Haftek M, Falson F, Pirot F (2011) Development and in vitro assay of oxidative stress modifying formulations for wound healing promotion. Eur J Dermatol 2:52-62. [CrossRef]
- Barki A, Harpaz S, Karplus I (1997) Contradictory asymmetries in body and weapon size, and assessment in fighting male prawns, Macrobrachium rosenbergii. AggrBehav 23: 81-91. [CrossRef]
- Barve S (2013) Gut microbiota, intestinal barrier function, endotoxemia and alcoholic liver injury. Alcohol and Alcoholism 48(1):i17. [CrossRef]
- Belsare S, Dhaker HS, Pawase A, Joshi V, Shelke S (2017) Effect of dietary carbohydrate-lipid ratio on growth, body composition and digestive enzyme activities of juvenile goldfish (Carassius auratus). Animal Nutrition & Feed Technology 17(1):43. [CrossRef]
- Carani FR (2014) Expression of growth-related factors in skeletal muscle of pirarucu (Arapaima gigas) during growth. Journal of Aquaculture Research & Development 5:6. [CrossRef]
- Carlson BE, Langkilde T (2017) Body size variation in aquatic consumers causes pervasive community effects, independent of mean body size. EcolEvol 7:9978– 9990. [CrossRef]
- Chaiyapechara S, Rungrassamee W, Suriyachay I, Kuncharin Y, Klanchui A, Karoonuthaisiri N, Jiravanichpaisal P (2012) Bacterial community associated with the intestinal tract of P. monodon in commercial farms. Microb Ecol 63(4):938-53. [CrossRef]
- Chang Xu, Erchao Li, Yan Liu, Xiaodan Wang, Jian G. Qin, Liqiao Chen (2017) Comparative proteome analysis of the hepatopancreas from the Pacific white shrimp Litopenaeus vannamei under long-term low salinity stress. Journal of Proteomics 162:1-10. [CrossRef]
- Chong Li, Ganesan Hagilaa, Yong Chean, Tan Wen Siang, Ho KokLian (2019) Expression, purification and characterization of the dimeric protruding domain of Macrobrachium rosenbergii nodavirus capsid protein expressed in Escherichia coli. PLOS ONE 14:e0211740. [CrossRef]
- Christian Quast, ElmarPruesse, PelinYilmaz, Jan Gerken, Timmy Schweer, Pablo Yarza, JörgPeplies, Frank Oliver Glöckner (2019) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41(1):590–596. [CrossRef]
- Claudio Chávez Justo, Katsumi Aida, Isao Hanyu (1991) Effects of Photoperiod and Temperature on Molting, Reproduction and Growth of the Freshwater Prawn Macrobrachium rosenbergii. Bulletin of the Japanese Society of Scientific Fisheries (Japan) 57(2): 209-217. [CrossRef]
- Cornejo-Granados F, Lopez-Zavala AA, Gallardo-Becerra L, Mendoza-Vargas A, Sánchez F, Vichido R, Brieba LG, Viana MT, Sotelo-Mundo RR, Ochoa-Leyva A (2017) Microbiome of pacific white leg shrimp reveals differential bacterial community composition between wild, aquacultured and AHPND/EMS outbreak conditions. Sci Rep 7:11783. [CrossRef]
- Dan Cohen, Ziva Ra'anan (1983) The production of the freshwater prawn Macrobrachium rosenbergii in Israel: III. Density effect of all-male Tilapia hybrids on prawn yeld characters in polyculture. Aquaculture 35:57-71. [CrossRef]
- Davey AJH, Jellyman DJ. (2005). Sex determination in freshwater eels and management options for manipulation of sex. Rev Fish Biol Fisheries 15, 37–52. [CrossRef]
- Denis Ricque-Marie, Ma. IsabelAbdo-de La Parra, L.Elizabeth Cruz-Suarez, Gerard Cuzon, Marc Cousin, Ian H Pike (1998) Raw material freshness, a quality criterion for fish meal fed to shrimp. Aquaculture 165(1-2):95-109. [CrossRef]
- Dinh H, Nguyen NH (2014) Genetic inheritance of female and male morphotypes in giant freshwater prawn Macrobrachium rosenbergii. PLoS One 9(2):e90142. [CrossRef]
- Dong Yunwei, NiuCuijuan, Du Li (2001) Effects of dietary protein levels on growth and activity of digestive enzymes of giant fresh water prawn (Macrobrachium rosenbergii). Journal of Beijing Normal University (Natural Science) 37(1):96-99.
