Submitted:
17 July 2023
Posted:
18 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Cloning and Expression Vector Construction of NtHDR
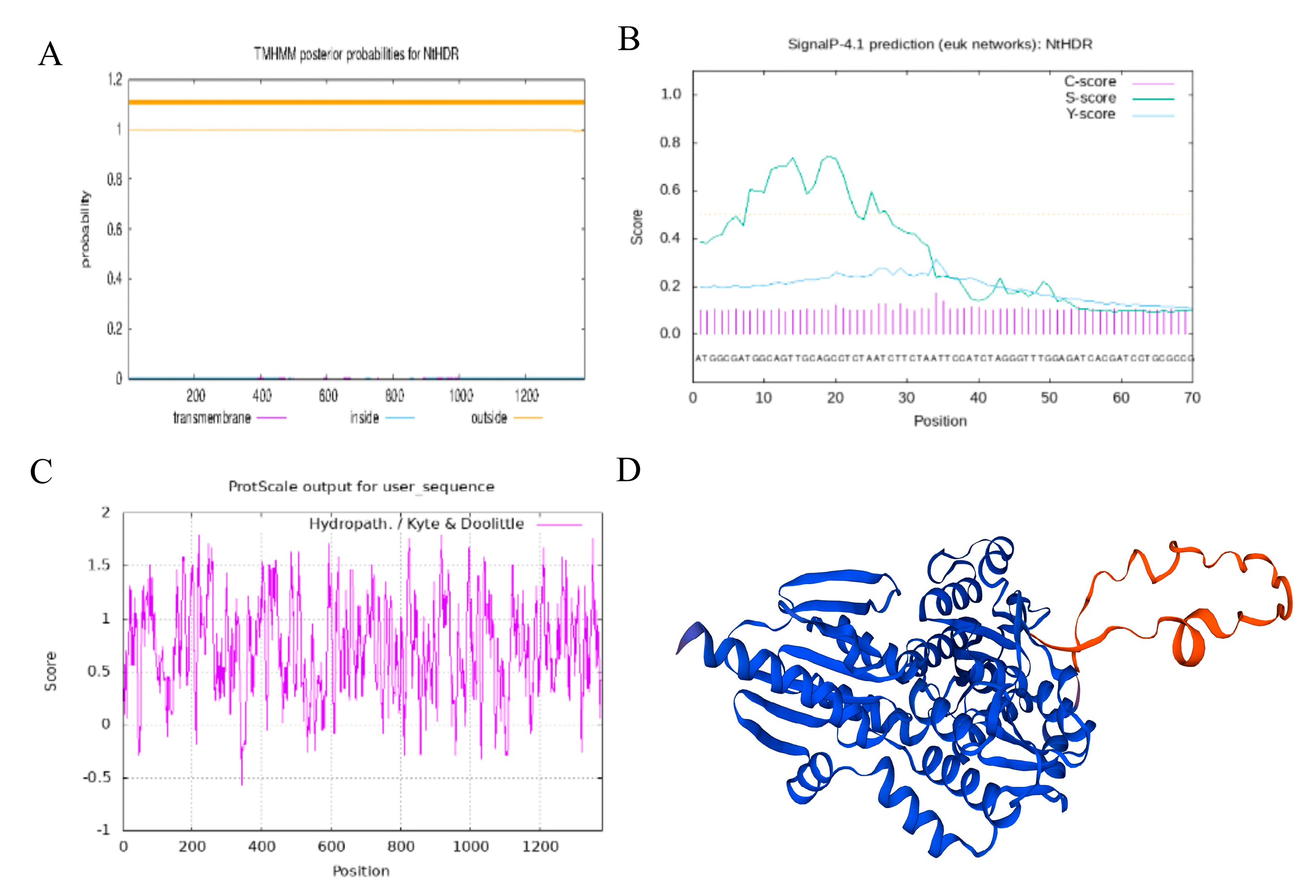
2.4. NtHDR Bioinformatics Analysis
2.5. Fluorescence Quantification of NtHDR in ‘Jinzhanyintai’
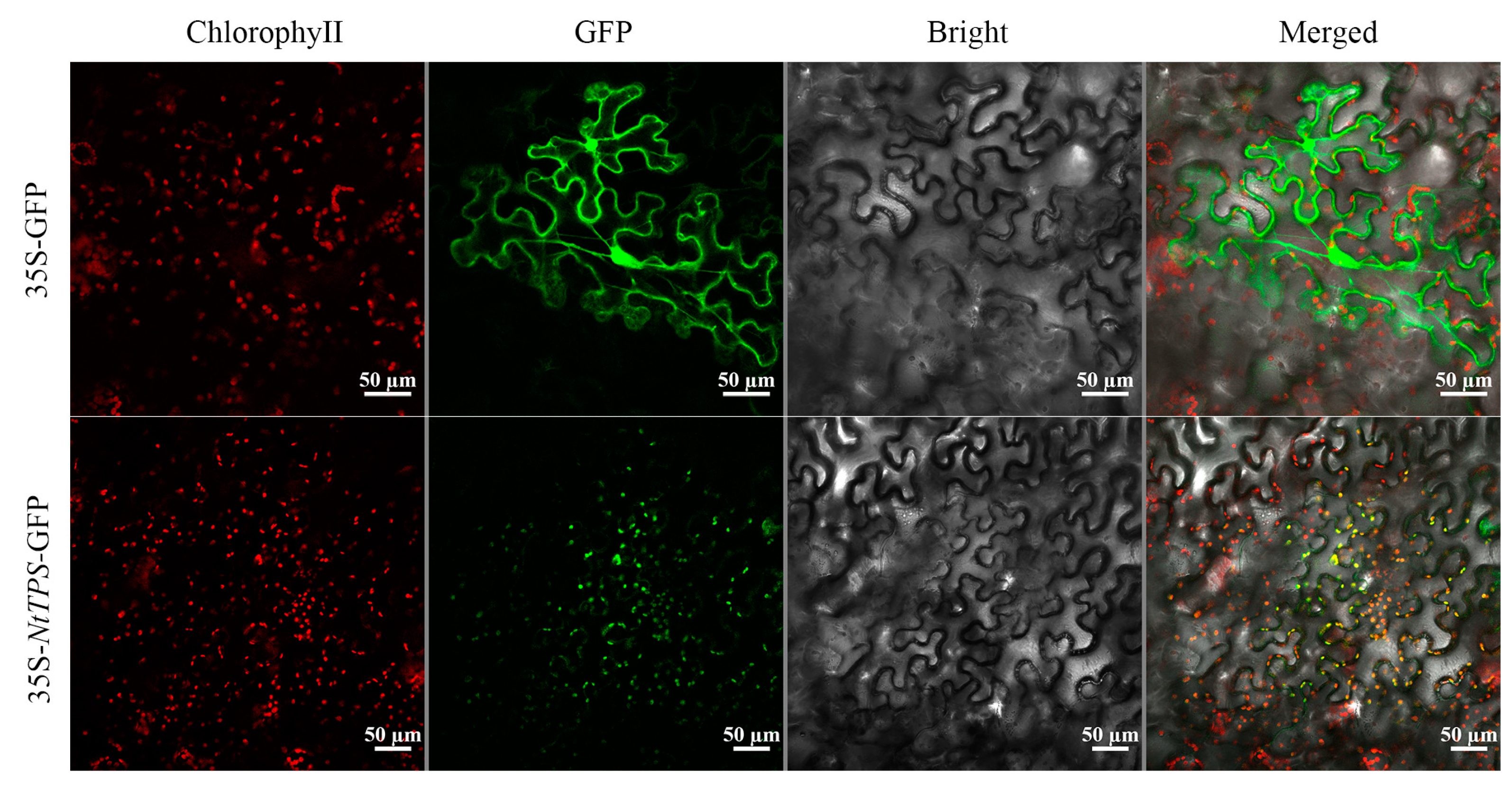
2.6. Subcellular Localization Assay of NtHDR Proteins in Tobacco
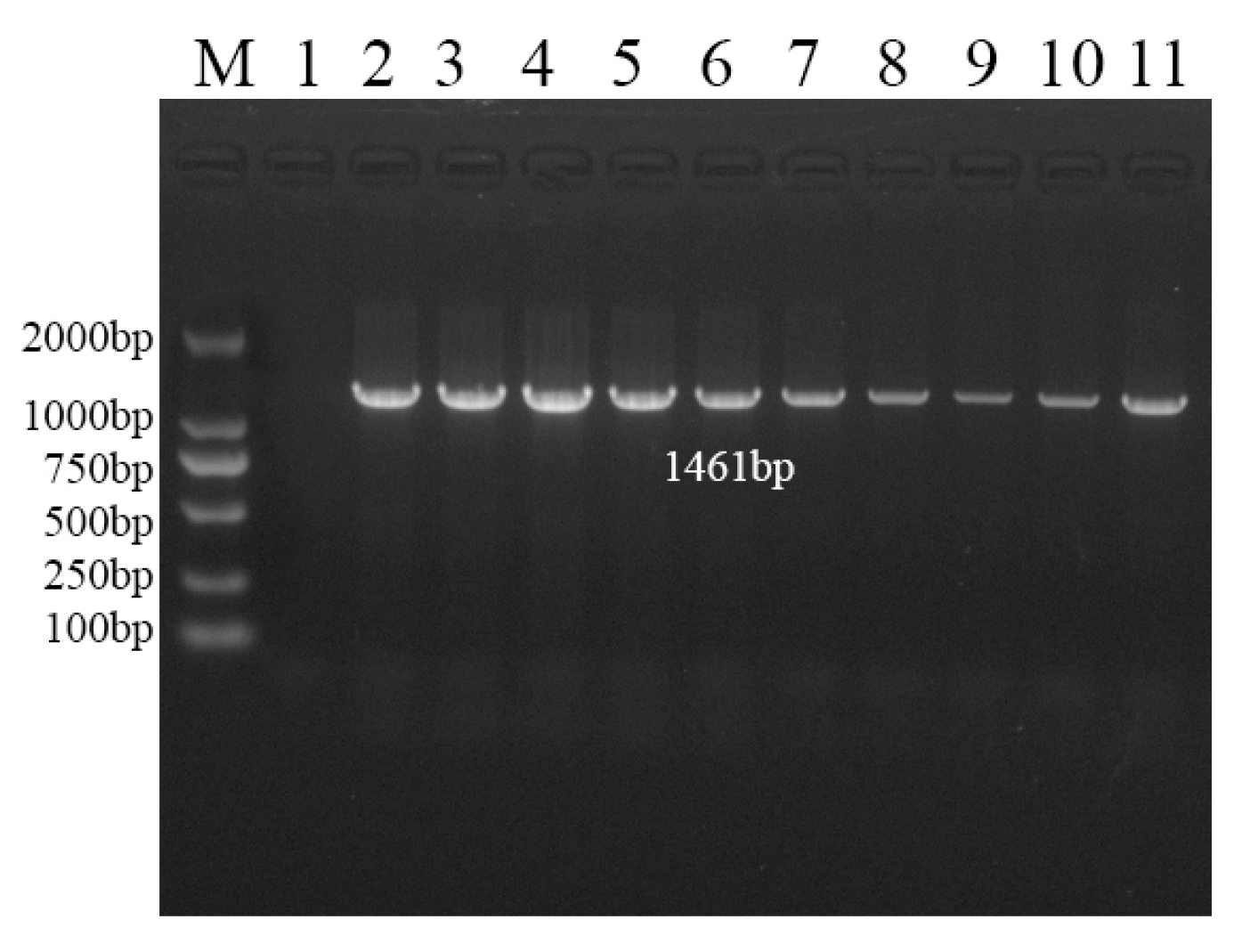
2.7. Agrobacterium-Mediated Transformation and Identification of N. benthamiana
2.8. Analysis of Volatile Components in Transgenic N. benthamiana Flowers
3. Results
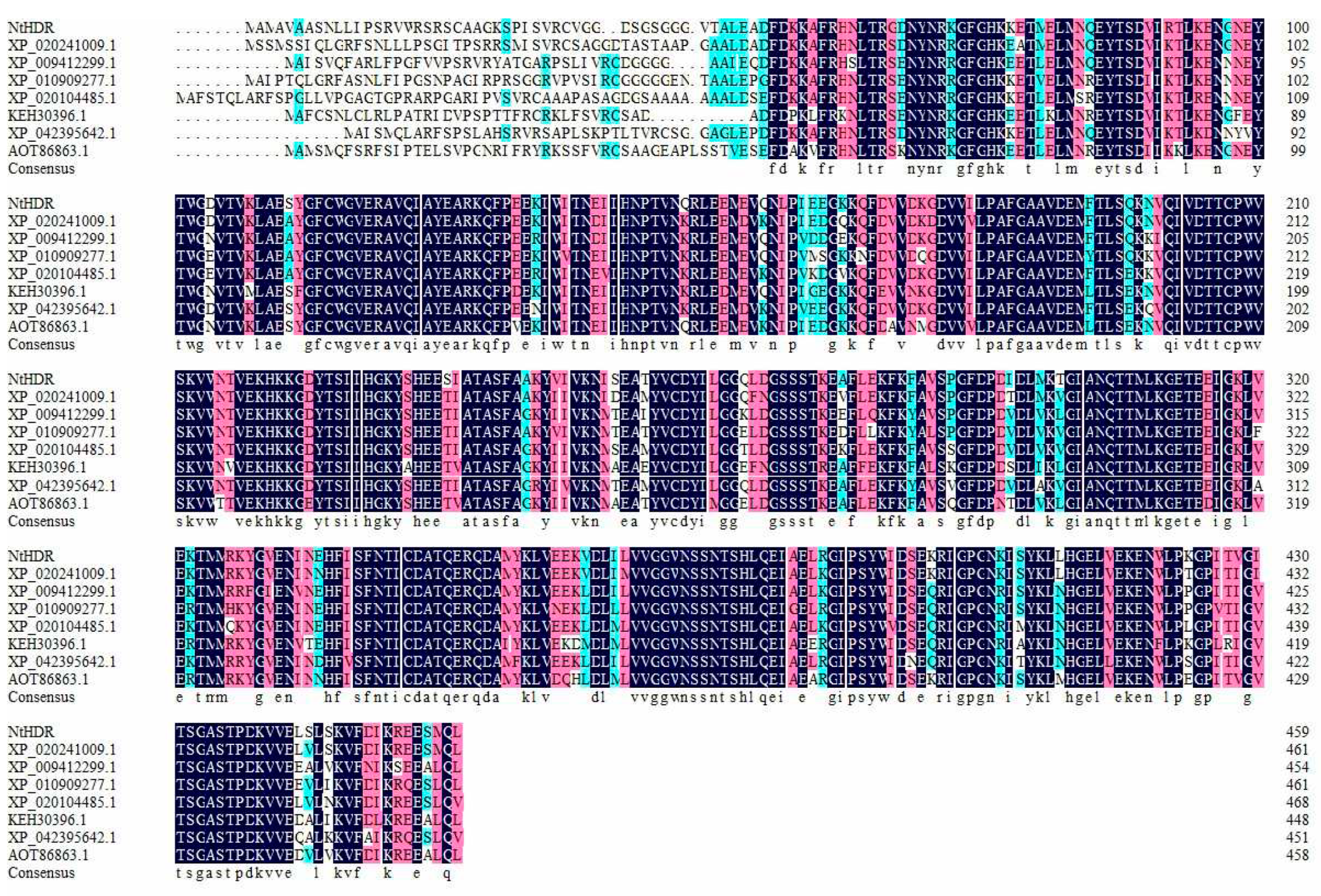
3.1. Characterization of NtHDR
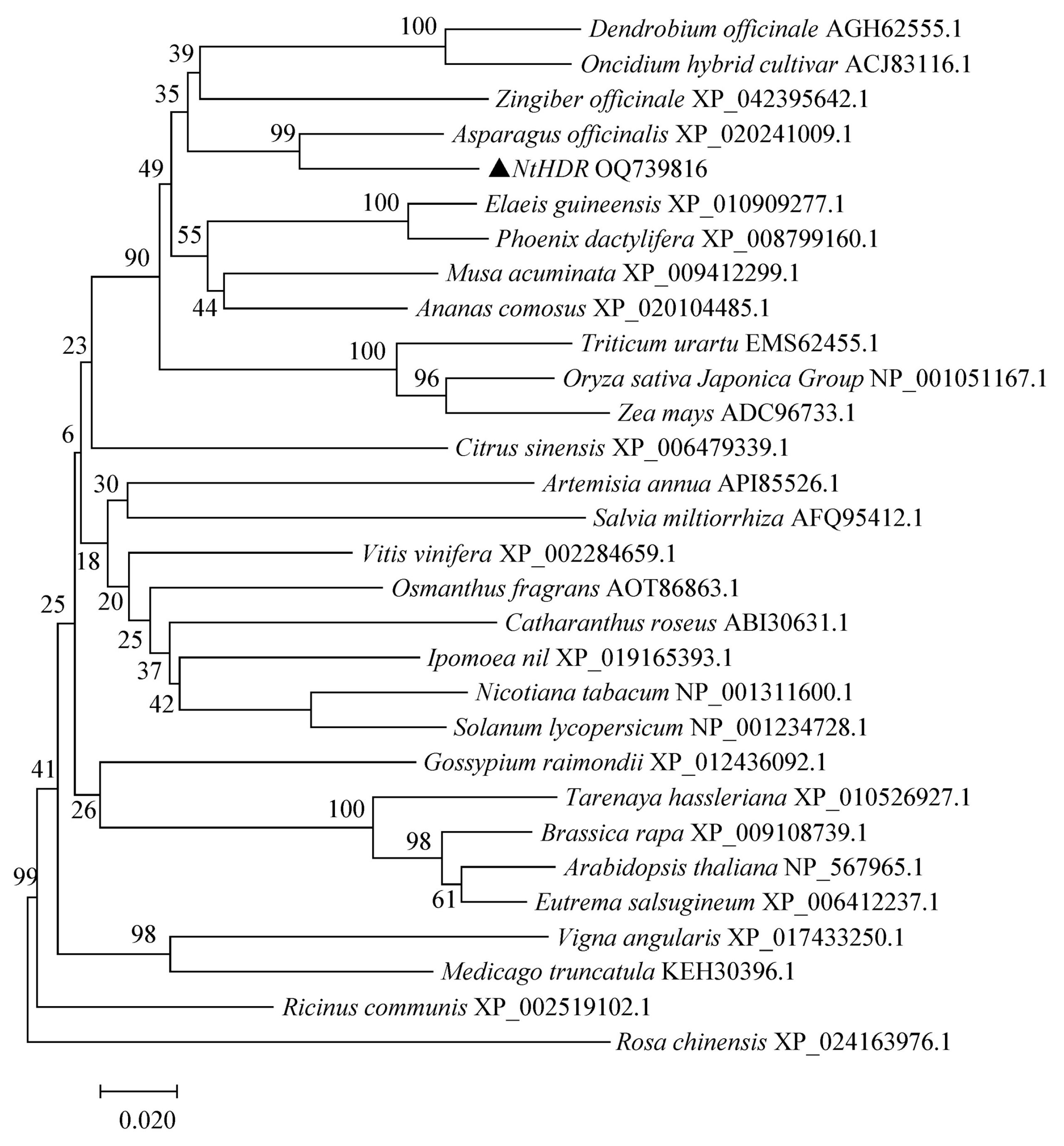
3.2. Phylogenetic Analysis and Sequence Alignment of NtHDR
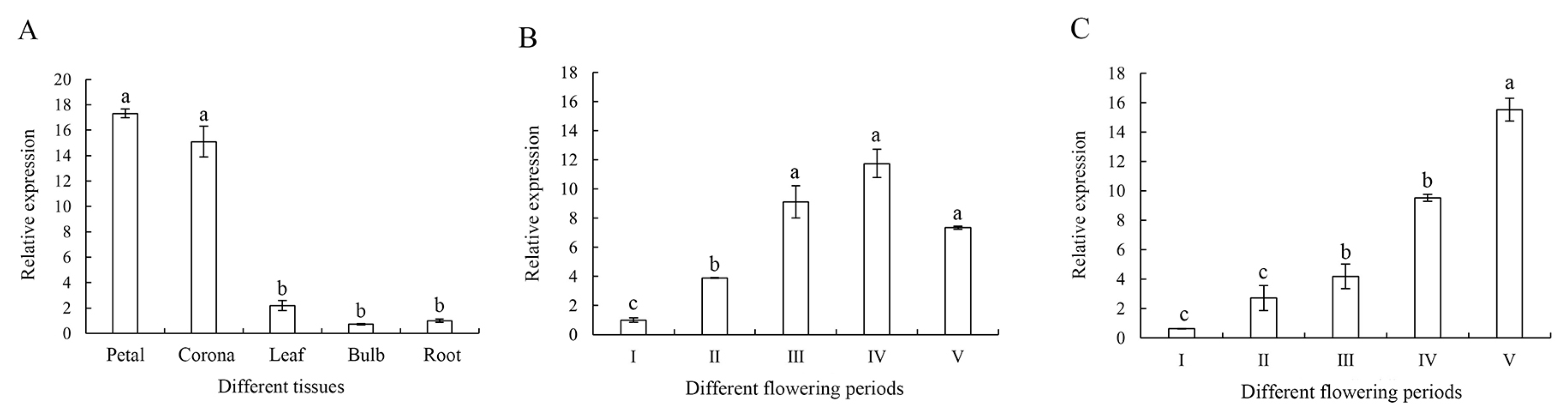
3.3. NtHDR Expression Specificity Analysis
3.4. NtHDR Subcellular Localization Analysis
3.5. Stable Expression of NtHDR in N. benthamiana
