Submitted:
24 July 2023
Posted:
24 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
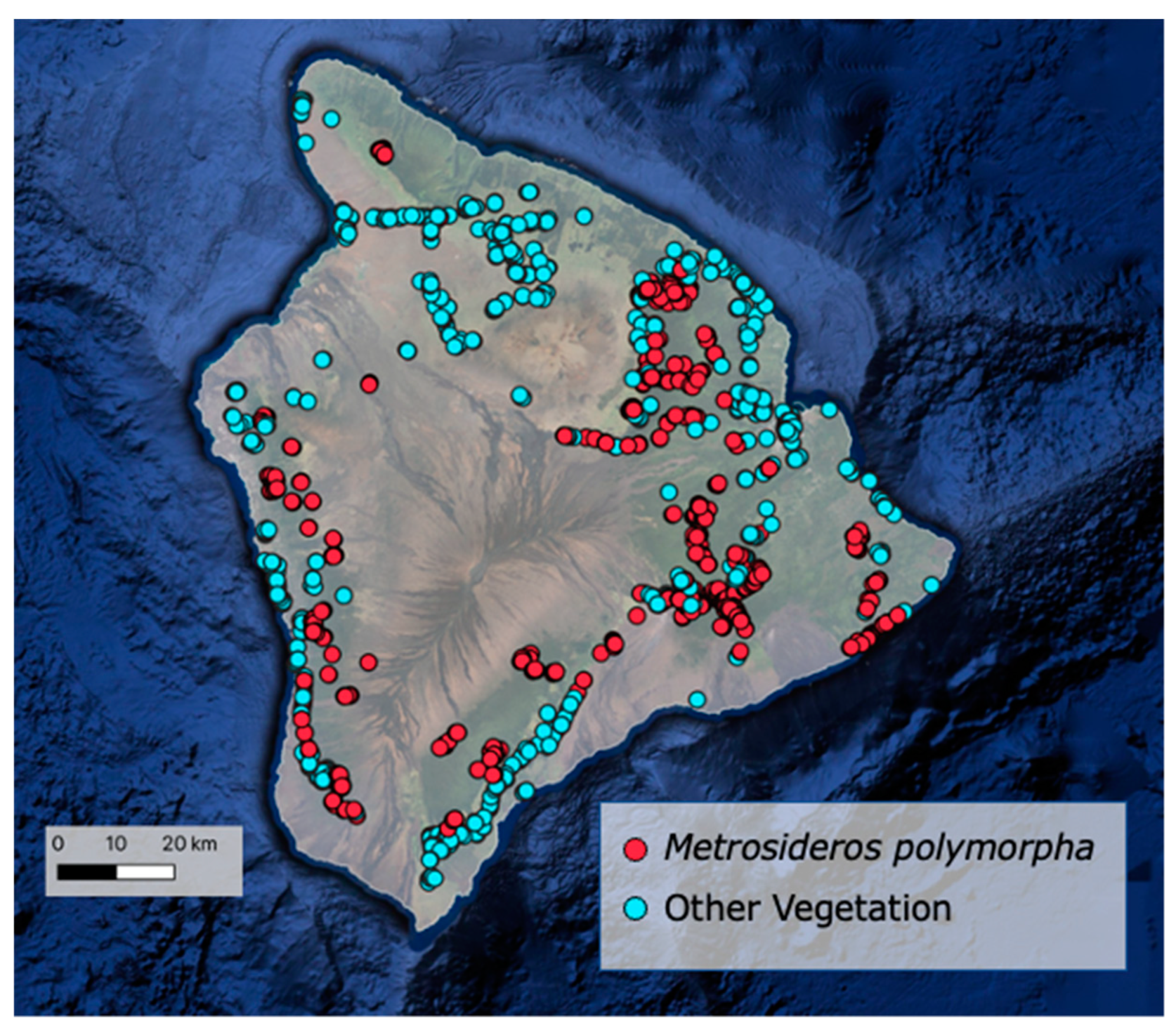
2.1. Training Data Collection
2.2. Species Classification
3. Results
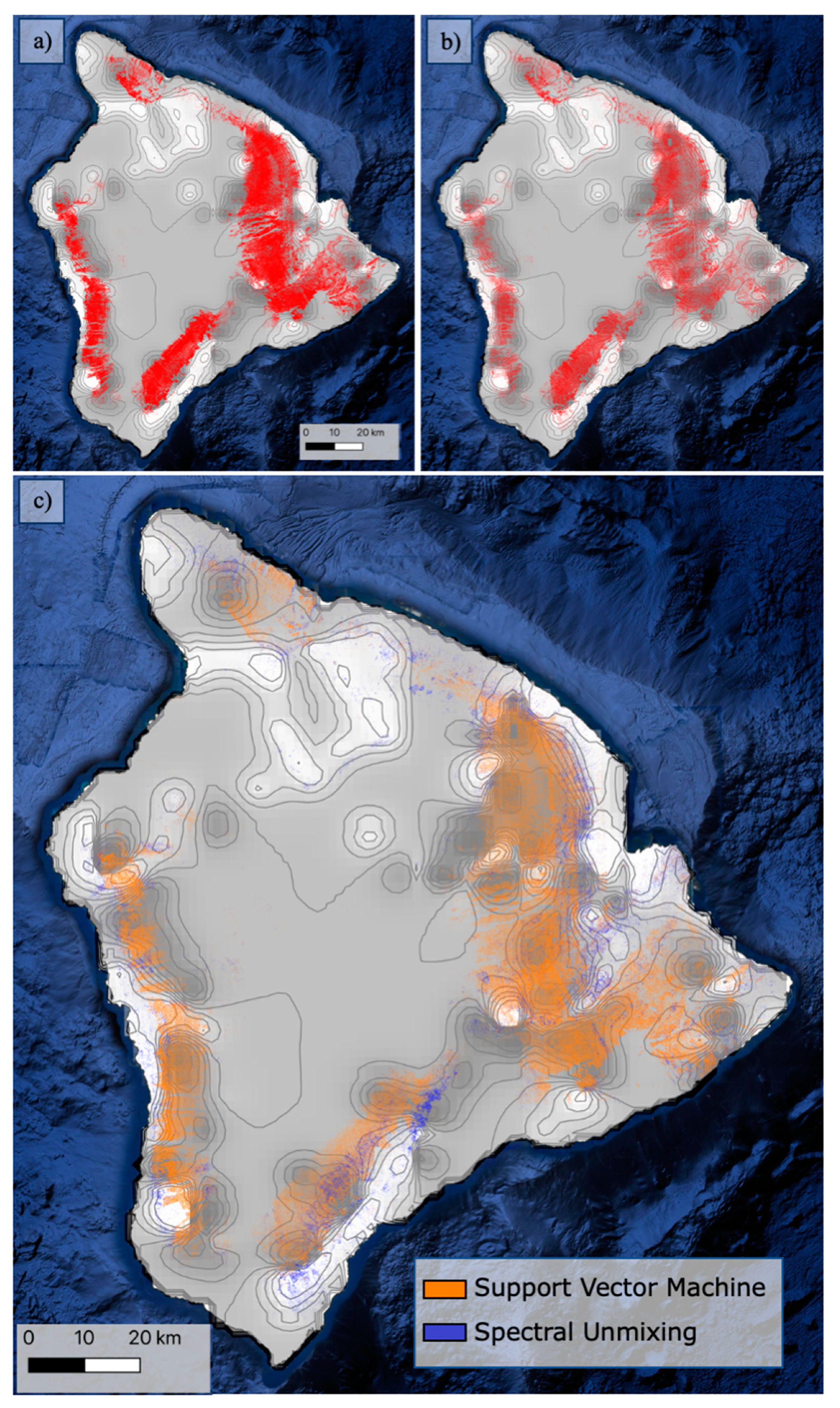
3.1. Model Performance
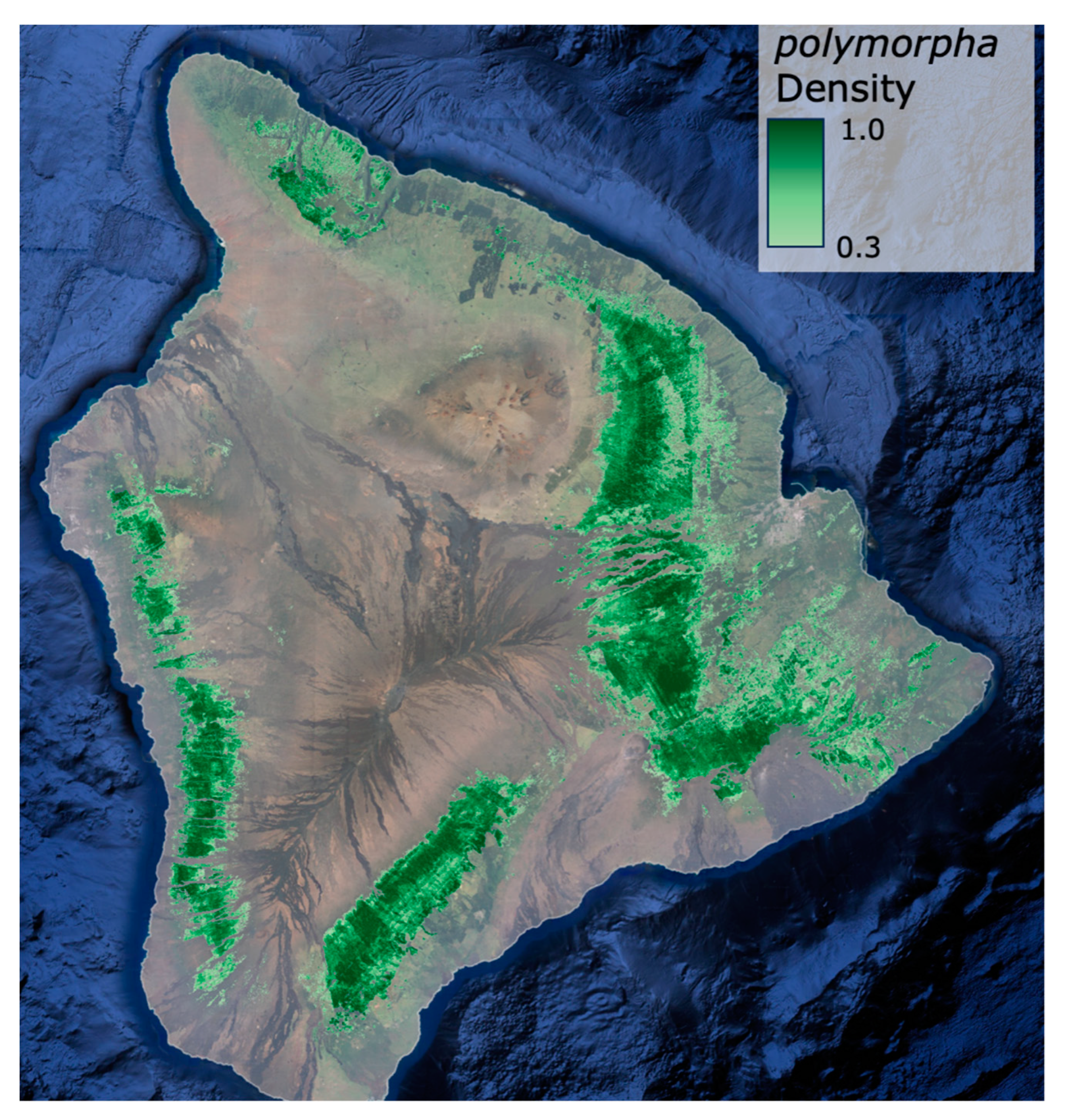
3.2. Metrosideros polymorpha Distribution
4. Discussion
4.1. High Resolution Model Comparison
4.2. Considerations for Future Large-Scale Modeling Efforts
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fassnacht FE, Latifi H, Stereńczak K, Modzelewska A, Lefsky M, Waser LT, et al. Review of studies on tree species classification from remotely sensed data. Remote Sensing of Environment. 2016 Dec 1;186:64–87. [CrossRef]
- Baldeck CA, Asner GP, Martin RE, Anderson CB, Knapp DE, Kellner JR, et al. Operational Tree Species Mapping in a Diverse Tropical Forest with Airborne Imaging Spectroscopy. Kumar L, editor. PLoS ONE. 2015 Jul 8;10(7):e0118403. [CrossRef]
- Roth KL, Roberts DA, Dennison PE, Alonzo M, Peterson SH, Beland M. Differentiating plant species within and across diverse ecosystems with imaging spectroscopy. Remote Sensing of Environment. 2015 Sep 15;167:135–51. [CrossRef]
