Submitted:
22 July 2023
Posted:
25 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. DNA Extraction and Next-Generation Sequencing
2.3. NGS data analysis and interpretation
3. Results
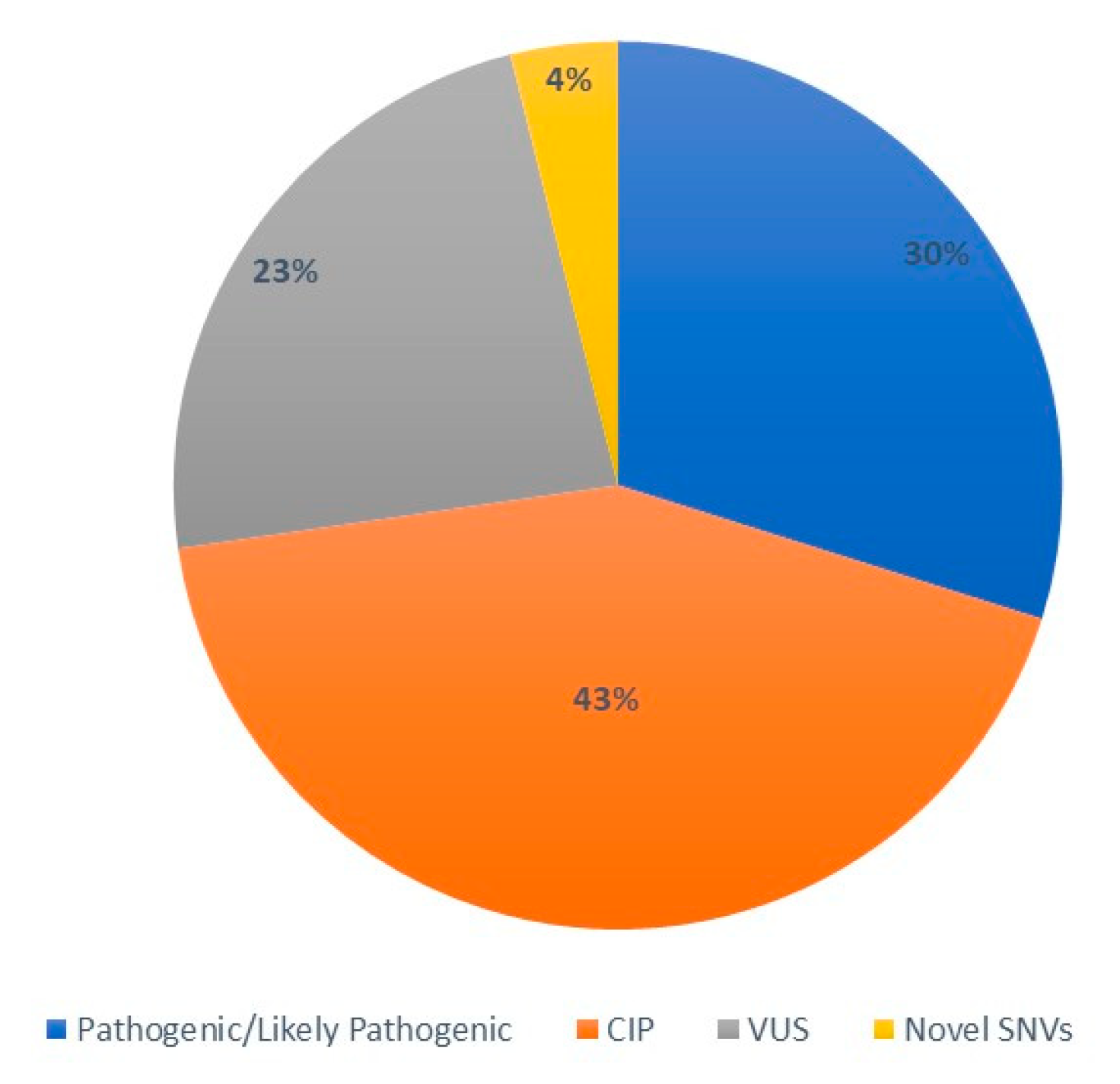
3.1. Overall description of CFTR mutational spectrum
