Submitted:
19 July 2023
Posted:
20 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Sample Collection and Preparation
2.3. PCR and ELISA Diagnostic
2.4. Virus Isolation and Sequencing
2.5. Data Analysis
3. Results
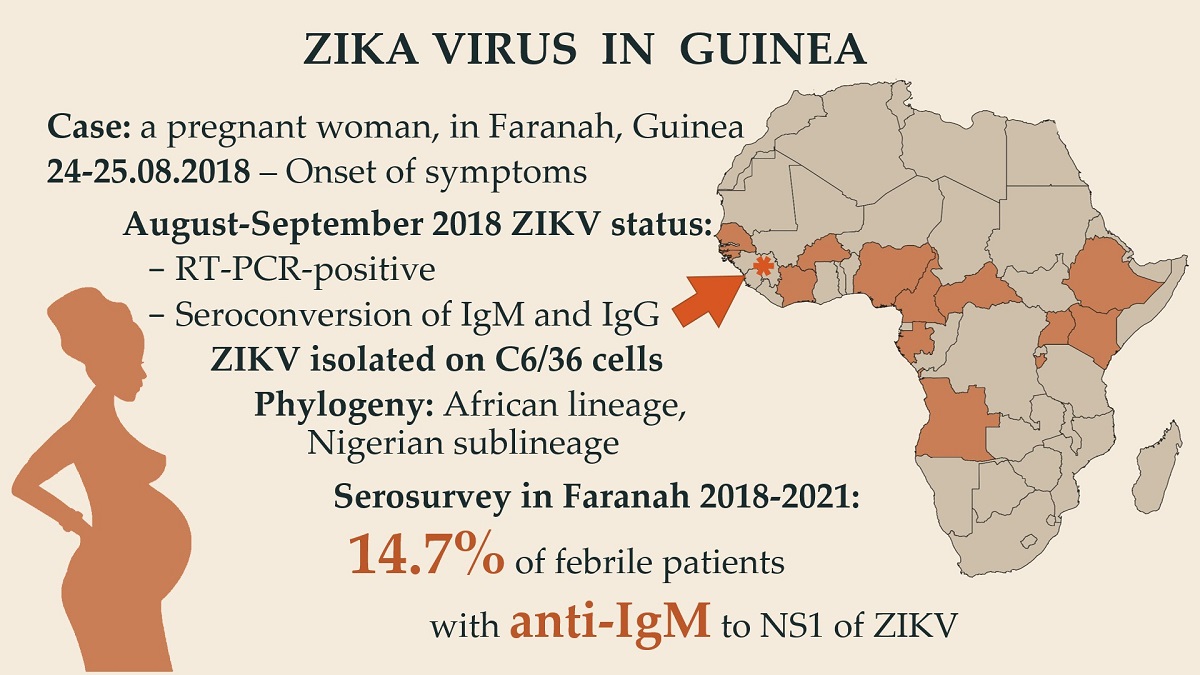
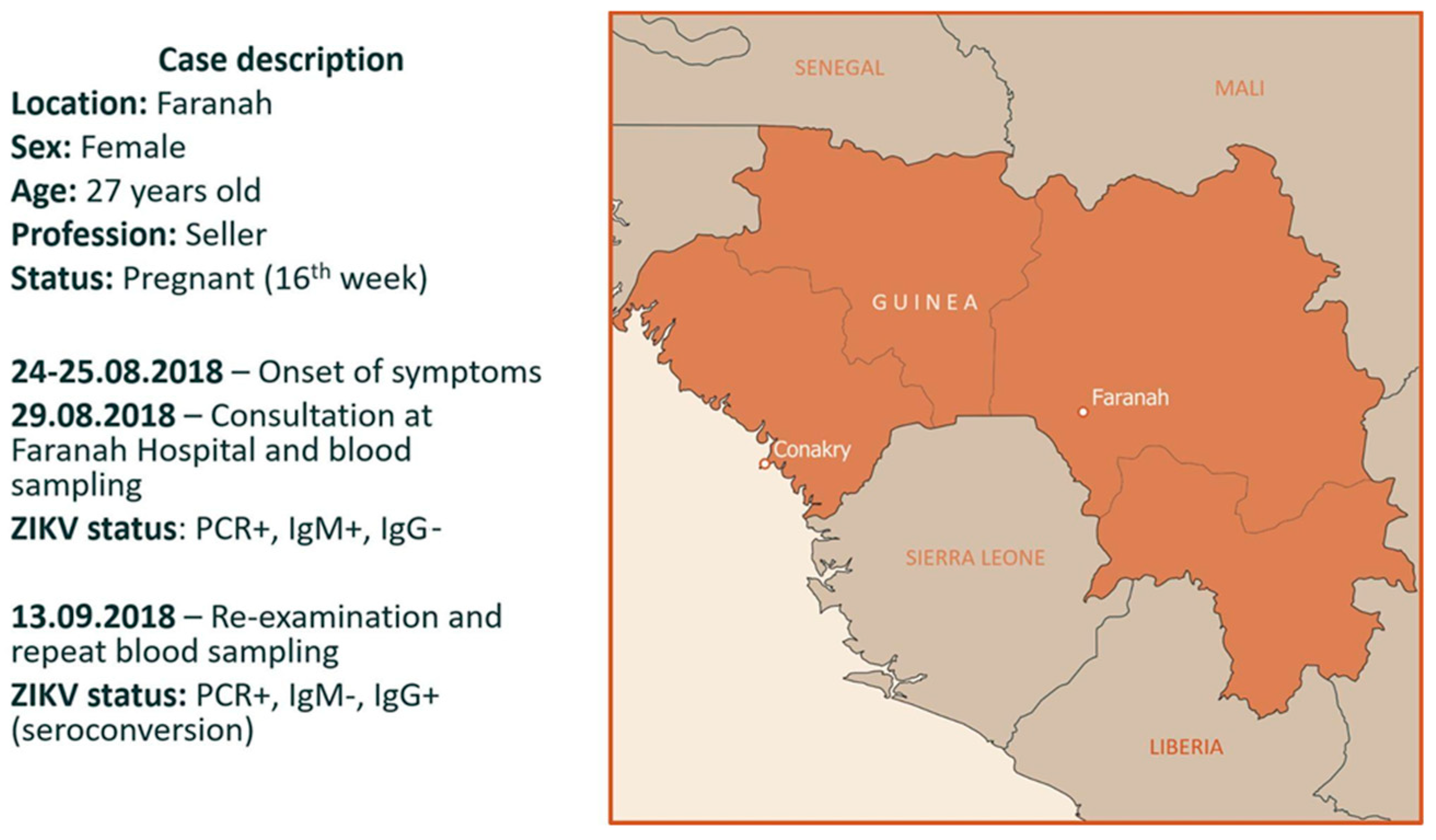
3.1. Case Description and Diagnostics
3.2. Serosurvey
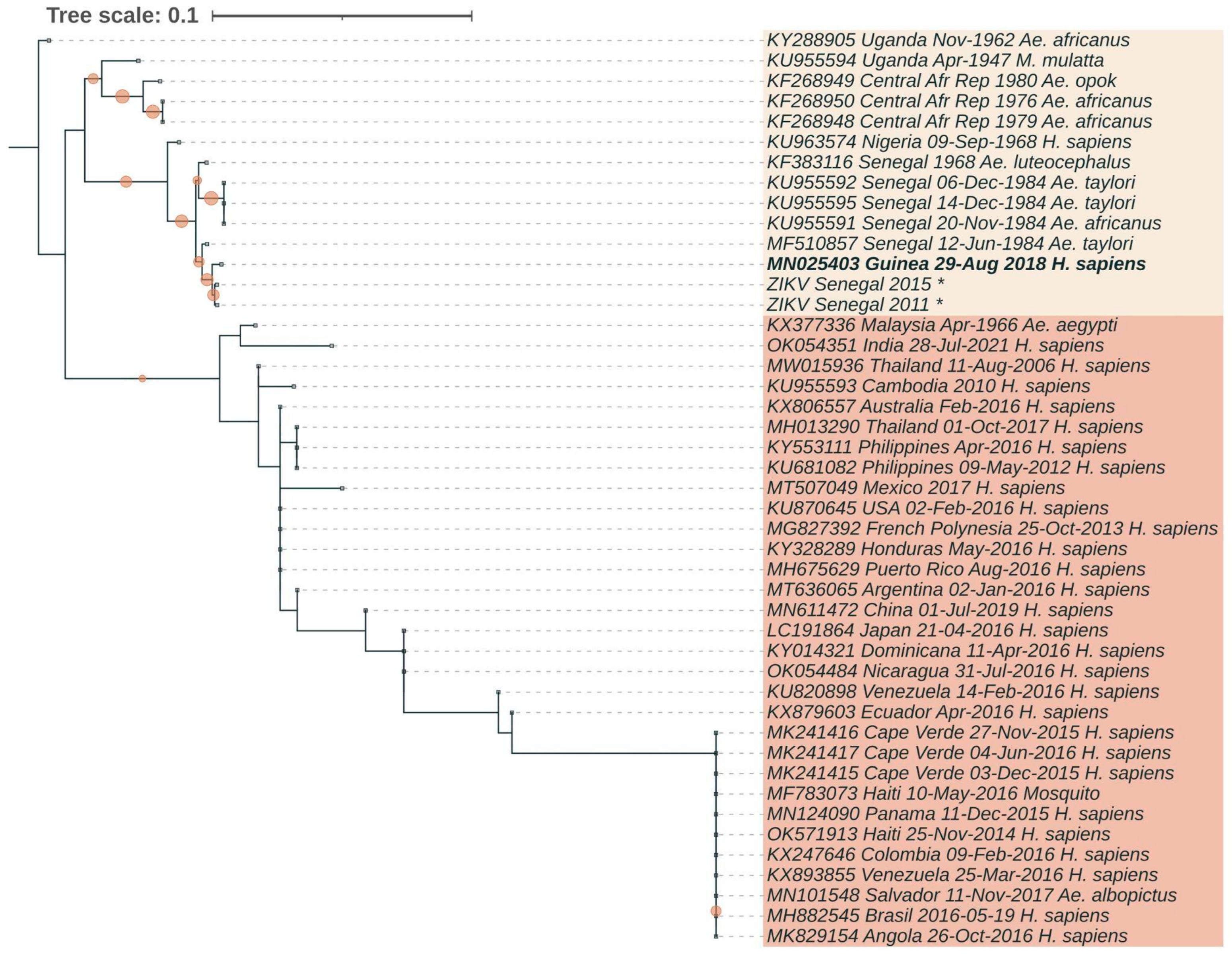
3.3. Phylogenetic Analysis
3.4. Mutation Profile
- Capsid protein C coding region, position 106 (hereinafter, the position in the amino acid sequence of the ZIKV polyprotein relative to the beginning of the open reading frame is indicated, GenBank accession number of the reference: ASR91936), amino acid substitution: A or T;
- Membrane glycoprotein precursor prM coding region, position 123, amino acid substitution: V or A;
- Membrane glycoprotein precursor prM coding region, position 139, amino acid substitution: S or N;
- Membrane glycoprotein precursor prM coding region, position 143, amino acid substitution: E or K;
- Envelope protein E coding region, position 763, amino acid substitution: V or M;
- Nonstructural protein NS1 coding region, position 982, amino acid substitution: A or V;
- RNA-dependent RNA polymerase NS5 coding region, position 3392, amino acid substitution: M or V.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Name | Nucleotide sequence 5”- 3′ | Length of the product, bp |
