Submitted:
22 July 2023
Posted:
24 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
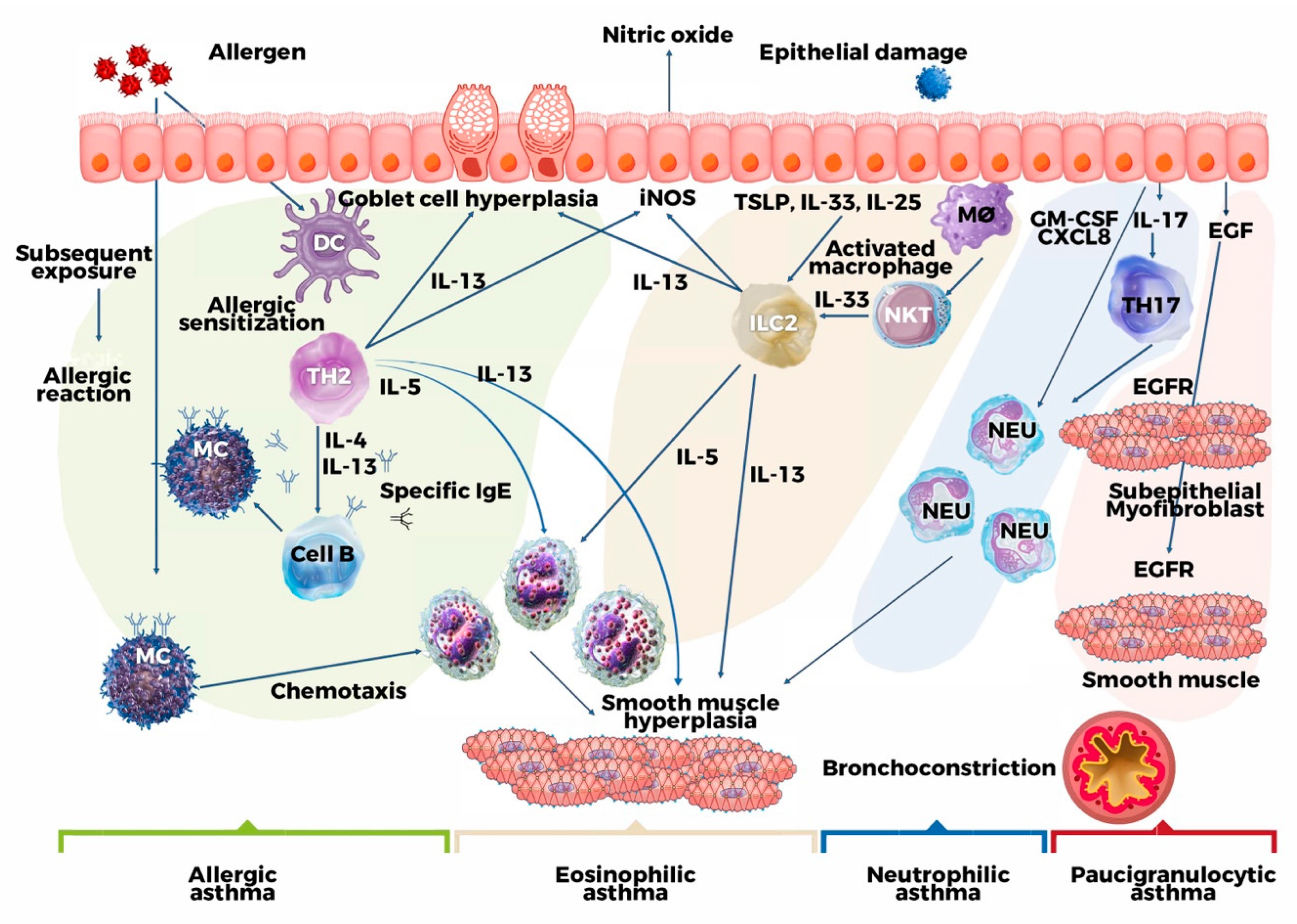
2. Pathogenetic Mechanism
3. Clinical and Molecular Phenotypes of Asthma
3.1. Mild-Early onset, allergic asthma
3.2. Late onset eosinophilic asthma
3.2.1. Non-steroidal anti-inflammatory drugs exacerbated respiratory disease (N-ERD)
3.2.2. Exercise-Induced Bronchoconstriction (EIB)
3.3. Non-eosinophilic neutrophilic asthma phenotypes
3.3.1. Obesity related asthma
4. Therapeutic targets in the era of personalized medicine
4.1. Therapeutic strategies for anti-IgE
4.2. Therapeutic strategies involving anti-IL5
4.3. Therapeutic strategies for anti-IL4-receptor alpha
4.4. Therapeutic strategies against TSLP
5. Tools that use machine learning to improve asthma care in the clinic
6. Conclusions
References
- Hopkin, J.M. The diagnosis of asthma, a clinical syndrome. Thorax 2012, 67, 660–662. [Google Scholar] [CrossRef]
- Wenzel, S.E. Emergence of Biomolecular Pathways to Define Novel Asthma Phenotypes. Type-2 Immunity and Beyond. Am. J. Respir. Cell Mol. Biol. 2016, 55, 1–4. [Google Scholar] [CrossRef]
- Svenningsen, S.; Nair, P. Asthma Endotypes and an Overview of Targeted Therapy for Asthma. Front. Med. 2017, 4, 158. [Google Scholar] [CrossRef]
- Kuruvilla, M.E.; Lee, F.E.-H.; Lee, G.B. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin. Rev. Allergy Immunol. 2018, 56, 219–233. [Google Scholar] [CrossRef]
- Hammad, H.; Lambrecht, B.N. The basic immunology of asthma. Cell 2021, 184, 1469–1485. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Hammad, H. The immunology of asthma. Nat. Immunol. 2014, 16, 45–56. [Google Scholar] [CrossRef]
- Agache, I.; Akdis, C.A. Precision medicine and phenotypes, endotypes, genotypes, regiotypes, and theratypes of allergic diseases. J. Clin. Investig. 2019, 129, 1493–1503. [Google Scholar] [CrossRef]
- Chang, H.S.; Lee, T.-H.; Jun, J.A.; Baek, A.R.; Park, J.-S.; Koo, S.-M.; Kim, Y.K.; Lee, H.S.; Park, C.-S. Neutrophilic inflammation in asthma: mechanisms and therapeutic considerations. Expert Rev. Respir. Med. 2016, 11, 29–40. [Google Scholar] [CrossRef]
- Salter, B.M.; Aw, M.; Sehmi, R. The role of type 2 innate lymphoid cells in eosinophilic asthma. J. Leukoc. Biol. 2019, 106, 889–901. [Google Scholar] [CrossRef]
- Pavord, I.D.; Korn, S.; Howarth, P.; Bleecker, E.R.; Buhl, R.; Keene, O.N.; Ortega, H.; Chanez, P. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet 2012, 380, 651–659. [Google Scholar] [CrossRef]
- Ortega, H.G.; Yancey, S.W.; Mayer, B.; Gunsoy, N.B.; Keene, O.N.; Bleecker, E.R.; E Brightling, C.; Pavord, I.D. Severe eosinophilic asthma treated with mepolizumab stratified by baseline eosinophil thresholds: a secondary analysis of the DREAM and MENSA studies. Lancet Respir. Med. 2016, 4, 549–556. [Google Scholar] [CrossRef]
- Bleecker, E.R.; FitzGerald, J.M.; Chanez, P.; Papi, A.; Weinstein, S.F.; Barker, P.; Sproule, S.; Gilmartin, G.; Aurivillius, M.; Werkström, V.; et al. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high-dosage inhaled corticosteroids and long-acting β2-agonists (SIROCCO): a randomised, multicentre, placebo-controlled phase 3 trial. Lancet 2016, 388, 2115–2127. [Google Scholar] [CrossRef]
- Castro, M.; Corren, J.; Pavord, I.D.; Maspero, J.; Wenzel, S.; Rabe, K.F.; Busse, W.W.; Ford, L.; Sher, L.; Fitzgerald, J.M.; et al. Dupilumab Efficacy and Safety in Moderate-to-Severe Uncontrolled Asthma. N. Engl. J. Med. 2018, 378, 2486–2496. [Google Scholar] [CrossRef]
- Maspero, J.F.; Katelaris, C.H.; Busse, W.W.; Castro, M.; Corren, J.; Chipps, B.E.; Peters, A.T.; Pavord, I.D.; Ford, L.B.; Sher, L.; et al. Dupilumab Efficacy in Uncontrolled, Moderate-to-Severe Asthma with Self-Reported Chronic Rhinosinusitis. J. Allergy Clin. Immunol. Pr. 2020, 8, 527–539. [Google Scholar] [CrossRef]
- E Wechsler, M.; Ford, L.B.; Maspero, J.F.; Pavord, I.D.; Papi, A.; Bourdin, A.; Watz, H.; Castro, M.; Nenasheva, N.M.; Tohda, Y.; et al. Long-term safety and efficacy of dupilumab in patients with moderate-to-severe asthma (TRAVERSE): an open-label extension study. Lancet Respir. Med. 2021, 10, 11–25. [Google Scholar] [CrossRef]
- Jonckheere, A.-C.; Bullens, D.M.; Seys, S.F. Innate lymphoid cells in asthma: pathophysiological insights from murine models to human asthma phenotypes. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 53–60. [Google Scholar] [CrossRef]
- Godar, M.; Blanchetot, C.; de Haard, H.; Lambrecht, B.N.; Brusselle, G. Personalized medicine with biologics for severe type 2 asthma: current status and future prospects. mAbs 2017, 10, 34–45. [Google Scholar] [CrossRef]
- Eberl, G.; Colonna, M.; Di Santo, J.P.; McKenzie, A.N.J. Innate lymphoid cells: A new paradigm in immunology. Science 2015, 348, aaa6566. [Google Scholar] [CrossRef]
- Cortez, V.S.; Robinette, M.L.; Colonna, M. Innate lymphoid cells: new insights into function and development. Curr. Opin. Immunol. 2015, 32, 71–77. [Google Scholar] [CrossRef]
- Roan, F.; Obata-Ninomiya, K.; Ziegler, S.F. Epithelial cell–derived cytokines: more than just signaling the alarm. J. Clin. Investig. 2019, 129, 1441–1451. [Google Scholar] [CrossRef]
- Varricchi, G.; Pecoraro, A.; Marone, G.; Criscuolo, G.; Spadaro, G.; Genovese, A.; Marone, G. Thymic Stromal Lymphopoietin Isoforms, Inflammatory Disorders, and Cancer. Front. Immunol. 2018, 9, 1595. [Google Scholar] [CrossRef]