- Laybros A, Schläpfer D, Féret JB, Descroix L, Bedeau C, Lefevre MJ, et al. Across Date Species Detection Using Airborne Imaging Spectroscopy. Remote Sensing. 2019 Jan;11(7):789. [CrossRef]
- Lopatin J, Dolos K, Kattenborn T, Fassnacht FE. How canopy shadow affects invasive plant species classification in high spatial resolution remote sensing. Remote Sensing in Ecology and Conservation. 2019;5(4):302–17. [CrossRef]
- Roth KL, Dennison PE, Roberts DA. Comparing endmember selection techniques for accurate mapping of plant species and land cover using imaging spectrometer data. Remote Sensing of Environment. 2012 Dec 1;127:139–52. [CrossRef]
- Comber A, Fisher P, Brunsdon C, Khmag A. Spatial analysis of remote sensing image classification accuracy. Remote Sensing of Environment. 2012 Dec 1;127:237–46. [CrossRef]
- Yu Q, Gong P, Tian Y, Pu R, Yang J. Factors Affecting Spatial Variation of Classification Uncertainty in an Image Object-based Vegetation Mapping. Photogrammetric Engineering and Remote Sensing. 2008 Mar 29;74:1007–18. [CrossRef]
- Mortenson LA, Hughes RF, Friday JB, Keith LM, Barbosa JM, Friday NJ, et al. Assessing spatial distribution, stand impacts and rate of Ceratocystis fimbriata induced ‘ōhi‘a (Metrosideros polymorpha) mortality in a tropical wet forest, Hawai‘i Island, USA. Forest Ecology and Management 377: 83-92. 2016;377:83–92. [CrossRef]
- Loope L, Hughes F, Keith L, Harrington R, Hauff R, Friday JB, et al. Guidance document for Rapid Ohia Death: background for the 2017-2019 ROD Strategic Response Plan. University of Hawaii: College of Tropical Agriculture and Human Resources. 2016.
- Pratt T, Atkinson C, Banko PC, Jacobi J, Woodworth B. Conservation Biology of Hawaiian Forest Birds [Internet]. Yale University Press; 2009 [cited 2022 Aug 4]. Available from: https://yalebooks.yale.edu/9780300141085/conservation-biology-of-hawaiian-forest-birds.
- Chow, ET. The Sovereign Nation of Hawai’i: Resistance in the Legacy of “Aloha 'Oe.” SUURJ: Seattle University Undergraduate Research Journal. 2018;2(15):15.
- Westervelt, WD. Hawaiian Legends of Volcanoes (mythology). Ellis Press; 1916. 284 p.