| ZK-1F | AGTTGTTGATCTGTGTGAGTCAGACTG | 799 bp |
| ZK-799R | GACCGCGTTTGCAACTTCC | |
| ZK-665F | GTCGATTGCTGGTGCAACACGAC | 564 bp |
| ZK-1229R | AGGTAGGCTTCACCTTGTGTT | |
| ZK-1147F | GAGGCATCGATATCGGACATGGCTT | 717 bp |
| ZK-1864R | GGCGACATTTCAAGTGGCCAGAGGA | |
| ZK-1774F | GGAGCTCTGGAGGCTGAGATGGATG | 498 bp |
| ZK-2272R | CAGGCTGTGTCTCCCAAGACT | |
| ZK-2191F | GCATTTGAAGCCACTGTGAGAG | 644 bp |
| ZK-2835R | GGGCAGCTCGTTCACAGGCA | |
| ZK-2733F | GACGGTCGTTGTGGGATCTGT | 854 bp |
| ZK-3587R | GGGAGAAGTGGTCCATGTGATCAGTT | |
| ZK-3456F | TGGCTGTTGGTATGGAATGGAGAT | 688 bp |
| ZK-4144R | TGGTAAGTTCTTCTTCACACTGCCTT | |
| ZK-4021F | GGCACACTGCTTGTGGCGTGGAGA | 686 bp |
| ZK-4707R | CGAGTCATTACTCTGTACACTCCA | |
| ZK-4598F | GGAGTGGTGCTCTATGGGATGT | 663 bp |
| ZK-5261R | TGGCTTCACGGACTATTTCAGGA | |
| ZK-5145F | GATGCTGAAGAAGAAGCAGCTAACTGT | 620 bp |
| ZK-5765R | TCCAGCCTTTGTCAGACAAGCTGCGA | |
| ZK-5632F | TGGAGCTCAGGCTTTGATTGGGTGA | 1051 bp |
| ZK-6683R | TCCCAGCGAGACTGTTCCCAGCA | |
| ZK-6143F | AGCCTCGCTCTATCGACCTGA | 540 bp |
| ZK-6683R | TCCCAGCGAGACTGTTCCCA | |
| ZK-6537F | ATTGACAACCTCGCCGTGCTC | 869 bp |
| ZK-7406R | GGGGTCTATTGTCATTGTGTCAATG | |
| ZK-7263F | GCGCACTACATGTACTTGATCCC | 667 bp |