- Liu, S.; Verma, M.; Michalec, L.; Liu, W.; Sripada, A.; Rollins, D.; Good, J.; Ito, Y.; Chu, H.; Gorska, M.M.; et al. Steroid resistance of airway type 2 innate lymphoid cells from patients with severe asthma: The role of thymic stromal lymphopoietin. J. Allergy Clin. Immunol. 2017, 141, 257–268. [Google Scholar] [CrossRef]
- Tsilingiri, K.; Fornasa, G.; Rescigno, M. Thymic Stromal Lymphopoietin: To Cut a Long Story Short. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 174–182. [Google Scholar] [CrossRef]
- Boita, M.; Garzaro, M.; Raimondo, L.; Riva, G.; Mazibrada, J.; Vizio, B.; Bellone, G.; Pecorari, G.; Bucca, C.; Rolla, G.; et al. The Expression of TSLP Receptor in Chronic Rhinosinusitis with and without Nasal Polyps. Int. J. Immunopathol. Pharmacol. 2011, 24, 761–768. [Google Scholar] [CrossRef]
- Boita, M.; Heffler, E.; Omedè, P.; Bellocchia, M.; Bussolino, C.; Solidoro, P.; Giorgis, V.; Guerrera, F.; Riva, G.; Brussino, L.; et al. Basophil Membrane Expression of Epithelial Cytokine Receptors in Patients with Severe Asthma. Int. Arch. Allergy Immunol. 2018, 175, 171–176. [Google Scholar] [CrossRef]
- Mantovani, A.; Dinarello, C.A.; Molgora, M.; Garlanda, C. Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 2019, 50, 778–795. [Google Scholar] [CrossRef]
- Peebles, R.S., Jr.; Aronica, M.A. Proinflammatory Pathways in the Pathogenesis of Asthma. Clin. Chest Med. 2019, 40, 29–50. [Google Scholar] [CrossRef]
- Yao, X.; Sun, Y.; Wang, W.; Sun, Y. Interleukin (IL)-25: Pleiotropic roles in asthma. Respirology 2015, 21, 638–647. [Google Scholar] [CrossRef]
- Kouzaki, H.; Tojima, I.; Kita, H.; Shimizu, T. Transcription of Interleukin-25 and Extracellular Release of the Protein Is Regulated by Allergen Proteases in Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Das, J.; Wenzel, S.E. Determining asthma endotypes and outcomes: Complementing existing clinical practice with modern machine learning. Cell Rep. Med. 2022, 3, 100857. [Google Scholar] [CrossRef]
- Zhu, J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine 2015, 75, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Tindemans, I.; Serafini, N.; Di Santo, J.P.; Hendriks, R.W. GATA-3 Function in Innate and Adaptive Immunity. Immunity 2014, 41, 191–206. [Google Scholar] [CrossRef]
- Varricchi, G.; Bagnasco, D.; Borriello, F.; Heffler, E.; Canonica, G.W. Interleukin-5 pathway inhibition in the treatment of eosinophilic respiratory disorders: evidence and unmet needs. Curr. Opin. Allergy Clin. Immunol. 2016, 16, 186–200. [Google Scholar] [CrossRef]
- Doran, E.; Cai, F.; Holweg, C.T.J.; Wong, K.; Brumm, J.; Arron, J.R. Interleukin-13 in Asthma and Other Eosinophilic Disorders. Front. Med. 2017, 4, 139–139. [Google Scholar] [CrossRef]
- Gandhi, N.A.; Pirozzi, G.; Graham, N.M.H. Commonality of the IL-4/IL-13 pathway in atopic diseases. Expert Rev. Clin. Immunol. 2017, 13, 425–437. [Google Scholar] [CrossRef]
- Maspero, J.; Adir, Y.; Al-Ahmad, M.; Celis-Preciado, C.A.; Colodenco, F.D.; Giavina-Bianchi, P.; Lababidi, H.; Ledanois, O.; Mahoub, B.; Perng, D.-W.; et al. Type 2 inflammation in asthma and other airway diseases. ERJ Open Res. 2022, 8. [Google Scholar] [CrossRef]
- Pelaia, C.; Pelaia, G.; Maglio, A.; Tinello, C.; Gallelli, L.; Lombardo, N.; Terracciano, R.; Vatrella, A. Pathobiology of Type 2 Inflammation in Asthma and Nasal Polyposis. J. Clin. Med. 2023, 12, 3371. [Google Scholar] [CrossRef]
- Walker, C.; Bode, E.; Boer, L.; Hansel, T.T.; Blaser, K.; Virchow, J.-C. Allergic and Nonallergic Asthmatics Have Distinct Patterns of T-Cell Activation and Cytokine Production in Peripheral Blood and Bronchoalveolar Lavage. Am. Rev. Respir. Dis. 1992, 146, 109–115. [Google Scholar] [CrossRef]
- Yao, W.; Zhang, Y.; Jabeen, R.; Nguyen, E.T.; Wilkes, D.S.; Tepper, R.S.; Kaplan, M.H.; Zhou, B. Interleukin-9 Is Required for Allergic Airway Inflammation Mediated by the Cytokine TSLP. Immunity 2013, 38, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Ricciardolo, F.L.; Sorbello, V.; Folino, A.; Gallo, F.; Massaglia, G.M.; Favatà, G.; Conticello, S.; Vallese, D.; Gani, F.; Malerba, M.; et al. Identification of IL-17F/frequent exacerbator endotype in asthma. J. Allergy Clin. Immunol. 2017, 140, 395–406. [Google Scholar] [CrossRef]
- Willis, C.R.; Siegel, L.; Leith, A.; Mohn, D.; Escobar, S.; Wannberg, S.; Misura, K.; Rickel, E.; Rottman, J.B.; Comeau, M.R.; et al. IL-17RA Signaling in Airway Inflammation and Bronchial Hyperreactivity in Allergic Asthma. Am. J. Respir. Cell Mol. Biol. 2015, 53, 810–821. [Google Scholar] [CrossRef]
- Ota, K.; Kawaguchi, M.; Matsukura, S.; Kurokawa, M.; Kokubu, F.; Fujita, J.; Morishima, Y.; Huang, S.-K.; Ishii, Y.; Satoh, H.; et al. Potential Involvement of IL-17F in Asthma. J. Immunol. Res. 2014, 2014, 602846. [Google Scholar] [CrossRef]
- Nalbant, A.; Eskier, D. Genes associated with T helper 17 cell differentiation and function. Front. Biosci. 2016, 8, 427–435. [Google Scholar] [CrossRef] [PubMed]
- I Wan, Y.; Shrine, N.R.G.; Artigas, M.S.; Wain, L.V.; Blakey, J.D.; Moffatt, M.F.; Bush, A.; Chung, K.F.; Cookson, W.O.C.M.; Strachan, D.P.; et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax 2012, 67, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Shrine, N.; A Portelli, M.; John, C.; Artigas, M.S.; Bennett, N.; Hall, R.; Lewis, J.; Henry, A.P.; Billington, C.K.; Ahmad, A.; et al. Moderate-to-severe asthma in individuals of European ancestry: a genome-wide association study. Lancet Respir. Med. 2019, 7, 20–34. [Google Scholar] [CrossRef] [PubMed]