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792-1797. [CrossRef]
- Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996-998. [CrossRef]
- Elin Videvall, Se JinSong, Hanna Bensch, Maria Strandh, Anel Engelbrecht, Naomi Serfontein, Olof Hellgren, Adriaan Olivier, Schalk Cloete, Rob Knight, Charlie K (2018) The development of gut microbiota in ostriches and its association with juvenile growth. Molecular Ecology 270017. [CrossRef]
- Giuseppe Blaiotta, Carmelina Pennacchia, DaniloErcolini, Giancarlo Moschetti, Francesco Villani (2003) Combining denaturing gradient gel electrophoresis of 16S rDNA V3 region and 16S–23S rDNA spacer region polymorphism analyses for the identification of staphylococci from Italian fermented sausages. Systematic and Applied Microbiology 26(3): 423-433. [CrossRef]
- Gregory Caporaso J, Justin Kuczynski, Jesse Stombaugh, Kyle Bittinger, Frederic D Bushman, Elizabeth K Costello, Noah Fierer, Antonio Gonzalez Peña, Julia K Goodrich, Jeffrey I Gordon, Gavin A Huttley, Scott T Kelley, Dan Knights, Jeremy E Koenig, Ruth E Ley, Catherine A Lozupone, Daniel McDonald, Brian D Muegge, Meg Pirrung, Jens Reeder, Joel R Sevinsky, Peter J Turnbaugh, William A Walters, Jeremy Widmann, Tanya Yatsunenko, Jesse Zaneveld, Rob Knight (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. [CrossRef]
- Guo S, Geng W, Chen S, Wang L, Rong X, Wang S, Wang T, Xiong L, Huang J, Pang X, Lu Y (2021) Ginger alleviates DSS-induced ulcerative colitis severity by improving the diversity and function of gut microbiota. Front Pharmacol 12:632569. [CrossRef]
- Habte-Tsion HM, Bo L, Ge X, Xie J, Chen R (2013) Effects of dietary protein level on growth performance, muscle composition, blood composition, and digestive enzyme activity of wuchang bream (Megalobrama amblycephala) fry. The Israeli journal of aquaculture Bamidgeh, 65: 9.
- Holmes DE, O'Neil RA, Vrionis HA, N'guessan LA, Ortiz-Bernad I, Larrahondo MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP, Chavan MA, Johnson JP, Long PE, Lovley DR (2007) Subsurface clade of geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J 1(8):663-77. [CrossRef]
- Hossain MM, Chakraborty SC (2017) Growth and economic analysis of freshwater prawn, Macrobrachium rosenbergii(de Man), produced with feeds substituting sunflower cake for fish meal, soya bean meal and mustard oil cake. Aquaculture Research 48:5418–5429. [CrossRef]
- Huang Z, Li X, Wang L, Shao Z (2016) Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac Res 47:1737-1746. [CrossRef]
- Jun-jie Yao, Yun-long Zhao, Qun Wang, Zhong-liang Zhou, Xian-cheng Hu, Xiao-weiDuan, Chuan-guang An (2006) Biochemical compositions and digestive enzyme activities during the embryonic development of prawn, Macrobrachium rosenbergii. Aquaculture 253:573-582. [CrossRef]
- Kumar JSS, Nagarathinam N, Sundararaj V (2000) Production characteristics of Macrobrachium rosenbergii and M. malcolmsonii under controlled monoculture system. Journal of Aquaculture in the tropics 15(3):207-217.