| ZK-7930R | CAGCCCCCTCTGCCACATC | |
| ZK-7854F | CCATGCTGTGTCCCGAGGAA | 751 bp |
| ZK-8605R | TCCTATATGGGTGGTTCTCGTCAA | |
| ZK-8445F | GCCAGTGAAATATGAGGAGGATGT | 599 bp |
| ZK-9044R | TGGCACTCTCCTCTCAGGTGGT | |
| ZK-8956F | TGGAAGACTGCAGTGGAAGCTGTGA | 724 bp |
| ZK-9680R | GCCATTCGTTTGAGCCTATCCCA | |
| ZK-9572F | TGGAGGCTGAGGAAGTTCTAGAGAT | 601 bp |
| ZK-10173R | GTGGTCGTTCTCCTCAATCCACACT | |
| ZK-10087F | TGGTCAATCCATGGAAAGGGAGAA | 644 bp |
| ZK-10731R | CTGGTCTTTCCCAGCGTCAATATGCT |
References
- McCrae, A.W.R.; Kirya, B.G. Yellow Fever and Zika Virus Epizootics and Enzootics in Uganda. - PubMed - NCBI. Trans R Soc Trop Med Hyg 1982, 76, 552–562.
- Ayres, C.F.J. Identification of Zika Virus Vectors and Implications for Control. Lancet Infect Dis 2016, 16, 278–279. [CrossRef]
- Epelboin, Y.; Talaga, S.; Epelboin, L.; Dusfour, I. Zika Virus: An Updated Review of Competent or Naturally Infected Mosquitoes. PLoS Negl Trop Dis 2017, 11, 1–22. [CrossRef]
- Ioos, S.; Mallet, H.P.; Leparc Goffart, I.; Gauthier, V.; Cardoso, T.; Herida, M. Current Zika Virus Epidemiology and Recent Epidemics. Med Mal Infect 2014.
- Vasilakis, N.; Weaver, S.C. Flavivirus Transmission Focusing on Zika. Curr Opin Virol 2017.