3.2. CFTR pathogenic/likely pathogenic variants
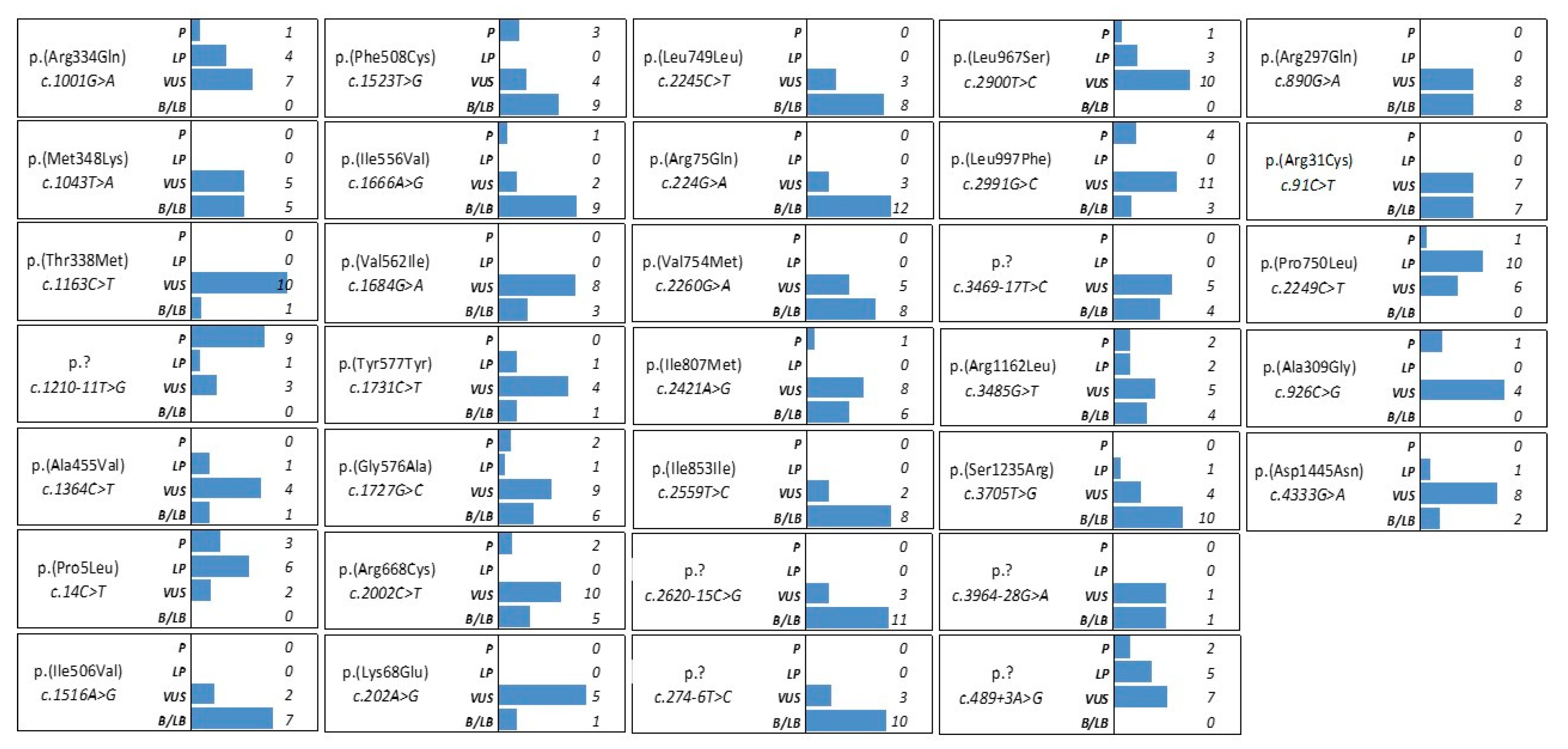
3.3. CFTR variants with conflicting interpretation of pathogenicity and variants of uncertain significance
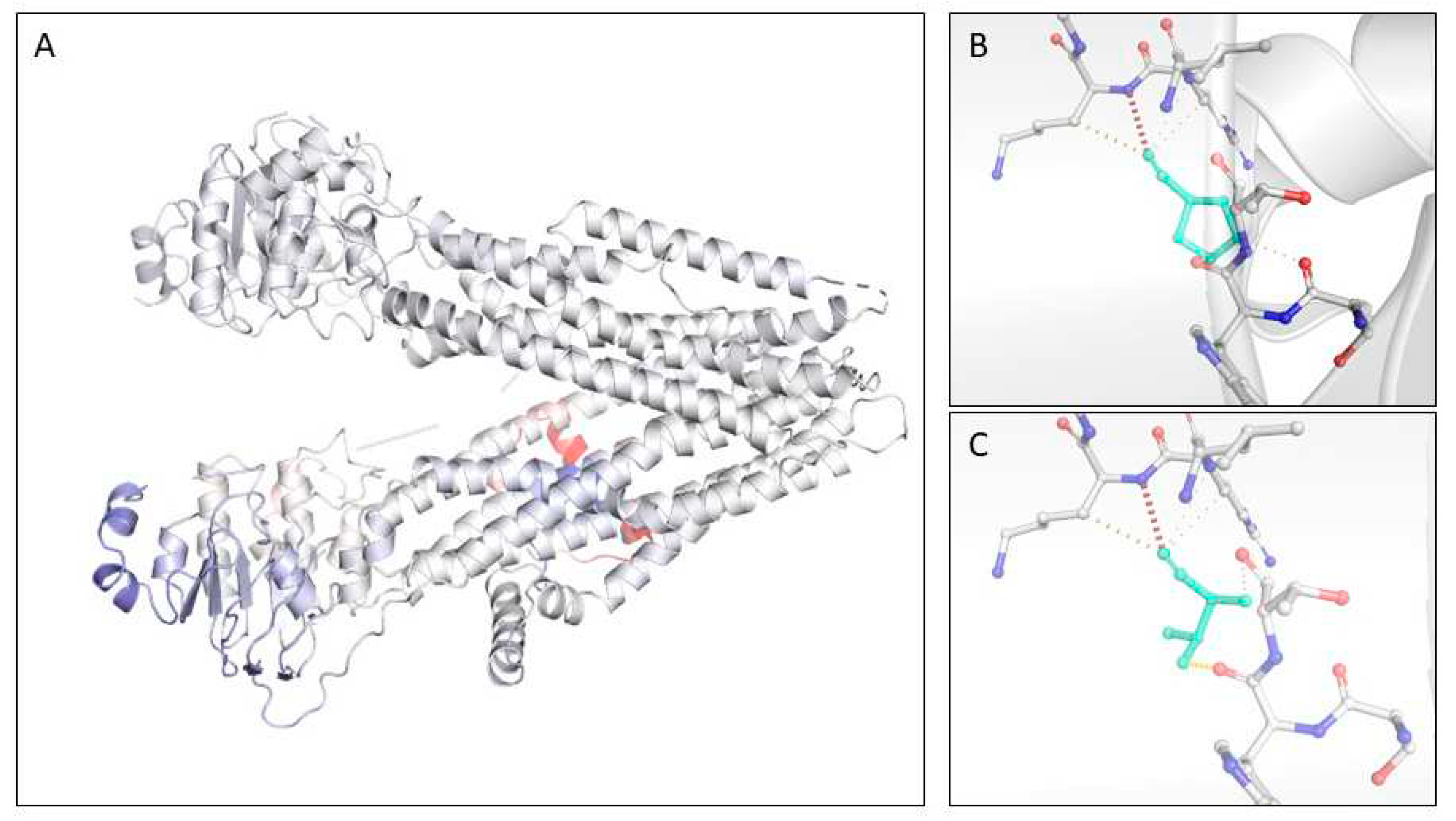
3.4. CFTR novel variants
4. Discussion
Author Contributions
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savant, A.; Lyman, B.; Bojanowski, C.; Upadia, J.; Adam, M.P.; Mirzaa G.M.; et al. Cystic Fibrosis. In: Mirzaa, G.M.; Pagon, R.A.; Wallace, S.E.; Bean, L.J.H.; Gripp, K.W.; Amemiya, A.; eds. GeneReviews®. Seattle (WA): University of Washington, Seattle; March 26, 2001.