- Hekking, P.-P.; Loza, M.J.; Pavlidis, S.; de Meulder, B.; Lefaudeux, D.; Baribaud, F.; Auffray, C.; Wagener, A.H.; Brinkman, P.; Lutter, R.; et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J. Allergy Clin. Immunol. 2018, 141, 1280–1290. [Google Scholar] [CrossRef]
- Bigler, J.; Boedigheimer, M.; Schofield, J.P.R.; Skipp, P.J.; Corfield, J.; Rowe, A.; Sousa, A.R.; Timour, M.; Twehues, L.; Hu, X.; et al. A Severe Asthma Disease Signature from Gene Expression Profiling of Peripheral Blood from U-BIOPRED Cohorts. Am. J. Respir. Crit. Care Med. 2017, 195, 1311–1320. [Google Scholar] [CrossRef]
- Modena, B.D.; Bleecker, E.R.; Busse, W.W.; Erzurum, S.C.; Gaston, B.M.; Jarjour, N.N.; Meyers, D.A.; Milosevic, J.; Tedrow, J.R.; Wu, W.; et al. Gene Expression Correlated with Severe Asthma Characteristics Reveals Heterogeneous Mechanisms of Severe Disease. Am. J. Respir. Crit. Care Med. 2017, 195, 1449–1463. [Google Scholar] [CrossRef]
- Singhania, A.; Rupani, H.; Jayasekera, N.; Lumb, S.; Hales, P.; Gozzard, N.; Davies, D.E.; Woelk, C.H.; Howarth, P.H. Altered Epithelial Gene Expression in Peripheral Airways of Severe Asthma. PLOS ONE 2017, 12, e0168680. [Google Scholar] [CrossRef]
- Gautam, Y.; Johansson, E.; Mersha, T.B. Multi-Omics Profiling Approach to Asthma: An Evolving Paradigm. J. Pers. Med. 2022, 12, 66. [Google Scholar] [CrossRef]
- Tyler, S.R.; Bunyavanich, S. Leveraging -omics for asthma endotyping. J. Allergy Clin. Immunol. 2019, 144, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, L.; Chen, D.; Feng, M.; Lu, Y.; Chen, R.; Qiu, C.; Li, J. The application of proteomics in the diagnosis and treatment of bronchial asthma. Ann. Transl. Med. 2020, 8, 132–132. [Google Scholar] [CrossRef]
- Park, C.-S.; Rhim, T. Application of proteomics in asthma research. Expert Rev. Proteom. 2011, 8, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Zhang, X.; Chen, X.; Brown, A.P.; Weirauch, M.T.; Guilbert, T.W.; Hershey, G.K.K.; Biagini, J.M.; Ji, H. Nasal DNA methylation differentiates severe from non-severe asthma in African-American children. Allergy 2020, 76, 1836–1845. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Mao, Z.-D.; Shi, Y.-J.; Qian, Y.; Liu, Z.-G.; Yin, X.-W.; Zhang, Q. Comprehensive analysis of miRNA–mRNA–lncRNA networks in severe asthma. Epigenomics 2019, 11, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.C.; Meyers, D.A.; Wenzel, S.E.; Teague, W.G.; Li, H.; Li, X.; D’Agostino, R., Jr.; Castro, M.; Curran-Everett, D.; Fitzpatrick, A.M.; et al. Identification of Asthma Phenotypes Using Cluster Analysis in the Severe Asthma Research Program. Am. J. Respir. Crit. Care Med. 2010, 181, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Haldar, P.; Pavord, I.D.; Shaw, D.E.; Berry, M.A.; Thomas, M.; Brightling, C.E.; Wardlaw, A.J.; Green, R.H. Cluster Analysis and Clinical Asthma Phenotypes. Am. J. Respir. Crit. Care Med. 2008, 178, 218–224. [Google Scholar] [CrossRef]
- Lefaudeux, D.; De Meulder, B.; Loza, M.J.; Peffer, N.; Rowe, A.; Baribaud, F.; Bansal, A.T.; Lutter, R.; Sousa, A.R.; Corfield, J.; et al. U-BIOPRED clinical adult asthma clusters linked to a subset of sputum omics. J. Allergy Clin. Immunol. 2017, 139, 1797–1807. [Google Scholar] [CrossRef]
- Yan, X.; Chu, J.-H.; Gomez, J.; Koenigs, M.; Holm, C.; He, X.; Perez, M.F.; Zhao, H.; Mane, S.; Martinez, F.D.; et al. Noninvasive Analysis of the Sputum Transcriptome Discriminates Clinical Phenotypes of Asthma. Am. J. Respir. Crit. Care Med. 2015, 191, 1116–1125. [Google Scholar] [CrossRef]
- Kaur, R.; Chupp, G. Phenotypes and endotypes of adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019, 144, 1–12. [Google Scholar] [CrossRef]
- Khalaf, K.; Paoletti, G.; Puggioni, F.; Racca, F.; De Luca, F.; Giorgis, V.; Canonica, G.W.; Heffler, E. Asthma from immune pathogenesis to precision medicine. Semin. Immunol. 2019, 46, 101294. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Taunk, S.T.; Cardet, J.C.; Ledford, D.K. Clinical implications of asthma endotypes and phenotypes. Allergy Asthma Proc. 2022, 43, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Makrinioti, H.; Tiotiu, A.; Gonzalez-Barcala, F.-J. Severe asthma patients’ and physicians’ perspectives of disease burden: do they match? ERJ Open Res. 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, B.; Chatburn, E.; Bansal, A.T.; Fulton, O.; Hamerlijnck, D.; Coleman, C.; Eger, K.; Hyland, M.; Holmes, J.; Heaney, L.; et al. What bothers severe asthma patients most? A paired patient-clinician study across seven European countries. ERJ Open Res. 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Porsbjerg, C.; Menzies-Gow, A. Co-morbidities in severe asthma: Clinical impact and management. Respirology 2017, 22, 651–661. [Google Scholar] [CrossRef]
- Bucca, C.; Culla, B.; Brussino, L.; Ricciardolo, F.L.; Cicolin, A.; Heffler, E.; Bugiani, M.; Rolla, G. Effect of iron supplementation in women with chronic cough and iron deficiency. Int. J. Clin. Pr. 2012, 66, 1095–1100. [Google Scholar] [CrossRef]