- Weaver, S.C.; Costa, F.; Garcia-Blanco, M.A.; Ko, A.I.; Ribeiro, G.S.; Saade, G.; Shi, P.Y.; Vasilakis, N. Zika Virus: History, Emergence, Biology, and Prospects for Control. Antiviral Res 2016.
- Althaus, C.L.; Low, N. How Relevant Is Sexual Transmission of Zika Virus? PLoS Med 2016, 13. [CrossRef]
- Vasquez, A.M.; Sapiano, M.R.P.; Basavaraju, S. V.; Kuehnert, M.J.; Rivera-Garcia, B. Survey of Blood Collection Centers and Implementation of Guidance for Prevention of Transfusion-Transmitted Zika Virus Infection — Puerto Rico, 2016. MMWR Morb Mortal Wkly Rep 2016, 65. [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects — Reviewing the Evidence for Causality. New England Journal of Medicine 2016, 374. [CrossRef]
- Besnard, M.; Lastère, S.; Teissier, A.; Cao-Lormeau, V.M.; Musso, D. Evidence of Perinatal Transmission of Zika Virus, French Polynesia, December 2013 and February 2014. Eurosurveillance 2014. [CrossRef]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Rus, K.R.; Vipotnik, T.V.; Vodušek, V.F.; et al. Zika Virus Associated with Microcephaly. New England Journal of Medicine 2016. [CrossRef]
- Oliveira Melo, A.S.; Malinger, G.; Ximenes, R.; Szejnfeld, P.O.; Alves Sampaio, S.; Bispo De Filippis, A.M. Zika Virus Intrauterine Infection Causes Fetal Brain Abnormality and Microcephaly: Tip of the Iceberg? Ultrasound in Obstetrics and Gynecology 2016.