- Kerem, B.; Rommens, J.M.; Buchanan, J.A.; Markiewicz, D.; Cox, T.K.; Chakravarti, A.; et al. Identification of the cystic fibrosis gene: genetic analysis. Science 1989; 245, 1073-1080. [CrossRef]
- Hanssens, L.S.; Duchateau, J.; Casimir, G.J. CFTR Protein: Not Just a Chloride Channel? Cells. 2021, 10-2844. [CrossRef]
- Trezise, A.E.O. Exquisite and Multilevel Regulation of CFTR Expression. In: Bush, A.; Alton, E.W.F.W.; Davies, J.C.; Griesenbach, U.; Jaffe, A. eds. Prog Respir Res.Cystic Fibrosis in the 21st Century. Basel, Karger, 2006.
- Vukovic, V.; Agodi, A.; Assael, B.; Calabro', G.; Campanella, P.; Castellani, C.; et al. VP103 Health Technology Assessment Of Genetic Tests For Cystic Fibrosis Carrier Screening In Italy. International Journal of Technology Assessment in Health Care 2017 33(S1), 197-197. [CrossRef]
- Castellani, C.; Picci, L.; Tridello, G.; Casati, E.; Tamanini, A.; Bartoloni, L.; et al. Cystic fibrosis carrier screening effects on birth prevalence and newborn screening. Genet Med. 2016, 18, 145–151. [Google Scholar] [CrossRef]
- Radhakrishnan, M.; van Gool, K.; Hall, J.; Delatycki, M.; Massie, J. Economic evaluation of cystic fibrosis screening: a review of the literature. Health Policy 2008, 85, 133–147. [Google Scholar] [CrossRef]
- Picci, L.; Cameran, M.; Marangon, O.; Marzenta, D.; Ferrari, S.; Frigo, A.C.; et al. A 10-year large-scale cystic fibrosis carrier screening in the Italian population. J Cyst Fibros. 2010, 9, 29–35. [Google Scholar] [CrossRef]
- Grody, W.W.; Cutting, G.R. , Watson, M.S. The Cystic Fibrosis mutation "arms race": when less is more. Genet Med. 2007, 9, 739–744. [Google Scholar]
- Norman, R.; van Gool, K.; Hall, J.; Delatycki, M.; Massie, J. Cost-effectiveness of carrier screening for cystic fibrosis in Australia. J Cyst Fibros. 2012, 11, 281–287. [Google Scholar] [CrossRef]
- Castellani, C.; Picci, L.; Tamanini, A.; Girardi, P.; Rizzotti, P.; Assael, B.M. Association between carrier screening and incidence of cystic fibrosis. JAMA. 2009, 302, 2573–2579. [Google Scholar] [CrossRef]
- Doherty, R.A. National Institutes of Health consensus development conference statement on genetic testing for cystic fibrosis. J Med Screen. 1997, 4, 179–180. [Google Scholar]
- Castellani, C.; Massie, J. Newborn screening and carrier screening for cystic fibrosis: alternative or complementary? Eur Respir J. 2014, 43, 20–23. [Google Scholar]
- Bergougnoux, A.; Taulan-Cadars, M.; Claustres, M.; Raynal, C. Current and future molecular approaches in the diagnosis of cystic fibrosis. Expert Rev Respir Med. 2018, 12, 415–426. [Google Scholar] [CrossRef]
- 15.ClinVar, National Cencer for Biotechnology Information. https://www.ncbi.nlm.nih.gov/clinvar/ (accessed June 2023).