- Kagawa A, Sack L, Duarte K, James S. Hawaiian native forest conserves water relative to timber plantation: species and stand traits influence water use. Ecol Appl. 2009 Sep;19(6):1429–43. [CrossRef]
- Vaughn NR, Asner GP, Brodrick PG, Martin RE, Heckler JW, Knapp DE, et al. An Approach for High-Resolution Mapping of Hawaiian Metrosideros Forest Mortality Using Laser-Guided Imaging Spectroscopy. Remote Sensing. 2018 Apr;10(4):502. [CrossRef]
- Wagner WL, Herbst DR, Sohmer SH. Manual of the Flowering Plants of Hawai’i [Internet]. Manual of the Flowering Plants of Hawai’i. University of Hawaii Press; [cited 2023 Apr 20]. Available from: https://www.degruyter.com/document/isbn/9780824885779/html?lang=en.
- Somers B, Asner GP. Invasive Species Mapping in Hawaiian Rainforests Using Multi-Temporal Hyperion Spaceborne Imaging Spectroscopy. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing. 2013 Apr;6(2):351–9. [CrossRef]
- Vitousek, PM. Nutrient Cycling and Limitation: Hawai’i as a Model System [Internet]. Princeton University Press; 2004 [cited 2022 Sep 23]. Available from: http://www.jstor.org/stable/j.ctv39x77c.