- Robinson, D.; Humbert, M.; Buhl, R.; Cruz, A.A.; Inoue, H.; Korom, S.; Hanania, N.A.; Nair, P. Revisiting Type 2-high and Type 2-low airway inflammation in asthma: current knowledge and therapeutic implications. Clin. Exp. Allergy 2017, 47, 161–175. [Google Scholar] [CrossRef]
- Gao, J.; Wu, F.; Wu, S.; Yang, X. Inflammatory Subtypes in Classic Asthma and Cough Variant Asthma. J. Inflamm. Res. 2020, ume 13, 1167–1173. [Google Scholar] [CrossRef]
- Matsuoka, H.; Niimi, A.; Matsumoto, H.; Takemura, M.; Ueda, T.; Yamaguchi, M.; Jinnai, M.; Inoue, H.; Ito, I.; Chin, K.; et al. Inflammatory Subtypes in Cough-Variant Asthma: association with maintenance doses of inhaled corticosteroids. Chest 2010, 138, 1418–1425. [Google Scholar] [CrossRef]
- Chen, M.; Shepard, K.; Yang, M.; Raut, P.; Pazwash, H.; Holweg, C.T.; Choo, E. Overlap of allergic, eosinophilic and type 2 inflammatory subtypes in moderate-to-severe asthma. Clin. Exp. Allergy 2020, 51, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.; Oppenheimer, J. Elucidating asthma phenotypes and endotypes: progress towards personalized medicine. Ann. Allergy, Asthma Immunol. 2016, 116, 394–401. [Google Scholar] [CrossRef] [PubMed]
- de Kleer, I.M.; Kool, M.; de Bruijn, M.J.; Willart, M.; van Moorleghem, J.; Schuijs, M.J.; Plantinga, M.; Beyaert, R.; Hams, E.; Fallon, P.G.; et al. Perinatal Activation of the Interleukin-33 Pathway Promotes Type 2 Immunity in the Developing Lung. Immunity 2016, 45, 1285–1298. [Google Scholar] [CrossRef]
- Castro-Rodriguez, J.A.; Saglani, S.; Rodriguez-Martinez, C.E.; Oyarzun, M.A.; Fleming, L.; Bush, A. The relationship between inflammation and remodeling in childhood asthma: A systematic review. Pediatr. Pulmonol. 2018, 53, 824–835. [Google Scholar] [CrossRef]
- Lang, D.M. Severe asthma: Epidemiology, burden of illness, and heterogeneity. Allergy Asthma Proc. 2015, 36, 418–424. [Google Scholar] [CrossRef]
- Larenas-Linnemann, D.; Salas-Hernández, J.; Del Río-Navarro, B.E.; Luna-Pech, J.A.; Navarrete-Rodríguez, E.M.; Gochicoa, L.; Cano-Salas, M.d.C.; García-Ramírez, U.N.; López-Estrada, E.d.C.; Ortega-Martell, J.A.; et al. MIA 2021, Manejo Integral del Asma. Lineamientos para México. 2021, 68, s1–s122. [Google Scholar] [CrossRef]
- Pakkasela, J.; Ilmarinen, P.; Honkamäki, J.; Tuomisto, L.E.; Andersén, H.; Piirilä, P.; Hisinger-Mölkänen, H.; Sovijärvi, A.; Backman, H.; Lundbäck, B.; et al. Age-specific incidence of allergic and non-allergic asthma. BMC Pulm. Med. 2020, 20, 1–9. [Google Scholar] [CrossRef]
- Bachert, C.; Marple, B.; Schlosser, R.J.; Hopkins, C.; Schleimer, R.P.; Lambrecht, B.N.; Bröker, B.M.; Laidlaw, T.; Song, W.-J. Adult chronic rhinosinusitis. Nat. Rev. Dis. Prim. 2020, 6, 1–19. [Google Scholar] [CrossRef]
- Peters, M.C.; Ringel, L.; Dyjack, N.; Herrin, R.; Woodruff, P.G.; Rios, C.; O’connor, B.; Fahy, J.V.; Seibold, M.A. A Transcriptomic Method to Determine Airway Immune Dysfunction in T2-High and T2-Low Asthma. Am. J. Respir. Crit. Care Med. 2019, 199, 465–477. [Google Scholar] [CrossRef]
- Hirano, T.; Matsunaga, K. Late-onset asthma: current perspectives. J. Asthma Allergy 2018, ume 11, 19–27. [Google Scholar] [CrossRef]
- Ozyigit, L.P.; Morita, H.; Akdis, M. Innate lymphocyte cells in asthma phenotypes. Clin. Transl. Allergy 2015, 5, 23–8. [Google Scholar] [CrossRef]
- Kowalski, M.L.; Agache, I.; Bavbek, S.; Bakirtas, A.; Blanca, M.; Bochenek, G.; Bonini, M.; Heffler, E.; Klimek, L.; Laidlaw, T.M.; et al. Diagnosis and management of NSAID -Exacerbated Respiratory Disease (N- ERD )—a EAACI position paper. Allergy 2018, 74, 28–39. [Google Scholar] [CrossRef]
- Woo, S.-D.; Luu, Q.Q.; Park, H.-S. NSAID-Exacerbated Respiratory Disease (NERD): From Pathogenesis to Improved Care. Front. Pharmacol. 2020, 11, 1147. [Google Scholar] [CrossRef] [PubMed]
- Bonini, M.; Silvers, W. Exercise-Induced Bronchoconstriction: Background, Prevalence, and Sport Considerations. Immunol. Allergy Clin. North Am. 2018, 38, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Malewska-Kaczmarek, K.; Podlecka, D.; Mańkowski, T.; Jerzyńska, J.; Stelmach, I. Exercise-Induced Bronchoconstriction in Children: A Comparison between Athletes and Non-Athletes. Healthcare 2023, 11, 1349. [Google Scholar] [CrossRef]
- Vollsæter, M.; Stensrud, T.; Maat, R.; Halvorsen, T.; Røksund, O.D.; Sandnes, A.; Clemm, H. Exercise Related Respiratory Problems in the Young—Is It Exercise-Induced Bronchoconstriction or Laryngeal Obstruction? Front. Pediatr. 2022, 9, 800073. [Google Scholar] [CrossRef] [PubMed]
- Tikkakoski, A.P.; Tikkakoski, A.; Sipilä, K.; Kivistö, J.E.; Huhtala, H.; Kähönen, M.; Karjalainen, J.; Lehtimäki, L. Exercise-induced bronchoconstriction is associated with air humidity and particulate matter concentration in preschool children. Pediatr. Pulmonol. 2023, 58, 996–1003. [Google Scholar] [CrossRef]
- Klain, A.; Indolfi, C.; Dinardo, G.; Contieri, M.; Decimo, F.; Del Giudice, M.M. Exercise-Induced Bronchoconstriction in Children. Front. Med. 2022, 8, 814976. [Google Scholar] [CrossRef]
- Tliba, O.; Panettieri, R.A., Jr. Paucigranulocytic asthma: Uncoupling of airway obstruction from inflammation. J. Allergy Clin. Immunol. 2019, 143, 1287–1294. [Google Scholar] [CrossRef]
- Ray, A.; Kolls, J.K. Neutrophilic Inflammation in Asthma and Association with Disease Severity. Trends Immunol. 2017, 38, 942–954. [Google Scholar] [CrossRef]
- Seys, S.F.; Lokwani, R.; Simpson, J.L.; Bullens, D.M. New insights in neutrophilic asthma. Curr. Opin. Pulm. Med. 2019, 25, 113–120. [Google Scholar] [CrossRef]