- Cystic Fibrosis Mutation Database. http://www.genet.sickkids.on.ca/ (accessed June 2023).
- CFTR-France database. https://cftr.iurc.montp.inserm.fr/cftr/ (accessed June 2023).
- CFTR 2. https://cftr2.org/ (accessed June 2023).
- LOVD v.3.0 - Leiden Open Variation Database, https://databases.lovd.nl/shared/genes/CFTR (accessed June 2023).
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; et al. VarSome: the human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Quan, L.; Kai, W. InterVar: Clinical interpretation of genetic variants by ACMG-AMP 2015 guideline. The American Journal of Human Genetics 2017, 100, 1–14. [Google Scholar]
- Cystic Fibrosis Missense Analysis. https://cysma.iurc.montp.inserm.fr/cysma/ (accessed June 2023).
- Sasorith, S.; Baux, D.; Bergougnoux, A. The CYSMA web server: An example of integrative tool for in silico analysis of missense variants identified in Mendelian disorders. Hum Mutat. 2020, 41, 375–386. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank. https://www.rcsb.org/ (accessed June 2023).
- SWISS-MODEL. https://wissmodel.expasy.org/ (accessed June 2023).
- DynaMut 2. https://biosig.lab.uq.edu.au/dynamut2/ (accessed June 2023).
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, B.P. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Science. 2021, 30, 60–69. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Poly-Phen2. http://genetics.bwh.harvard.edu/pph2/dokuwiki/about (accessed June 2023).
- Human Splicing Finder. https://www.genomnis.com/access-hsf (accessed June 2023).
- MobyDetails. https://mobidetails.iurc.montp.inserm.fr/MD/ (accessed June 2023).
- Chamayou, S.; Sicali, M.; Lombardo, D.; Maglia, E.; Liprino, A.; Cardea, C.; et al. The true panel of cystic fibrosis mutations in the Sicilian population. BMC Med Genet. 2020, 21, 89. [Google Scholar] [CrossRef]
- Bareil, C.; Bergougnoux, A. CFTR gene variants, epidemiology and molecular pathology. Arch Pediatr. 2020, 27 Suppl 1:eS8-eS12. [CrossRef]
- Registro Italiano Fibrosi Cistica. https://www.registroitalianofibrosicistica.it/servizi-36-rapporti_e_pubblicazioni (accessed June 2023).
- Dell'Edera, D.; Benedetto, M.; Gadaleta, G.; Carone, D.; Salvatore, D.; Angione, A.; et al. Analysis of cystic fibrosis gene mutations in children with cystic fibrosis and in 964 infertile couples within the region of Basilicata, Italy: a research study. J Med Case Rep. 2014, 8, 39. [Google Scholar] [CrossRef]
- WHO Human Genetics Programme. The molecular genetic epidemiology of cystic fibrosis: report of a joint meeting of WHO/IECFTN/ICF(M)A/ECFS, Genoa, Italy, 19 June 2002. World Health Organization (2004). https://apps.who.int/iris/handle/10665/68702.
- Società Italiana per lo studio della fibrosi cistica. Raccomandazioni sul test del portatore di mutazioni del gene CFTR. https://www.sifc.it/documento/raccomandazioni-sul-test-del-portatore-di-mutazioni-del-gene-ctfr/.