- Cordell S, Goldstein G, Mueller-Dombois D, Webb D, Vitousek PM. Physiological and morphological variation in Metrosideros polymorpha, a dominant Hawaiian tree species, along an altitudinal gradient: the role of phenotypic plasticity. Oecologia. 1998 Jan 1;113(2):188–96. [CrossRef]
- Joel G, Aplet G, Vitousek PM. Leaf Morphology Along Environmental Gradients in Hawaiian Metrosideros Polymorpha. Biotropica. 1994;26(1):17–22. [CrossRef]
- Martin RE, Asner GP, Sack L. Genetic variation in leaf pigment, optical and photosynthetic function among diverse phenotypes of Metrosideros polymorpha grown in a common garden. Oecologia. 2007 Mar 1;151(3):387–400. [CrossRef]
- Stacy EA, Johansen JB, Sakishima T, Price DK, Pillon Y. Incipient radiation within the dominant Hawaiian tree Metrosideros polymorpha. Heredity. 2014 Oct;113(4):334–42. [CrossRef]
- Stacy EA, Johansen JB, Sakishima T, Price DK. Genetic analysis of an ephemeral intraspecific hybrid zone in the hypervariable tree, Metrosideros polymorpha, on Hawai‘i Island. Heredity. 2016 Sep;117(3):173–83. [CrossRef]
- Seeley MM, Stacy EA, Martin RE, Asner GP. Foliar functional and genetic variation in a keystone Hawaiian tree species estimated through spectroscopy. Oecologia [Internet]. 2023 May 12 [cited 2023 May 17]. [CrossRef]
- U.S. Geological Survey Gap Analysis Program. GAP/LANDFIRE National Terrestrial Ecosystems 2011: U.S. Geological Survey [Internet]. 2011. (20160513). [CrossRef]
- Pascual A, Giardina CP, Povak NA, Hessburg PF, Asner GP. Integrating ecosystem services modeling and efficiencies in decision-support models conceptualization for watershed management. Ecological Modelling. 2022 Apr 1;466:109879.
- Pascual A, Giardina CP, Povak NA, Hessburg PF, Heider C, Salminen E, et al. Optimizing invasive species management using mathematical programming to support stewardship of water and carbon-based ecosystem services. Journal of Environmental Management. 2022 Jan 1;301:113803. [CrossRef]
- Povak NA, Hessburg PF, Giardina CP, Reynolds KM, Heider C, Salminen E, et al. A watershed decision support tool for managing invasive species on Hawai‘i Island, USA. Forest Ecology and Management. 2017 Sep;400:300–20. [CrossRef]
- Boardman JW, Green RO. Exploring the spectral variability of the Earth as measured by AVIRIS in 1999. 2000 Dec [cited 2023 Jan 3]; Available from: https://trs.jpl.nasa.gov/handle/2014/16602.
- Green RO, Boardman JW. Exploration of the relationship between information content and signal-to-noise ratio and spatial resolution in AVIRIS spectral data. Spectrum. 2000;7(8).