- Sheridan, M.A.; Balaraman, V.; Schust, D.J.; Ezashi, T.; Michael Roberts, R.; Franz, A.W.E. African and Asian Strains of Zika Virus Differ in Their Ability to Infect and Lyse Primitive Human Placental Trophoblast. PLoS One 2018, 13. [CrossRef]
- Dick, G.W.A. Zika Virus (I). Isolations and Serological Specificity. Trans R Soc Trop Med Hyg 1952, 46. [CrossRef]
- Grard, G.; Caron, M.; Mombo, I.M.; Nkoghe, D.; Mboui Ondo, S.; Jiolle, D.; Fontenille, D.; Paupy, C.; Leroy, E.M. Zika Virus in Gabon (Central Africa) - 2007: A New Threat from Aedes Albopictus? PLoS Negl Trop Dis 2014, 8, 1–6. [CrossRef]
- Moi, M.L.; Nguyen, T.T.T.; Nguyen, C.T.; Vu, T.B.H.; Tun, M.M.N.; Pham, T.D.; Pham, N.T.; Tran, T.; Morita, K.; Le, T.Q.M.; et al. Zika Virus Infection and Microcephaly in Vietnam. Lancet Infect Dis 2017.
- Moore, D.L.; Causey, O.R.; Carey, D.E.; Reddy, S.; Cooke, A.R.; Akinkugbe, F.M.; David-West, T.S.; Kemp, G.E. Arthropod-Borne Viral Infections of Man in Nigeria, 1964-1970. Ann Trop Med Parasitol 1975, 69. [CrossRef]
- Olson, J.G.; Ksiazek, T.G.; Suhandiman, G.; Triwibowo, V. Zika Virus, a Cause of Fever in Central Java, Indonesia. Trans R Soc Trop Med Hyg 1981. [CrossRef]
- Pond, W.L. Arthropod-Borne Virus Antibodies in Sera from Residents of South-East Asia. Trans R Soc Trop Med Hyg 1963. [CrossRef]
- Simpson, D.I.H. Zika Virus Infection in Man. Trans R Soc Trop Med Hyg 1964, 58. [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and Serologic Properties of Zika Virus Associated with an Epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 2008. [CrossRef]
- Cao-Lormeau, V.M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.L.; Mallet, H.P.; Sall, A.A.; Musso, D. Zika Virus, French Polynesia, South Pacific, 2013. Emerg Infect Dis 2014.
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg Infect Dis 2015, 21, 1885–1886. [CrossRef]
- Zammarchi, L.; Tappe, D.; Fortuna, C.; Remoli, M.E.; Günther, S.; Venturi, G.; Bartoloni, A.; Schmidt-Chanasit, J. Zika Virus Infection in a Traveller Returning to Europe from Brazil, March 2015. Eurosurveillance 2015, 20, 21153. [CrossRef]
- Bogoch, I.I.; Brady, O.J.; Kraemer, M.U.G.; German, M.; Creatore, M.I.; Brent, S.; Watts, A.G.; Hay, S.I.; Kulkarni, M.A.; Brownstein, J.S.; et al. Potential for Zika Virus Introduction and Transmission in Resource-Limited Countries in Africa and the Asia-Pacific Region: A Modelling Study. Lancet Infect Dis 2016, 16, 1237–1245. [CrossRef]
- World Health Organization Zika Epidemiology Update. July 2019. Who 2019.
- Kindhauser, M.K.; Allen, T.; Frank, V.; Santhana, R.S.; Dye, C. Zika: The Origin and Spread of a Mosquito-Borne Virus. Bull World Health Organ 2016. [CrossRef]
- Diallo, D.; Sall, A.A.; Diagne, C.T.; Faye, O.; Faye, O.; Ba, Y.; Hanley, K.A.; Buenemann, M.; Weaver, S.C.; Diallo, M. Zika Virus Emergence in Mosquitoes in Southeastern Senegal, 2011. PLoS One 2014, 9, 4–11. [CrossRef]
- Fagbami, A.H. Zika Virus Infections in Nigeria: Virological and Seroepidemiological Investigations in Oyo State. Journal of Hygiene 1979, 83, 213–219. [CrossRef]
- Robin, Y.; Mouchet, J. Enquête Sérologique et Entomologique Sur La Fièvre Jaune En Sierra Leone. Serological and Entomological Study on Yellow Fever in Sierra Leone. Bull Soc Pathol Exot Filiales 1975, 68.