- Çolak, Y.; Nordestgaard, B.G.; Afzal, S. Morbidity and mortality in carriers of the cystic fibrosis mutation CFTR Phe508del in the general population. Eur Respir J. 2020, 56, 2000558. [Google Scholar] [CrossRef]
- Miller, A.C.; Comellas, A.P.; Hornick, D.B.; Stoltz, D.A.; Cavanaugh, J.E.; Gerke, A.K.; et al. Cystic fibrosis carriers are at increased risk for a wide range of cystic fibrosis-related conditions. Proc Natl Acad Sci U S A. 2020, 117, 1621–1627. [Google Scholar] [CrossRef]
- Martin, C.; Burgel, P.R. Carriers of a single CFTR mutation are asymptomatic: an evolving dogma? Eur Respir J. 2020, 56, 2002645. [Google Scholar]
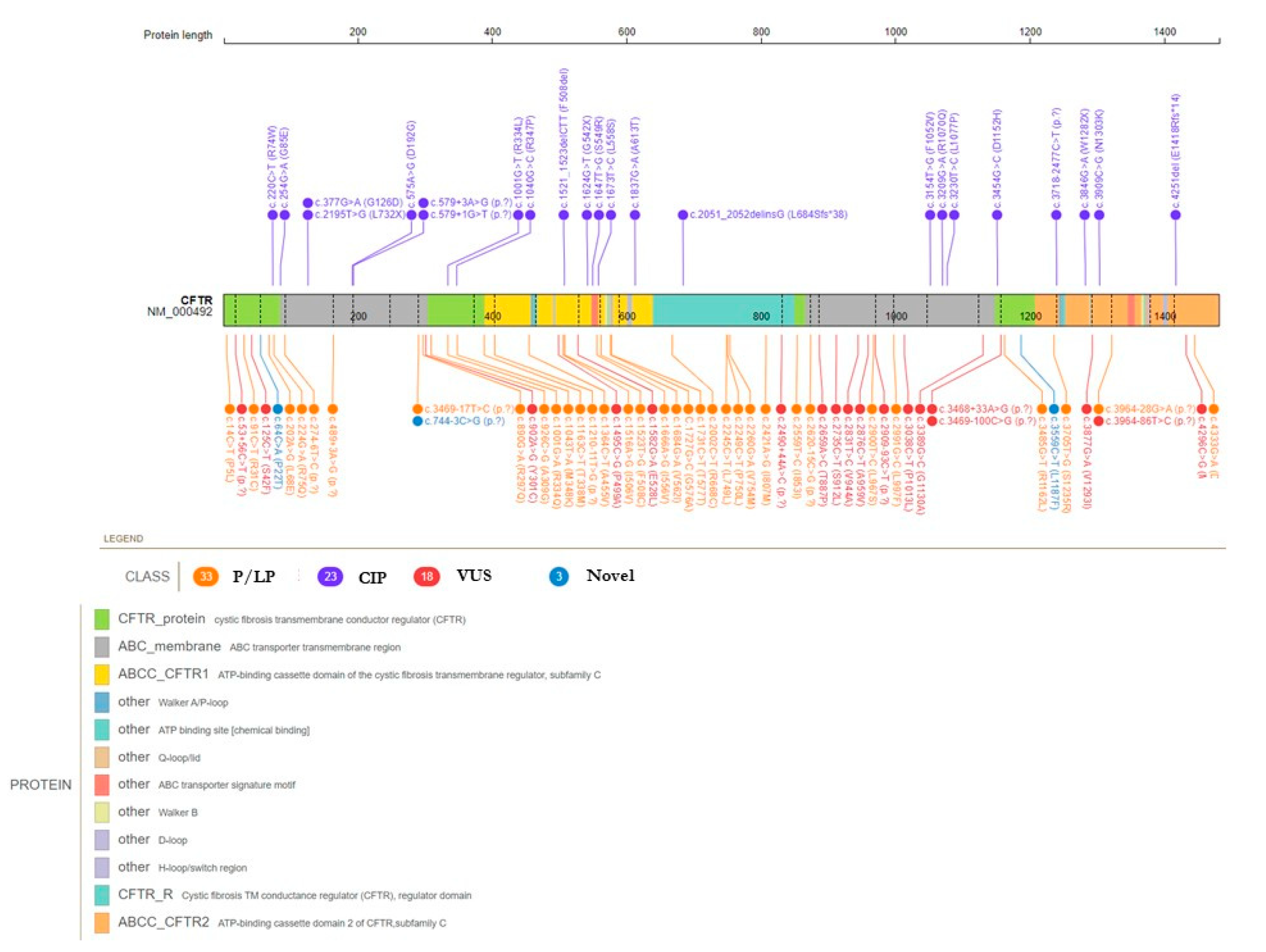

| HGVS cDNA change | Protein change | dbSNP |
|---|---|---|
| c.220C>T | p.(Arg74Trp) | rs115545701 |
| c.254G>A | p.(Gly85Glu) | rs75961395 |
| c.377G>A | p.(Gly126Asp) | rs397508609 |
| c.575A>G | p.(Asp192Gly) | rs397508758 |
| c.579+1G>T | p.(?) | rs77188391 |
| c.579+3A>G | p.(?) | rs397508761 |
| c.1001G>T | p.(Arg334Leu) | rs397508137 |
| c.1040G>C | p.(Arg347Pro) | rs77932196 |
| c.1521_1523delCTT | p.(Phe508del) | rs113993960 |
| c.1624G>T | p.(Gly542Ter) | rs113993959 |
| c.1647T>G | p.(Ser549Arg) | rs121909005 |
| c.1673T>C | p.(Leu558Ser) | rs193922504 |
| c.1837G>A | p.(Ala613Thr) | rs201978662 |
| c.2051_2052delinsG | p.(Lys684Serfs*38) | rs121908799 |
| c.2195T>G | p.(Leu732Ter) | rs397508609 |
| c.3154T>G | p.(Phe1052Val) | rs150212784 |
| c.3209G>A | p.(Arg1070Gln) | rs78769542 |
| c.3230T>C | p.(Leu1077Pro) | rs139304906 |