- Strzelak, A.; Ratajczak, A.; Adamiec, A.; Feleszko, W. Tobacco Smoke Induces and Alters Immune Responses in the Lung Triggering Inflammation, Allergy, Asthma and Other Lung Diseases: A Mechanistic Review. Int. J. Environ. Res. Public Health 2018, 15, 1033. [Google Scholar] [CrossRef]
- Gonzalez-Uribe, V.; Martinez-Tenopala, R.; Baro-Alvarez, P.; Mojica-Gonzalez, Z. Frequency of ADIPOQ 276 and ADIPOQ 45 Polymorphisms in Obese and Eutrophic Adolescents with and without Asthma and their Relationship with Serum Adiponectin Levels. Med Res. Arch. 2022, 10. [Google Scholar] [CrossRef]
- Hanania, N.A.; King, M.J.; Braman, S.S.; Saltoun, C.; Wise, R.A.; Enright, P.; Falsey, A.R.; Mathur, S.K.; Ramsdell, J.W.; Rogers, L.; et al. Asthma in the elderly: Current understanding and future research needs—a report of a National Institute on Aging (NIA) workshop. J. Allergy Clin. Immunol. 2011, 128, S4–S24. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.L.; Ruwanpathirana, A.; Lewis, B.W.; Britt, R.D. Aging-Related Mechanisms Contribute to Corticosteroid Insensitivity in Elderly Asthma. Int. J. Mol. Sci. 2023, 24, 6347. [Google Scholar] [CrossRef] [PubMed]
- Khosa, J.K.; Louie, S.; Moreno, P.L.; Abramov, D.; Rogstad, D.K.; Alismail, A.; Matus, M.J.; Tan, L.D. Asthma Care in the Elderly: Practical Guidance and Challenges for Clinical Management - A Framework of 5 “Ps”. J. Asthma Allergy 2023, ume 16, 33–43. [Google Scholar] [CrossRef]
- Jartti, T.; Saarikoski, L.; Jartti, L.; Lisinen, I.; Jula, A.; Huupponen, R.; Viikari, J.; Raitakari, O.T. Obesity, adipokines and asthma. Allergy 2009, 64, 770–777. [Google Scholar] [CrossRef]
- Yuksel, H.; Sogut, A.; Yilmaz, O.; Onur, E.; Dinç, G. Role of Adipokines and Hormones of Obesity in Childhood Asthma. Allergy Asthma Immunol. Res. 2012, 4, 98–103. [Google Scholar] [CrossRef]
- Leija-Martínez, J.J.; Giacoman-Martínez, A.; Del-Río-Navarro, B.E.; Sanchéz-Muñoz, F.; Hernández-Diazcouder, A.; Muñoz-Hernández, O.; Romero-Nava, R.; Villafaña, S.; Marchat, L.A.; Hong, E.; et al. Promoter methylation status of RORC, IL17A, and TNFA in peripheral blood leukocytes in adolescents with obesity-related asthma. Heliyon 2022, 8, e12316. [Google Scholar] [CrossRef]
- Vieira, C.P.; de Oliveira, L.P.; Da Silva, M.B.; Andre, D.M.; Tavares, E.B.G.; Pimentel, E.R.; Antunes, E. Role of metalloproteinases and TNF-α in obesity-associated asthma in mice. Life Sci. 2020, 259, 118191. [Google Scholar] [CrossRef]
- Leija-Martínez, J.J.; Del-Río-Navarro, B.E.; Sanchéz-Muñoz, F.; Muñoz-Hernández, O.; Hong, E.; Giacoman-Martínez, A.; Romero-Nava, R.; Patricio-Román, K.L.; Hall-Mondragon, M.S.; Espinosa-Velazquez, D.; et al. Associations of TNFA, IL17A, and RORC mRNA expression levels in peripheral blood leukocytes with obesity-related asthma in adolescents. Clin. Immunol. 2021, 229, 108715. [Google Scholar] [CrossRef]
- Yon, C.; Thompson, D.A.; Jude, J.A.; Panettieri, R.A.; Rastogi, D. Crosstalk between CD4+ T Cells and Airway Smooth Muscle in Pediatric Obesity-related Asthma. Am. J. Respir. Crit. Care Med. 2023, 207, 461–474. [Google Scholar] [CrossRef]
- Martínez-Aguilar, N.E.; Del Río-Navarro, B.E.; Navarro-Olivos, E.; García-Ortíz, H.; Orozco, L.; Jiménez-Morales, S. SPINK5andADRB2haplotypes are risk factors for asthma in Mexican pediatric patients. J. Asthma 2014, 52, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhu, Z.; Xiao, Q.; Li, J.; Hong, X.; Wang, X.; Hasegawa, K.; Camargo, C.A.; Liang, L. Obesity-related biomarkers underlie a shared genetic architecture between childhood body mass index and childhood asthma. Commun. Biol. 2022, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Everaere, L.; Yahia, S.A.; Bouté, M.; Audousset, C.; Chenivesse, C.; Tsicopoulos, A. Innate lymphoid cells at the interface between obesity and asthma. Immunology 2017, 153, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.E.; Peters, U. The effect of obesity on lung function. Expert Rev. Respir. Med. 2018, 12, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Spathopoulos, D.; Paraskakis, E.; Trypsianis, G.; Tsalkidis, A.; Arvanitidou, V.; Emporiadou, M.; Bouros, D.; Chatzimichael, A. The effect of obesity on pulmonary lung function of school aged children in Greece. Pediatr. Pulmonol. 2009, 44, 273–280. [Google Scholar] [CrossRef]
- Mahadev, S.; Salome, C.M.; Berend, N.; King, G.G. The effect of low lung volume on airway function in obesity. Respir. Physiol. Neurobiol. 2013, 188, 192–199. [Google Scholar] [CrossRef]
- Starr, S.; Wysocki, M.; DeLeon, J.D.; Silverstein, G.; Arcoleo, K.; Rastogi, D.; Feldman, J.M. Obesity-related pediatric asthma: relationships between pulmonary function and clinical outcomes. J. Asthma 2022, 60, 1418–1427. [Google Scholar] [CrossRef]
- Bhatawadekar, S.A.; Peters, U.; Walsh, R.R.; Daphtary, N.; MacLean, E.S.; Mori, V.; Hodgdon, K.; Kinsey, C.M.; Kaminsky, D.A.; Bates, J.H.; et al. Central airway collapse is related to obesity independent of asthma phenotype. Respirology 2021, 26, 334–341. [Google Scholar] [CrossRef]