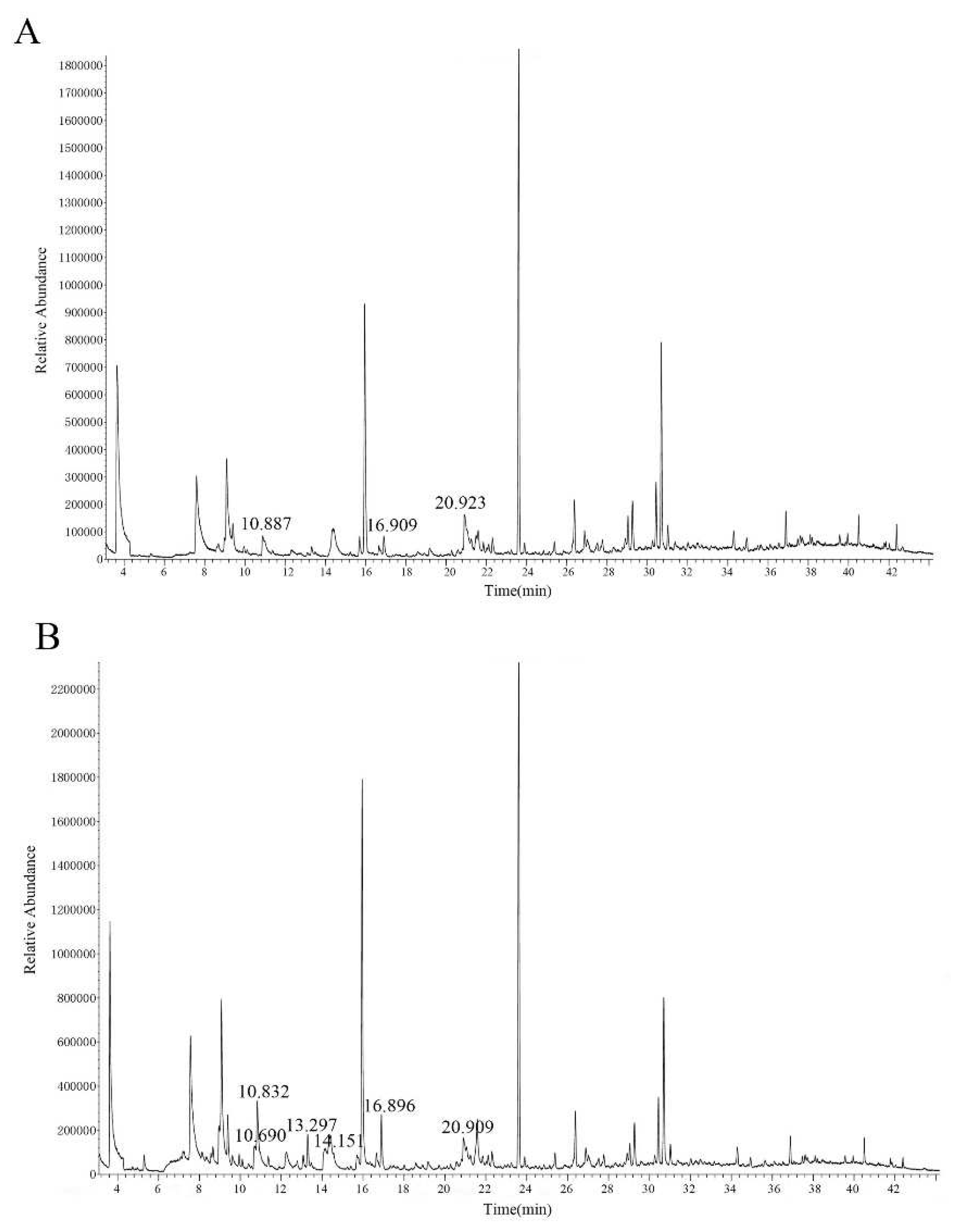
3.6. Determination and Analysis of Floral Volatiles of Transgenic N. benthamiana
| Main volatile components | Retention time/min | Relative content/% | |||||
|---|---|---|---|---|---|---|---|
| A Wild-type N. benthamiana | B Transgenic N. benthamiana | ||||||
| CK1 | CK2 | CK3 | H1 | H2 | H3 | ||
| Benzyl alcohol | 10.690 | — | — | — | — | 1.288 | 1.687 |
| Phenylacetaldehyde | 10.832 | 1.682 | 1.001 | — | 2.652 | 4.247 | 1.105 |
| Linalool | 13.297 | — | — | — | 0.551 | 1.086 | 0.777 |
| Phenethyl alcohol | 14.151 | — | — | — | — | 1.812 | — |
| Decamethylcyclopentasiloxane | 15.963 | 10.766 | 10.488 | 7.715 | 16.885 | 12.974 | 11.461 |
| 2-Isobutyl-3-methoxypyrazine | 16.893 | 0.870 | — | — | 1.003 | 1.940 | — |
| Cinnamic aldehyde | 20.923 | 3.203 | — | 0.233 | 4.047 | — | 4.133 |
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pichersky, E.; Noel, J.P.; Dudareva, N. Biosynthesis of plant volatiles: Nature’s diversity and ingenuity. Science, 2006, 311, 808–811. [CrossRef]