- Asner GP, Martin RE. Airborne spectranomics: mapping canopy chemical and taxonomic diversity in tropical forests. Frontiers in Ecology and the Environment. 2009;7(5):269–76. [CrossRef]
- Balzotti CS, Asner GP, Adkins ED, Parsons EW. Spatial drivers of composition and connectivity across endangered tropical dry forests. Journal of Applied Ecology. 2020;57(8):1593–604. [CrossRef]
- Chakravortty S, Shah E, Chowdhury AS. Application of Spectral Unmixing Algorithm on Hyperspectral Data for Mangrove Species Classification. In: Gupta P, Zaroliagis C, editors. Applied Algorithms. Cham: Springer International Publishing; 2014. p. 223–36. (Lecture Notes in Computer Science). [CrossRef]
- Pontius J, Hanavan RP, Hallett RA, Cook BD, Corp LA. High spatial resolution spectral unmixing for mapping ash species across a complex urban environment. Remote Sensing of Environment. 2017 Sep 15;199:360–9. [CrossRef]
- Shang X, Chisholm LA. Classification of Australian Native Forest Species Using Hyperspectral Remote Sensing and Machine-Learning Classification Algorithms. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing. 2014 Jun;7(6):2481–9. [CrossRef]
- Somers B, Asner GP. Tree species mapping in tropical forests using multi-temporal imaging spectroscopy: Wavelength adaptive spectral mixture analysis. International Journal of Applied Earth Observation and Geoinformation. 2014 Sep 1;31:57–66. [CrossRef]
- Torabzadeh H, Leiterer R, Hueni A, Schaepman ME, Morsdorf F. Tree species classification in a temperate mixed forest using a combination of imaging spectroscopy and airborne laser scanning. Agricultural and Forest Meteorology. 2019 Dec 15;279:107744. [CrossRef]
- Asner GP, Knapp DE, Boardman J, Green RO, Kennedy-Bowdoin T, Eastwood M, et al. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sensing of Environment. 2012 Sep 1;124:454–65. [CrossRef]
- Féret JB, Asner GP. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecological Applications. 2014;24(6):1289–96. [CrossRef]
- Miller, CJ. Performance assessment of ACORN atmospheric correction algorithm. In: Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery VIII [Internet]. SPIE; 2002 [cited 2021 Dec 21]. p. 438–49. Available from: https://www.spiedigitallibrary.org/conference-proceedings-of-spie/4725/0000/Performance-assessment-of-ACORN-atmospheric-correction-algorithm/10.1117/12.478777.full.
- Schaepman-Strub G, Schaepman M, Martonchik J, Schaaf C. Whats in a Satellite Albedo Product? IEEE. 2006;2848–52.
- Colgan MS, Baldeck CA, Féret JB, Asner GP. Mapping Savanna Tree Species at Ecosystem Scales Using Support Vector Machine Classification and BRDF Correction on Airborne Hyperspectral and LiDAR Data. Remote Sensing. 2012 Nov;4(11):3462–80. [CrossRef]
- Feilhauer H, Asner GP, Martin RE, Schmidtlein S. Brightness-normalized Partial Least Squares Regression for hyperspectral data. Journal of Quantitative Spectroscopy and Radiative Transfer. 2010 Aug 1;111(12):1947–57. [CrossRef]
- Asner GP, Knapp DE, Kennedy-Bowdoin T, Jones MO, Martin RE, Boardman JW, et al. Carnegie Airborne Observatory: in-flight fusion of hyperspectral imaging and waveform light detection and ranging for three-dimensional studies of ecosystems. JARS. 2007;1(1):013536. [CrossRef]
- Weingarten E, Martin R, Hughes F, Vaughn N, Schafron E, Asner GP. Early Detection of a Tree Pathogen using Airborne Remote Sensing. Ecological Applications. 2021;21(2). [CrossRef]
- Crabbé AH, Somers B, Roberts DA, Halligan K, Dennison P, Dudley K. MESMA QGIS Plugin [Internet]. 2020. Available from: https://bitbucket.org/kul-reseco/mesma.
- Roberts DA, Gardner M, Church R, Ustin S, Scheer G, Green RO. Mapping Chaparral in the Santa Monica Mountains Using Multiple Endmember Spectral Mixture Models. Remote Sensing of Environment. 1998 Sep 1;65(3):267–79. [CrossRef]
- Feret JB, Asner GP. Tree Species Discrimination in Tropical Forests Using Airborne Imaging Spectroscopy. IEEE Transactions on Geoscience and Remote Sensing. 2013 Jan;51(1):73–84. [CrossRef]
- Camps-Valls G, Gomez-Chova L, Calpe-Maravilla J, Martin-Guerrero JD, Soria-Olivas E, Alonso-Chorda L, et al. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Transactions on Geoscience and Remote Sensing. 2004 Jul;42(7):1530–42. [CrossRef]
- Melgani F, Bruzzone L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Transactions on Geoscience and Remote Sensing. 2004 Aug;42(8):1778–90. [CrossRef]
- Dalponte M, Bruzzone L, Gianelle D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sensing of Environment. 2012 Aug 1;123:258–70. [CrossRef]
- Dalponte M, Ørka HO, Gobakken T, Gianelle D, Næsset E. Tree Species Classification in Boreal Forests With Hyperspectral Data. IEEE Transactions on Geoscience and Remote Sensing. 2013 May;51(5):2632–45. [CrossRef]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. MACHINE LEARNING IN PYTHON. :6.
- Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3(1):32–5. [CrossRef]
- Rasmussen, CE. Gaussian Processes in Machine Learning. In: Bousquet O, von Luxburg U, Rätsch G, editors. Advanced Lectures on Machine Learning [Internet]. Berlin, Heidelberg: Springer Berlin Heidelberg; 2004 [cited 2023 Apr 19]. p. 63–71. (Lecture Notes in Computer Science; vol. 3176). Available from: http://link.springer.com/10.1007/978-3-540-28650-9_4. [CrossRef]
- Culver T, Rydeen A, Dix M, Camello M, Gallaher M, Lapidus D, et al. SBG user needs and valuation study. RTI Innovation Advisors; 2020.
- Iwasaki A, Tanii J, Kashimura O, Ito Y. Prelaunch Status of Hyperspectral Imager Suite (Hisui). In: IGARSS 2019 - 2019 IEEE International Geoscience and Remote Sensing Symposium. 2019. p. 5887–90.
- Lopinto E, Ananasso C. The Prisma Hyperspectral Mission. :12.
- Müller R, Alonso K, Krawczyk H, Bachmann M, Cerra D, Krutz D, et al. Overview and Status of the DESIS Mission. In Amsterdam, Netherlands; 2018 [cited 2021 Jan 5]. Available from: https://elib.dlr.de/123992/.
- Martin RE, Asner GP. Leaf Chemical and Optical Properties of Metrosideros polymorpha across Environmental Gradients in Hawaii. Biotropica. 2009;41(3):292–301. [CrossRef]
- Seeley MM, Martin RE, Vaughn NR, Thompson DR, Dai J, Asner GP. Quantifying the Variation in Reflectance Spectra of Metrosideros polymorpha Canopies across Environmental Gradients. Remote Sensing. 2023 Jan;15(6):1614. [CrossRef]
- Jensen RR, Hardin PJ, Hardin AJ. Classification of urban tree species using hyperspectral imagery. Geocarto International. 2012 Aug 1;27(5):443–58. [CrossRef]
- Callaghan J, McAlpine C, Mitchell D, Thompson J, Bowen M, Rhodes J, et al. Ranking and mapping koala habitat quality for conservation planning on the basis of indirect evidence of tree-species use: a case study of Noosa Shire, south-eastern Queensland. Wildl Res. 2011 Apr 20;38(2):89–102. [CrossRef]
- Fremout T, Thomas E, Gaisberger H, Van Meerbeek K, Muenchow J, Briers S, et al. Mapping tree species vulnerability to multiple threats as a guide to restoration and conservation of tropical dry forests. Global Change Biology. 2020;26(6):3552–68. [CrossRef]
- Jonsson M, Bengtsson J, Gamfeldt L, Moen J, Snäll T. Levels of forest ecosystem services depend on specific mixtures of commercial tree species. Nature Plants. 2019 Feb;5(2):141–7. [CrossRef]
- Maciel EA, Martins FR. Rarity patterns and the conservation status of tree species in South American savannas. Flora. 2021 Dec 1;285:151942. [CrossRef]
- Roth KL, Roberts DA, Dennison PE, Peterson SH, Alonzo M. The impact of spatial resolution on the classification of plant species and functional types within imaging spectrometer data. Remote Sensing of Environment. 2015 Dec 15;171:45–57. [CrossRef]
| Predicted | |||||
|---|---|---|---|---|---|
| Spectral Unmixing | SVM | ||||
| M. polymorpha | Other Vegetation | M. polymorpha | Other Vegetation | ||
| Actual | M. polymorpha | 478 | 46 | 497 | 27 |
| Other Vegetation | 61 | 1025 | 44 | 1042 | |
| High M. polymorpha Likelihood | Low M. polymorpha Likelihood | Medium M. polymorpha Likelihood | |
|---|---|---|---|
| SVM | 47.5 | 13.9 | 38.9 |
| Spectral Unmixing | 29.4 | 30.6 | 39.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