- Reyes-Angel, J.; Kaviany, P.; Rastogi, D.; Forno, E. Obesity-related asthma in children and adolescents. Lancet Child Adolesc. Heal. 2022, 6, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Hudler, A.C.; Díaz, I.R.R.; Sharma, S.; Holguin, F. Gaps and Future Directions in Clinical Research on Obesity-Related Asthma. Pulm. Ther. 2023, 9, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Eguiluz-Gracia, I.; Cojanu, C.; Laculiceanu, A.; del Giacco, S.; Zemelka-Wiacek, M.; Kosowska, A.; Akdis, C.A.; Jutel, M. Advances and highlights in asthma in 2021. Allergy 2021, 76, 3390–3407. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.B.; Abrams, E.M. Asthma guidelines: the Global Initiative for Asthma in relation to national guidelines. Curr. Opin. Allergy Clin. Immunol. 2017, 17, 99–103. [Google Scholar] [CrossRef]
- Global Initiative for Asthma. 2023. Accessed 30th April 2023.
- Fukunaga, K. [ASTHMA PREVENTION AND MANAGEMENT GUIDELINES 2021]. 2023, 72, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Makoni, M. Guidelines might help reduce the burden of asthma in African children. Lancet Respir. Med. 2022, 10, e83–e84. [Google Scholar] [CrossRef]
- GEMA 5.1. Guía Española para el Manejo del Asma. 2023. Accessed 6 May 2023. www.gemasma.com.
- Cloutier, M.M.; Baptist, A.P.; Blake, K.V.; Brooks, E.G.; Bryant-Stephens, T.; DiMango, E.; Dixon, A.E.; Elward, K.S.; Hartert, T.; Krishnan, J.A.; et al. 2020 Focused Updates to the Asthma Management Guidelines: A Report from the National Asthma Education and Prevention Program Coordinating Committee Expert Panel Working Group. J. Allergy Clin. Immunol. 2020, 146, 1217–1270. [Google Scholar] [CrossRef]
- “International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma.” Kian Fan Chung, Sally E. Wenzel, Jan L. Brozek, et al. Eur Respir J 2014; 43: 343–373.. Eur. Respir. J. 2022, 59, 1362020. [CrossRef]
- Canonica, G.W.; Ferrando, M.; Baiardini, I.; Puggioni, F.; Racca, F.; Passalacqua, G.; Heffler, E. Asthma: personalized and precision medicine. Curr. Opin. Allergy Clin. Immunol. 2018, 18, 51–58. [Google Scholar] [CrossRef]
- Pelaia, C.; Calabrese, C.; Terracciano, R.; de Blasio, F.; Vatrella, A.; Pelaia, G. Omalizumab, the first available antibody for biological treatment of severe asthma: more than a decade of real-life effectiveness. Ther. Adv. Respir. Dis. 2018, 12. [Google Scholar] [CrossRef]
- Genentech USA INPC. Xolair® (omalizumab) Prescribing Information. 2023. Accessed 05/23, 2023. https://www.xolairhcp.com.
- Riccio, A.M.; Negro, R.W.; Micheletto, C.; De Ferrari, L.; Folli, C.; Chiappori, A.; Canonica, G.W. Omalizumab Modulates Bronchial Reticular Basement Membrane Thickness and Eosinophil Infiltration in Severe Persistent Allergic Asthma Patients. Int. J. Immunopathol. Pharmacol. 2012, 25, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, G.; Brookman, L.; Fox, H.; Johnson, C.; Matthews, J.; Ren, S.; Deniz, Y. Pharmacodynamics of omalizumab: implications for optimised dosing strategies and clinical efficacy in the treatment of allergic asthma. Curr. Med Res. Opin. 2003, 19, 491–499. [Google Scholar] [CrossRef]
- Casale, T.B.; Luskin, A.T.; Busse, W.; Zeiger, R.S.; Trzaskoma, B.; Yang, M.; Griffin, N.M.; Chipps, B.E. Omalizumab Effectiveness by Biomarker Status in Patients with Asthma: Evidence From PROSPERO, A Prospective Real-World Study. J. Allergy Clin. Immunol. Pr. 2019, 7, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Canonica, G.W.; Rottoli, P.; Bucca, C.; Zappa, M.C.; Michetti, G.; Macciocchi, B.; Caruso, C.; Santus, P.; Bartezaghi, M.; Rigoni, L. Improvement of patient-reported outcomes in severe allergic asthma by omalizumab treatment: the real life observational PROXIMA study. World Allergy Organ. J. 2018, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Bhutani, M.; Yang, W.H.; Hébert, J.; de Takacsy, F.; Stril, J.-L. The real world effect of omalizumab add on therapy for patients with moderate to severe allergic asthma: The ASTERIX Observational study. PLOS ONE 2017, 12, e0183869–e0183869. [Google Scholar] [CrossRef] [PubMed]
- Barnes, N.; Menzies-Gow, A.; Mansur, A.H.; Spencer, D.; Percival, F.; Radwan, A.; Niven, R.; Frcp.; Frcp.; (Hons)., M.B.; et al. Effectiveness of Omalizumab in Severe Allergic Asthma: A Retrospective UK Real-World Study. J. Asthma 2013, 50, 529–536. [CrossRef]
- Bousquet, J.; Humbert, M.; Gibson, P.G.; Kostikas, K.; Jaumont, X.; Pfister, P.; Nissen, F. Real-World Effectiveness of Omalizumab in Severe Allergic Asthma: A Meta-Analysis of Observational Studies. J. Allergy Clin. Immunol. Pr. 2021, 9, 2702–2714. [Google Scholar] [CrossRef]
- Faulkner, K.M.; MacDonald, K.; Abraham, I.; Alhossan, A.; Lee, C.S. ‘Real-world’ effectiveness of omalizumab in adults with severe allergic asthma: a meta-analysis. Expert Rev. Clin. Immunol. 2020, 17, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Torres-Duque, C.A.; Ocampo-Gómez, J.; Castillo, M.M.; Cano-Rosales, D.; Giraldo-Montoya, .; Rodríguez, F.; Palacios-Ortega, I.; Durán-Silva, M.; Reynales, H.; García, E.; et al. Real-world effectiveness of omalizumab for severe allergic asthma treatment in Colombia. BMC Pulm. Med. 2022, 22, 1–10. [CrossRef]
- Braunstahl, G.-J.; Chen, C.-W.; Maykut, R.; Georgiou, P.; Peachey, G.; Bruce, J. The eXpeRience registry: The ‘real-world’ effectiveness of omalizumab in allergic asthma. Respir. Med. 2013, 107, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Kirchnerová, O.R.; Valena, T.; Novosad, J.; Teřl, M. Real-world effectiveness and safety of omalizumab in patients with uncontrolled severe allergic asthma from the Czech Republic. Adv. Dermatol. Allergol. 2019, 36, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Zhi, L.; Liu, F.; Wang, Y.; Zhang, Q.; Liu, X.; Wang, X.; Hao, G.; Zhang, X.; Hu, Q.; et al. Real-World Safety and Effectiveness of Omalizumab in Moderate to Severe Allergic Asthma Patients in China: A Post-Authorization Study. J. Asthma Allergy 2023, ume 16, 625–636. [Google Scholar] [CrossRef]