- Kessler, A.; Baldwin, I.T. Defensive function of herbivore-induced plant volatile emissions in nature. Science, 2001, 291, 2141–2144. [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J, 2011, 66, 212–229. 66. [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol, 2013, 198, 16–32. [CrossRef]
- Kitaoka, N.; Lu, X.; Yang, B.; Peters, R. J. The application of synthetic biology to elucidation of plant mono-, sesqui-, and diterpenoid metabolism. Mol. Plant, 2015, 8, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.T.; Cui, J.B.; Hu, H.L.; Xue, J.Y.; Yang, J.J.; Xu, J. Integrated four comparative-omics reveals the mechanism of the terpenoid biosynthesis in two different overwintering Cryptomeria fortunei phenotypes. Front. Plant Sci, 2021, 12, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Adam, P.; Hecht, S.; Eisenreich, W.; Kaiser, J.; Gräwert, T.; Arigoni, D.; Bacher, A.; Rohdich, F. Biosynthesis of terpenes: Studies on 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Proc. Natl. Acad. Sci. U. S. A, 2002, 99, 12108–12113. [CrossRef]
- Altincicek, B.; Duin, E.C.; Reichenberg, A.; Hedderich, R.; Kollas, A.K.; Hintz, M.; Wagner, S.; Wiesner, J.; Beck, E.; Jomaa, H. LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett, 2002, 532, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Wolff, M.; Seemann, M.; Tse Sum Bui, B.; Frapart, Y.; Tritsch, D.; Garcia Estrabot, A.; Rodríguez-Concepción, M.; Boronat, A.; Marquet, A.; Rohmer, M. Isoprenoid biosynthesis via the methylerythritol phosphate pathway: The (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase (LytB/IspH) from Escherichia coli is a [4Fe-4S] protein. FEBS Lett, 2003, 541, 115–120. [CrossRef]
- Botella-Pavía, P.; Besumbes, Ó.; Phillips, M.A.; Carretero-Paulet, L.; Boronat, A.; Rodríguez-Concepción, M. Regulation of carotenoid biosynthesis in