- Landsman A, St-Pierre B, Rosales-Leija M, Brown M, Gibbons W (2019) Impact of aquaculture practices on intestinal bacterial profiles of pacific white leg shrimp Litopenaeus vannamei. Microorganisms 7(4):93. [CrossRef]
- Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957-2963. [CrossRef]
- Martínez-Alarcón D, Harms L, Hagen W, Saborowski R (2019) Transcriptome analysis of the mid gut gland of the brown shrimp Crangon crangon indicates high polymorphism in digestive enzymes. Mar Genomics 43:1-8. [CrossRef]
- Martinez-Cordova LR, Villarreal-Colmenares H, Porchas-Cornejo MA, Naranjo-Paramo J, Aragon-Noriega A (1997) Effect of aeration rate on growth, survival and yield of white shrimp Litopenaeus vannamei in low water exchange ponds. Aquacultural Engineering 16(1-2):85-90. [CrossRef]
- Moss SM, LeaMaster BR, Sweeney JN (2000) Relative abundance and species composition of gram-negative, aerobic bacteria associated with the gut of juvenile white shrimp Litopenaeus vannamei reared in oligotrophic well water and eutrophic pond water. Journal of the World Aquaculture Society 31:255-263. [CrossRef]
- Nasir Kureshy, Allen Davis (2002) Protein requirement for maintenance and maximum weight gain for the Pacific white shrimp, Litopenaeus vannamei. Aquaculture 204(1–2):125-143. [CrossRef]
- Qiong Wang, George M Garrity, James M Tiedje, James R Cole (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 83(16):5261-5267. [CrossRef]
- Ren J, Na R, Chen H. Niu B, Lou B (2021) RNA sequencing and functional analysis of adult gonadal tissue to identify candidate key genes in Macrobrachium rosenbergii sex development. Aquacult Int 29:2805–2821. [CrossRef]
- Röling WF, Breukelen BM van, Braster M, Lin B, Verseveld HW van (2001) Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microbiol 67(10):4619-29. [CrossRef]
- Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N (2016) Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J Invertebr Pathol 133:12-9. [CrossRef]
- Seibert CH, Pinto AR (2012) Challenges in shrimp aquaculture due to viral diseases: distribution and biology of the five major penaeid viruses and interventions to avoidviral incidence and dispersion. Brazilian J Microbiol 43 (3):857–864. [CrossRef]
- Siddiqui AQ, Hinty HMA, Ali SA (1996) Evaluation of the production potential of Macrobrachium rosenbergii (de Man) in monoculture and in polyculture with Nile tilapia and common carp in Saudi Arabia. Aquaculture Research, 27: 515-521. [CrossRef]
- Silva MAS, Neto ME, Ramiro BO, Santos ITF, Guerra RR (2018) Histomorphologic characterization of the hepatopancreas of freshwater prawn Macrobrachium rosenbergii (De Man, 1879). ArquivoBrasileiro de MedicinaVeterinária e Zootecnia 70:1539-1546. [CrossRef]
- Simşek M, Naziroğlu M, Simşek H, Cay M, Aksakal M, Kumru S (1998) Blood plasma levels of lipoperoxides, glutathione peroxidase, beta carotene, vitamin A and E in women with habitual abortion. Cell Biochem Funct 16(4):227-31. [CrossRef]
- Sui J, Luan S, Yang G, Xia Z, Luo K, Tang Q, Lu X, Meng X, Kong J (2019) Genetic parameters and selection response for the harvest body weight of the giant freshwater prawn (Macrobrachium rosenbergii) in a breeding program in China. PloS one 14(8):e0218379. [CrossRef]
- Tang Q, Xia Z, Cai M, Xie J, Pan Y, Jingfen LI, Yang G (2019) Correlation and difference of phenotypic traits among cultured groups of the giant freshwater prawn Macrobrachium rosenbergii. Journal of Fishery Sciences of China 26(06):1075-1085.
- Wen-Fang Dai, Jin-Jie Zhang, Qiong-Fen Qiu, Jiong Chen, Wen Yang, Sui Ni, Jin-Bo Xiong (2018) Starvation stress affects the interplay among shrimp gut microbiota, digestion and immune activities. Fish & Shellfish Immunology 80: 191-199. [CrossRef]
- Wilhelm D, Pask AJ (2018) Genetic Mechanisms of sex determination - ScienceDirect. Encyclopedia of Reproduction (Second Edition) 3:245-249. [CrossRef]
- Wilkins MJ, Callister SJ, Miletto M, Williams KH, Nicora CD, Lovley DR, Long PE and Lipton MS (2011) Development of a biomarker for Geobacter activity and strain composition; Proteogenomic analysis of the citrate synthase protein during bioremediation of U(VI). Microbial Biotechnology 4:55-63. [CrossRef]
- Xue MY, Xie YY, Zhong Y, Ma XJ, Sun HZ, Liu JX (2022) Integrated meta-omics reveals new ruminal microbial features associated with feed efficiency in dairy cattle. Microbiome 10(1):32. [CrossRef]
- Yilong Wang, Baojie Wang, Xuqing Shao, Jianchun Shao, Mei Liu, Mengqiang Wang, Lei Wang (2019) The effect of rearing density on immune responses of hepatopancreas and intestine in Litopenaeus vananmei against Vibrio paraheamolyticus E1 challenge. Fish & Shellfish Immunology 93:517-530. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).