- Emma, R.; Morjaria, J.B.; Fuochi, V.; Polosa, R.; Caruso, M. Mepolizumab in the management of severe eosinophilic asthma in adults: current evidence and practical experience. Ther. Adv. Respir. Dis. 2018, 12. [Google Scholar] [CrossRef]
- Chupp, G.L.; Bradford, E.S.; Albers, F.C.; Bratton, D.J.; Wang-Jairaj, J.; Nelsen, L.M.; Trevor, J.L.; Magnan, A.; Brinke, A.T. Efficacy of mepolizumab add-on therapy on health-related quality of life and markers of asthma control in severe eosinophilic asthma (MUSCA): a randomised, double-blind, placebo-controlled, parallel-group, multicentre, phase 3b trial. Lancet Respir. Med. 2017, 5, 390–400. [Google Scholar] [CrossRef]
- Bel, E.H.; Wenzel, S.E.; Thompson, P.J.; Prazma, C.M.; Keene, O.N.; Yancey, S.W.; Ortega, H.G.; Pavord, I.D. Oral Glucocorticoid-Sparing Effect of Mepolizumab in Eosinophilic Asthma. New Engl. J. Med. 2014, 371, 1189–1197. [Google Scholar] [CrossRef]
- Canonica, G.W.; Colombo, G.L.; Bruno, G.M.; Di Matteo, S.; Martinotti, C.; Blasi, F.; Bucca, C.; Crimi, N.; Paggiaro, P.; Pelaia, G.; et al. Shadow cost of oral corticosteroids-related adverse events: A pharmacoeconomic evaluation applied to real-life data from the Severe Asthma Network in Italy (SANI) registry. World Allergy Organ. J. 2019, 12, 100007. [Google Scholar] [CrossRef]
- Heffler, E.; Bagnasco, D.; Canonica, G.W. Strategies to reduce corticosteroid-related adverse events in asthma. Curr. Opin. Allergy Clin. Immunol. 2019, 19, 61–67. [Google Scholar] [CrossRef]
- Deeks, E.D. Mepolizumab: A Review in Eosinophilic Asthma. BioDrugs 2016, 30, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Sahota, J.; Robinson, D.S. Update on new biologics for intractable eosinophilic asthma: impact of reslizumab. Drug Des. Dev. Ther. 2018, ume 12, 1173–1181. [Google Scholar] [CrossRef]
- Castro, M.; Zangrilli, J.E.; Wechsler, M.E.; Bateman, E.D.; Brusselle, G.G.; Bardin, P.; Murphy, K.; Maspero, J.F.; O'Brien, C.; Korn, S. Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir. Med. 2015, 3, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Pelaia, C.; Calabrese, C.; Vatrella, A.; Busceti, M.T.; Garofalo, E.; Lombardo, N.; Terracciano, R.; Pelaia, G. Benralizumab: From the Basic Mechanism of Action to the Potential Use in the Biological Therapy of Severe Eosinophilic Asthma. BioMed Res. Int. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, A.; Trikha, A.; Calhoun, W.J. Benralizumab – a humanized mAb to IL-5Rα with enhanced antibody-dependent cell-mediated cytotoxicity – a novel approach for the treatment of asthma. Expert Opin. Biol. Ther. 2011, 12, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Menzella, F.; Latorre, M.; Ruggiero, P.; Bagnasco, D.; Heffler, E. Reduction of oral corticosteroids in patients with severe eosinophilic asthma treated with Benralizumab: could it represent a marker of treatment efficacy? Expert Opin. Biol. Ther. 2019, 19, 601–606. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, J.; Chen, Y. Efficacy and safety of treatment with benralizumab for eosinophilic asthma. Int. Immunopharmacol. 2022, 111, 109131. [Google Scholar] [CrossRef]
- Bagnasco, D.; Ferrando, M.; Varricchi, G.; Passalacqua, G.; Canonica, G.W. A Critical Evaluation of Anti-IL-13 and Anti-IL-4 Strategies in Severe Asthma. Int. Arch. Allergy Immunol. 2016, 170, 122–131. [Google Scholar] [CrossRef]
- Balboul, S.; Kahn, J.; Tracy, A.; Peacock, A.; Cline, A. The Application of Dupilumab to Pediatric Patients Aged 6–11yrs with Moderate-to-Severe Atopic Dermatitis Whose Disease is Not Adequately Controlled: The Clinical Data so Far. Drug Des. Dev. Ther. 2023, ume 17, 1323–1327. [Google Scholar] [CrossRef]
- Silverberg, J.I.; Yosipovitch, G.; Simpson, E.L.; Kim, B.S.; Wu, J.J.; Eckert, L.; Guillemin, I.; Chen, Z.; Ardeleanu, M.; Bansal, A.; et al. Dupilumab treatment results in early and sustained improvements in itch in adolescents and adults with moderate to severe atopic dermatitis: Analysis of the randomized phase 3 studies SOLO 1 and SOLO 2, AD ADOL, and CHRONOS. J. Am. Acad. Dermatol. 2020, 82, 1328–1336. [Google Scholar] [CrossRef]
- Senner, S.; Seegräber, M.; Frey, S.; Kendziora, B.; Eicher, L.; Wollenberg, A. Dupilumab for the treatment of adolescents with atopic dermatitis. Expert Rev. Clin. Immunol. 2020, 16, 641–650. [Google Scholar] [CrossRef]
- Ferrante, G.; Tenero, L.; Piazza, M.; Piacentini, G. Severe pediatric asthma therapy: Dupilumab. Front. Pediatr. 2022, 10, 963610. [Google Scholar] [CrossRef] [PubMed]
- Rabe, K.F.; Nair, P.; Brusselle, G.; Maspero, J.F.; Castro, M.; Sher, L.; Zhu, H.; Hamilton, J.D.; Swanson, B.N.; Khan, A.; et al. Efficacy and Safety of Dupilumab in Glucocorticoid-Dependent Severe Asthma. N. Engl. J. Med. 2018, 378, 2475–2485. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; Sailer, M.M.; Capitani, F.; van Schaik, C.; Löwenheim, H.; Becker, S. Real-world evidence for the effectiveness and safety of dupilumab in patients with CRSwNP after 1 year of therapy. 2023, 16, 100780. [CrossRef]