plants: Evidence for a key role of hydroxymethylbutenyl diphosphate reductase in controlling the supply of plastidial isoprenoid precursors. Plant J, 2004, 40, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Page, J.E.; Hause, G.; Raschke, M.; Gao, W.Y.; Schmidt, J.; Zenk, M.H.; Kutchan, T.M. Functional analysis of the final steps of the 1-deoxy-D-xylulose 5-phosphate (DXP) pathway to isoprenoids in plants using virus-induced gene silencing. Plant Physiol, 2004, 134, 1401–1413. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.P.; Shi, R.J.; Tao, R.; Fang, Q.; Jiang, X.Y.; Ji, H.W.; Feng, L.; Huang, L.Q. Cloning, molecular characterization and functional analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (HDR) gene for diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza Bge. f. alba. Plant Physiol. Biochem, 2013, 70, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.J.; Wu, M.J.; Wang, X.; Lin, Y.; Cai Y.P.; Fan, H.H. Cloning and expression analysis of 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphatereductase gene in Dendrobium officinale. Chinese Traditional and Herbal Drugs, 2015, 46, 405–411. (in Chinese).
- Jiang, Z.; Peng, Z. Chinese Narcissus; China Forestry Publishing House: Beijing, China, 2014; ISBN 978-75-0386-911-2. [Google Scholar]
- Gao, J. The study on mutagenic effects and mechanism of 60CoY-ray on Chinese narcissus. Chinese Academy of Forestry, Beijing, 2000.
- Peng, A.M. Analysis on volatile components of Narcissus tazetta var. chinensis and influence factors. Chinese Academy of Forestry, Beijing, 2010. (in Chinese).
- Dai, L.; Yang, L.; Guo, Y.; Peng, Q. Study on the chemical composition of the essential oil of Narcissus tazetta var. chinensis. Chromatography, 1990, 6, 377–380. (in Chinese). [Google Scholar]
- Huang, Q.Q.; Feng, J.Y. Study of aroma changes in daffodils by adsorption filament/chromatography/mass spectrometry. Chinese Journal of Analytical Chemistry, 2003, 31, 1408. (in Chinese). [Google Scholar]
- Huang, Q.Q.; Feng, J.Y. Study on the variation in narcissus aroma composition during blossoming. Journal of Instrumental Analysis, 2004, 23, 110–113. (in Chinese).
- Dou, Y.J.; Zhai, J.; Hou, F.M.; Leng, P.S.; Wang, W.F.; Hu, Z.H. Effect of Different Light Intensities on the Floral Aroma Emitted from Chinese Daffodil (Narcissus tazetta L. var. chinensis Roem). Acta Agriculturae Boreali-Occidentalis Sinica 2014, 23, 85–91. (in Chinese). [Google Scholar]
- Song, G.; Xiao, J.; Deng, C.; Zhang, X.; Hu, Y. Use of solid-phase microextraction as a sampling technique for the characterization of volatile compounds emitted from Chinese daffodil flowers. J. Anal. Chem, 2007, 62, 674–679. [Google Scholar] [CrossRef]
- Wu, Y. Study on Cloning of HDS gene and resistant transcription factor of Narcissus tazetta. Fujian Agriculture and Forestry University, Fuzhou, 2016. (in Chinese).