- Jan, C.; Languillat, G.; Renaudet, J.; Robin, Y. [A serological survey of arboviruses in Gabon]. Bull Soc Pathol Exot Filiales 1978, 71, 140–146.
- Akoua-Koffi, C.; Diarrassouba, S.; Bénié, V.B.; Ngbichi, J.M.; Bozoua, T.; Bosson, A.; Akran, V.; Carnevale, P.; Ehouman, A. [Investigation surrounding a fatal case of yellow fever in Côte d’Ivoire in 1999]. Bull Soc Pathol Exot 2001, 94, 227–230.
- Althouse, B.M.; Hanley, K.A.; Diallo, M.; Sall, A.A.; Ba, Y.; Faye, O.; Diallo, D.; Watts, D.M.; Weaver, S.C.; Cummings, D.A.T. Impact of Climate and Mosquito Vector Abundance on Sylvatic Arbovirus Circulation Dynamics in Senegal. American Journal of Tropical Medicine and Hygiene 2015, 92, 88–97. [CrossRef]
- Diouf, B.; Gaye, A.; Diagne, C.T.; Diallo, M.; Diallo, D. Zika Virus in Southeastern Senegal: Survival of the Vectors and the Virus during the Dry Season. BMC Infect Dis 2020, 20, 1–9. [CrossRef]
- Monlun, E.; Zeller, H.; Le Guenno, B.; Traoré-Lamizana, M.; Hervy, J.P.; Adam, F.; Ferrara, L.; Fontenille, D.; Sylla, R.; Mondo, M. Surveillance of the Circulation of Arbovirus of Medical Interest in the Region of Eastern Senegal. Bull Soc Pathol Exot 1993, 86.
- Butenko, A.M. Arbovirus Circulation in the Republic of Guinea [Izuchenie Tsirkuliatsii Arbovirusov v Gvineiskoi Respublike.]. Med Parazitol (Mosk) 1996.
- Carlson, R. V.; Boyd, K.M.; Webb, D.J. The Revision of the Declaration of Helsinki: Past, Present and Future. Br J Clin Pharmacol 2004, 57.
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol 2018, 35. [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol 2018, 35. [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol 2015, 32. [CrossRef]
- Aubry, F.; Jacobs, S.; Darmuzey, M.; Lequime, S.; Delang, L.; Fontaine, A.; Jupatanakul, N.; Miot, E.F.; Dabo, S.; Manet, C.; et al. Recent African Strains of Zika Virus Display Higher Transmissibility and Fetal Pathogenicity than Asian Strains. Nat Commun 2021, 12, 1–14. [CrossRef]
- Shan, C.; Xia, H.; Haller, S.L.; Azar, S.R.; Liu, Y.; Liu, J.; Muruato, A.E.; Chen, R.; Rossi, S.L.; Wakamiya, M.; et al. A Zika Virus Envelope Mutation Preceding the 2015 Epidemic Enhances Virulence and Fitness for Transmission. Proc Natl Acad Sci U S A 2020, 117. [CrossRef]
- Liu, Y.; Liu, J.; Du, S.; Shan, C.; Nie, K.; Zhang, R.; Li, X.F.; Zhang, R.; Wang, T.; Qin, C.F.; et al. Evolutionary Enhancement of Zika Virus Infectivity in Aedes Aegypti Mosquitoes. Nature 2017, 545. [CrossRef]
- Lin, C.-S.; Li, W.-J.; Liao, C.-Y.; Kan, J.-Y.; Kung, S.-H.; Huang, S.-H.; Lai, H.-C.; Lin, C.-W. A Reverse Mutation E143K within the PrM Protein of Zika Virus Asian Lineage Natal RGN Strain Increases Infectivity and Cytopathicity. Viruses 2022, 14, 1572. [CrossRef]
- Musso, D.; Gubler, D.J. Zika Virus. Clin Microbiol Rev 2016, 29, 487–524. [CrossRef]
- Akoua-Koffi, C.; Diarrassouba, S.; Bénié, V.B.; Ngbichi, J.M.; Bozoua, T.; Bosson, A.; Akran, V.; Carnevale, P.; Ehouman, A. [Investigation Surrounding a Fatal Case of Yellow Fever in Côte d’Ivoire in 1999]. Bull Soc Pathol Exot 2001, 94.