- De Corso, E.; Pasquini, E.; Trimarchi, M.; La Mantia, I.; Pagella, F.; Ottaviano, G.; Garzaro, M.; Pipolo, C.; Torretta, S.; Seccia, V.; et al. Dupilumab in the treatment of severe uncontrolled chronic rhinosinusitis with nasal polyps (CRSwNP): A multicentric observational Phase IV real-life study (DUPIREAL). Allergy 2023. [Google Scholar] [CrossRef]
- Greuter, T.; Schoepfer, A.M. Dupilumab in Patients with Eosinophilic Esophagitis. New Engl. J. Med. 2023, 388, 955–957. [Google Scholar] [CrossRef]
- Dellon, E.S.; Rothenberg, M.E.; Collins, M.H.; Hirano, I.; Chehade, M.; Bredenoord, A.J.; Lucendo, A.J.; Spergel, J.M.; Aceves, S.; Sun, X.; et al. Dupilumab in Adults and Adolescents with Eosinophilic Esophagitis. New Engl. J. Med. 2022, 387, 2317–2330. [Google Scholar] [CrossRef]
- Maspero, J.F.; Cardona, G.; Schonffeldt, P.; Tolcachier, A.; González-Diaz, S.N.; Yañez, A.; Galvao, C.E.; Msihid, J.; Gall, R.; Siddiqui, S.; et al. Dupilumab efficacy and safety in Latin American patients with uncontrolled, moderate-to-severe asthma: phase 3 LIBERTY ASTHMA QUEST study. J. Asthma 2022, 60, 981–990. [Google Scholar] [CrossRef]
- Hopkins, C.; Buchheit, K.M.; Heffler, E.; A Cohen, N.; Olze, H.; Khan, A.H.; Msihid, J.; Siddiqui, S.; Nash, S.; A Jacob-Nara, J.; et al. Improvement in Health-Related Quality of Life with Dupilumab in Patients with Moderate-to-Severe Asthma with Comorbid Chronic Rhinosinusitis with/without Nasal Polyps: An Analysis of the QUEST Study. J. Asthma Allergy 2022, ume 15, 767–773. [Google Scholar] [CrossRef]
- Busse, W.W.; Pavord, I.D.; Siddiqui, S.; Khan, A.H.; Praestgaard, A.; Nash, S.; Jacob-Nara, J.A.; Rowe, P.J.; Deniz, Y. Dupilumab Improves Outcomes in Patients with Chronic Rhinosinusitis with Nasal Polyps and Coexisting Asthma Irrespective of Baseline Asthma Characteristics. J. Asthma Allergy 2023, ume 16, 411–419. [Google Scholar] [CrossRef]
- Dinardo, G.; Indolfi, C.; Klain, A.; Decimo, F.; DEL Giudice, M.M. Treatment of severe asthma: fast action of dupilumab in the pediatric setting. Minerva Pediatr. 2023, 75, 312–313. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.G.; Szefler, S.J.; Bacharier, L.B.; Maspero, J.F.; Domingo, C.; Fiocchi, A.; Lee, J.K.; Daizadeh, N.; Lederer, D.J.; Hardin, M.; et al. Assessment of dupilumab in children with moderate-to-severe type 2 asthma with or without evidence of allergic asthma. Allergy 2023, 78, 2157–2167. [Google Scholar] [CrossRef]
- Yang, D.-Y.; Li, L.; Lu, T.; Jing, W.-W.; Liu, X.; Li, X.-L. Efficacy and safety of dupilumab in pediatric patients with moderate to severe atopic dermatitis: a real-world study. Arch. Dermatol. Res. 2022, 315, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.L.; Paller, A.S.; Siegfried, E.C.; Thaçi, D.; Wollenberg, A.; Cork, M.J.; Marcoux, D.; Huang, R.; Chen, Z.; Rossi, A.B.; et al. Dupilumab Demonstrates Rapid and Consistent Improvement in Extent and Signs of Atopic Dermatitis Across All Anatomical Regions in Pediatric Patients 6 Years of Age and Older. 2021, 11, 1643–1656. [Google Scholar] [PubMed]
- Corren, J.; Menzies-Gow, A.; Chupp, G.; Israel, E.; Korn, S.; Cook, B.; Ambrose, C.S.; Hellqvist, .; Roseti, S.L.; Molfino, N.A.; et al. Efficacy of Tezepelumab in Severe, Uncontrolled Asthma: Pooled Analysis of the PATHWAY and NAVIGATOR Clinical Trials. Am. J. Respir. Crit. Care Med. 2023, 208, 13–24. [CrossRef]
- Roy, P.; Rafa, Z.I.; Haque, S.N.; Tasha, T.; Arko, S.B.; Agrawal, H.; Razu, I.; Parisapogu, A.; Maisha, S.; A Siddique, M.; et al. The Impact of Tezepelumab in Uncontrolled Severe Asthma: A Systematic Review of Randomized Controlled Trials. Cureus 2022, 14, e32156. [Google Scholar] [CrossRef]
- Corren, J.; Parnes, J.R.; Wang, L.; Mo, M.; Roseti, S.L.; Griffiths, J.M.; van der Merwe, R. Tezepelumab in Adults with Uncontrolled Asthma. New Engl. J. Med. 2017, 377, 936–946. [Google Scholar] [CrossRef]
- Wechsler, M.E.; Colice, G.; Griffiths, J.M.; Almqvist, G.; Skärby, T.; Piechowiak, T.; Kaur, P.; Bowen, K.; Hellqvist. ; Mo, M.; et al. SOURCE: a phase 3, multicentre, randomized, double-blind, placebo-controlled, parallel group trial to evaluate the efficacy and safety of tezepelumab in reducing oral corticosteroid use in adults with oral corticosteroid dependent asthma. Respir. Res. 2020, 21, 1–10. [Google Scholar] [CrossRef]
- Menzies-Gow, A.; Bourdin, A.; Chupp, G.; Israel, E.; Hellqvist, .; Hunter, G.; Roseti, S.L.; Ambrose, C.S.; Llanos, J.-P.; Cook, B.; et al. Effect of tezepelumab on healthcare utilization in patients with severe, uncontrolled asthma. Ann. Allergy, Asthma Immunol. 2023. [CrossRef]
- Chagas, G.C.L.; Xavier, D.; Gomes, L.; Ferri-Guerra, J.; Oquet, R.E.H. Effects of Tezepelumab on Quality of Life of Patients with Moderate-to-Severe, Uncontrolled Asthma: Systematic Review and Meta-Analysis. Curr. Allergy Asthma Rep. 2023, 1–12. [Google Scholar] [CrossRef]
- Rappoport, N.; Shamir, R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res. 2018, 46, 10546–10562. [Google Scholar] [CrossRef] [PubMed]
- Dugourd, A.; Kuppe, C.; Sciacovelli, M.; Gjerga, E.; Gabor, A.; Emdal, K.B.; Vieira, V.; Bekker-Jensen, D.B.; Kranz, J.; Bindels, E.; et al. Causal integration of multi-omics data with prior knowledge to generate mechanistic hypotheses. Mol. Syst. Biol. 2021, 17, e9730. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