- Zhong, X. The study ofrelationship of B-glucosidase andvolatile aromatic components of Narcissus tazetta L. var. Chinensis Roem. Fujian Agriculture and Forestry University, Fuzhou, 2014. (in Chinese).
- Zheng, Q.B. Clone of the NtSAMT Gene in Narcissus tazetta L. Var. chinensis roem and transformation of Asarina procumbens. Northeast Agricultural University, Haerbin, 2016. (in Chinese).
- Li, K. Fragrance component analysis and genecloning in Narcissus tazetta. Fujian Agriculture and Forestry University, Fuzhou, 2014. (in Chinese).
- Ma, C. Cloning of DXPS, DXR and PAL related to floral fragrance from Narcissus tazetta L. var. chinensis Roem. Fujian Agriculture and Forestry University, Fuzhou, 2010. (in Chinese).
- Li, L. Analysis of genes related to the terpenoids synthesis pathway in Narcissus tazetta 'Yunxiang' based on Transcriptome data. Fujian Agriculture and Forestry University, Fuzhou, 2019. (in Chinese).
- Pang, H.D.; Xiang, L.; Zhao, K.G.; Li, X.; Yang, N.; Cheng, L.Q. Genetic transformation and functional characterization of Chimonanthus praecox SAMT gene in tobacco. Journal of Beijing Forestry University, 2014, 36, 117–122. (in Chinese). [Google Scholar]
- Zhang, T. Molecular cloning and functional analysis of the key enzyme LiDXS genes involved in monoterpene biosynthesis in lily. Beijing Forestry University, Beijing, 2017. (in Chinese).
- Chen, J.D. Founction research of odorant in chimonanthus praecox. Central China Agricultural University, Wuhan, 2016. (in Chinese).
- Altschup, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol, 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol, 1987, 4, 406–425. [CrossRef]
- Wang, G. Transcriptome analysis of flowers and functional studies of MYB genes offlavonoid biosynthetic pathway in Chinese narcissus (Narcissus tazetta var. chinensis). Fujian Agriculture and Forestry University, Fuzhou, 2018. (in Chinese).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods, 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Meng, N.; Liu, Y.L.; Dou, X.X.; Liu, H.L.; Li, F.Y. Transient gene expression in Phalaenopsis aphrodite petals via Agrobacterium tumefaciens infiltration. Acta Botanica Boreali-Occidentalia Sinica, 2018, 38, 1017–1023. (in Chinese). [Google Scholar]
- Zhang, C.G.; Ge, Y.Y.; Liu, H.H.; Zong, Y.X.; Wu, X.J.; Yang, L.C.; Li, H.G. Cloning and bioinformatics analysis of LtuHMGS in Liriodendron tulipifera. Journal of Central South University of Forestry & Technology, 2022, 42, 146–155. (in Chinese). [Google Scholar]
- Liu, G. Differential regulation of catechin metabolism in albino and normal tea (Camellia sinensis) leaves and tea linalool biosynthesis. Anhui Agricultural University, Hefei, 2017. (in Chinese).
- Sun, M. Cloning and functional analysis of TPS gene family in tree peony fragrance. Beijing Forestry University, Beijing, 2021. (in Chinese).


| Primer use | Primer name | Sequences (5’→3’) |
|---|---|---|
| Cloning the CDS | NtHDR-F | atggcgatggcagttgcag |
| NtHDR-R | cgccaactgcatagattcttcc | |
| Subcellular localization analysis | pBI121-NtHDR-F | tggagagaacacgggggactatggcgatggcagttgcag |
| pBI121-NtHDR-R | ataagggactgaccacccgccaactgcatagattcttcc | |
| Stable transformation of N. benthamiana | pBI121-NtHDR-R2 | ataagggactgaccaccttacgccaactgcatagattcttcc |
| Real-time PCR | NtActin-F | tgcccagaagtgctattccag |
| NtActin-R | gttgacccaccactaagaacaatg | |
| qPCR -NtHDR-F | tgacgaggggcgataactacaatc | |
| qPCR -NtHDR-R | gactctgctagcttcaccgttac | |
| Identification of positive transgenic N. benthamiana | 35s-F | gacgcacaatcccactatcc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