- Diarra, I.; Nurtop, E.; Sangaré, A.K.; Sagara, I.; Pastorino, B.; Sacko, S.; Zeguimé, A.; Coulibaly, D.; Fofana, B.; Gallian, P.; et al. Zika Virus Circulation in Mali. Emerg Infect Dis 2020, 26. [CrossRef]
- Tinto, B.; Kaboré, D.P.A.; Kania, D.; Kagoné, T.S.; Kiba-Koumaré, A.; Pinceloup, L.; Thaurignac, G.; Van de Perre, P.; Dabire, R.K.; Baldet, T.; et al. Serological Evidence of Zika Virus Circulation in Burkina Faso. Pathogens 2022, 11, 1–10. [CrossRef]
- Herrera, B.B.; Chang, C.A.; Hamel, D.J.; MBoup, S.; Ndiaye, D.; Imade, G.; Okpokwu, J.; Agbaji, O.; Bei, A.K.; Kanki, P.J. Continued Transmission of Zika Virus in Humans in West Africa, 1992-2016. Journal of Infectious Diseases 2017, 215, 1546–1550. [CrossRef]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, Cross-Reactivity, and Function of Antibodies Elicited by Zika Virus Infection. Science (1979) 2016. [CrossRef]
- Huestis, D.L.; Dao, A.; Diallo, M.; Sanogo, Z.L.; Samake, D.; Yaro, A.S.; Ousman, Y.; Linton, Y.M.; Krishna, A.; Veru, L.; et al. Windborne Long-Distance Migration of Malaria Mosquitoes in the Sahel. Nature 2019, 574. [CrossRef]
- Faye, O.; de Lourdes Monteiro, M.; Vrancken, B.; Prot, M.; Lequime, S.; Diarra, M.; Ndiaye, O.; Valdez, T.; Tavarez, S.; Ramos, J.; et al. Genomic Epidemiology of 2015-2016 Zika Virus Outbreak in Cape Verde. Emerg Infect Dis 2020, 26. [CrossRef]
- Hill, S.C.; Vasconcelos, J.; Neto, Z.; Jandondo, D.; Zé-Zé, L.; Aguiar, R.S.; Xavier, J.; Thézé, J.; Mirandela, M.; Micolo Cândido, A.L.; et al. Emergence of the Asian Lineage of Zika Virus in Angola: An Outbreak Investigation. Lancet Infect Dis 2019, 19, 1138–1147. [CrossRef]
- Kasprzykowski, J.I.; Fukutani, K.F.; Fabio, H.; Fukutani, E.R.; Costa, L.C.; Andrade, B.B.; Queiroz, A.T.L. A Recursive Sub-Typing Screening Surveillance System Detects the Appearance of the ZIKV African Lineage in Brazil: Is There a Risk of a New Epidemic? International Journal of Infectious Diseases 2020, 96. [CrossRef]
- Smith, D.R.; Sprague, T.R.; Hollidge, B.S.; Valdez, S.M.; Padilla, S.L.; Bellanca, S.A.; Golden, J.W.; Coyne, S.R.; Kulesh, D.A.; Miller, L.J.; et al. African and Asian Zika Virus Isolates Display Phenotypic Differences Both in Vitro and in Vivo. American Journal of Tropical Medicine and Hygiene 2018, 98. [CrossRef]
- Duggal, N.K.; Ritter, J.M.; McDonald, E.M.; Romo, H.; Guirakhoo, F.; Davis, B.S.; Chang, G.J.J.; Brault, A.C. Differential Neurovirulence of African and Asian Genotype Zika Virus Isolates in Outbred Immunocompetent Mice. American Journal of Tropical Medicine and Hygiene 2017, 97. [CrossRef]
- Liu, J.; Liu, Y.; Shan, C.; Nunes, B.T.D.; Yun, R.; Haller, S.L.; Rafael, G.H.; Azar, S.R.; Andersen, C.R.; Plante, K.; et al. Role of Mutational Reversions Andfitnessrestoration in Zika Virus Spread to the Americas. Nat Commun 2021, 12. [CrossRef]
- Xia, H.; Luo, H.; Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Medeiros, D.B.A.; Zou, J.; Xie, X.; Giraldo, M.I.; Vasconcelos, P.F.C.; et al. An Evolutionary NS1 Mutation Enhances Zika Virus Evasion of Host Interferon Induction. Nat Commun 2018, 9. [CrossRef]
- Yuan, L.; Huang, X.Y.; Liu, Z.Y.; Zhang, F.; Zhu, X.L.; Yu, J.Y.; Ji, X.; Xu, Y.P.; Li, G.; Li, C.; et al. A Single Mutation in the PrM Protein of Zika Virus Contributes to Fetal Microcephaly. Science (1979) 2017. [CrossRef]
- Zhao, F.; Xu, Y.; Lavillette, D.; Zhong, J.; Zou, G.; Long, G. Negligible Contribution of M2634V Substitution to ZIKV Pathogenesis in AG6 Mice Revealed by a Bacterial Promoter Activity Reduced Infectious Clone. Sci Rep 2018, 8. [CrossRef]
- Jaeger, A.S.; Murrieta, R.A.; Goren, L.R.; Crooks, C.M.; Moriarty, R. V.; Weiler, A.M.; Rybarczyk, S.; Semler, M.R.; Huffman, C.; Mejia, A.; et al. Zika Viruses of African and Asian Lineages Cause Fetal Harm in a Mouse Model of Vertical Transmission. PLoS Negl Trop Dis 2019, 13. [CrossRef]
- Rossi, S.L.; Ebel, G.D.; Shan, C.; Shi, P.Y.; Vasilakis, N. Did Zika Virus Mutate to Cause Severe Outbreaks? Trends Microbiol 2018, 26.
- Crooks, C.M.; Jaeger, A.S.; Weiler, A.M.; Rybarczyk, S.L.; Bliss, M.I.; Brown, E.A.; Simmons, H.A.; Hayes, J.M.; Mejia, A.; Yamamoto, K.; et al. African-Lineage Zika Virus Causes Placental Pathology in Pregnant Rhesus Macaques. American Journal of Tropical Medicine and Hygiene 2019, 101.
| Collection date | Season | Number of samples | |||||
|---|---|---|---|---|---|---|---|
| Studied through RT-PCR | RT-PCR positive | Studied through ELISA | IgM positive |
IgM positive, % (CI95%) |
|||
| 2018 | May | Rainy season | 20 | 0 | 18 | 1 | 5.6 (0.1–27.3) |
| 2018 | August | Rainy season | 69 | 1 | 35 | 6 | 17.1 (6.6–33.6) |
| 2019 | February–March | Dry season | 30 | 0 | 30 | 5 | 16.7 (5.6–34.7) |
| 2021 | July | Rainy season | 33 | 0 | 33 | 5 | 15.2 (5.1–31.9) |
| Total | 152 | 1 | 116 | 17 | 14.7 (8.8–22.4) |
||
| GBank №/ strain |
Country | Date | Source | C- T106A |
prM-V123A | prM-S139N | prM-E143K | E- V763M |
NS1-A982V | NS5-M3392V |
|---|---|---|---|---|---|---|---|---|---|---|
| KU955594 | Uganda | Apr-1947 | M. mulatta | A | A | S | K | V | V | V |
| KU963574 | Nigeria | 09-Sep-1968 | H. sapiens | A | A | S | K | V | V | V |
| KF268950 | Central Afr. Republic | 1976 | Ae. africanus | A | A | S | K | V | V | V |
| MF510857 | Senegal | 12-Jun-1984 | Ae. taylori | A | A | S | K | V | V | V |
| MK241415 | Cape Verde | 03-Dec-2015 | H. sapiens | A | A | N | E | M | V | V |
| Senegal 2011 | Senegal | 2011 | Mosquito pools | A | A | S | K | V | V | V |
| Senegal 2015 | Senegal | 2015 | Mosquito pools | A | A | S | K | V | V | V |
| MN025403 | Guinea | 29-Aug-2018 | H. sapiens | A | A | S | K | V | V | V |
| KX377336 | Malaysia | Apr-1966 | Ae. aegypti | A | V | S | K | V | A | V |
| MW015936 | Thailand | 11-Aug-2006 | H. sapiens | T | V | S | E | V | A | M |
| KU955593 | Cambodia | 2010 | H. sapiens | T | V | S | E | V | A | M |
| MG827392 | Fr. Polynesia | 25-Oct-2013 | H. sapiens | A | A | N | E | M | V | V |
| MH013290 | Thailand | 01-Oct-2017 | H. sapiens | A | A | S | E | M | V | V |
| OK054351 | India | 28-Jul-2021 | H. sapiens | A | V | S | E | V | I | V |
| AMK49492 | Indonesia | 30-Dec-2014 | H. sapiens | T | A | S | E | M | V | V |
| OK571913 | Haiti | 25-Nov-2014 | H. sapiens | A | A | N | E | M | V | V |
| MN124090 | Panama | 11-Dec-2015 | H. sapiens | A | A | N | E | M | V | V |
| MH882545 | Brasil | 19-May-2016 | H. sapiens | A | A | N | E | M | V | V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