| c.3454G>C | p.(Asp1152His) | rs75541969 |
| c.3718-2477C>T | p.(?) | rs75039782 |
| c.3846G>A | p.(Trp1282Ter) | rs77010898 |
| c.3909C>G | p.(Asn1303Lys) | rs80034486 |
| c.4251del | p.(Glu1418Argfs*14) | rs397508706 |
| HGVS cDNA change | Protein change | dbSNP |
|---|---|---|
| c.14C>T | p.(Pro5Leu) | rs193922501 |
| c.91C>T | p.(Arg31Cys) | rs1800073 |
| c.202A>G | p.(Lys68Glu) | rs397508332 |
| c.224G>A | p.(Arg75Gln) | rs1800076 |
| c.274-6T>C | p.(?) | rs371315549 |
| c.489+3A>G | p.(?) | rs377729736 |
| c.890G>A | p.(Arg297Gln) | rs143486492 |
| c.926C>G | p.(Ala309Gly) | rs397508818 |
| c.1001G>A | p.(Arg334Gln) | rs397508137 |
| c.1043T>A | p.(Met348Lys) | rs142920240 |
| c.1163C>T | p.(Thr338Met) | rs143860237 |
| c.1210-11T>G | p.(?) | rs73715573 |
| c.1364C>T | p.(Ala455Val) | rs74551128 |
| c.1516A>G | p.(Ile506Val) | rs1800091 |
| c.1523T>G | p.(Phe508Cys) | rs74571530 |
| c.1666A>G | p.(Ile556Val) | rs75789129 |
| c.1684G>A | p.(Val562Ile) | rs1800097 |
| c.1731C>T | p.(Tyr577=) | rs55928397 |
| c.1727G>C | p.(Gly576Ala) | rs1800098 |
| c.2002C>T | p.(Arg668Cys) | rs1800100 |
| c.2245C>T | p.(Leu749Leu) | rs151235408 |
| c.2249C>T | p.(Pro750Leu) | rs140455771 |
| c.2260G>A | p.(Val754Met) | rs150157202 |
| c.2421A>G | p.(Ile807Met) | rs1800103 |
| c.2559T>C | p.(Ile853Ile) | rs1800104 |
| c.2620-15C>G | p.(?) | rs139379077 |
| c.2900T>C | p.(Leu967Ser) | rs1800110 |
| c.2991G>C | p.(Leu997Phe) | rs1800111 |
| c.3469-17T>C | p.(?) | rs79718042 |
| c.3485G>T | p.(Arg1162Leu) | rs1800120 |
| c.3705T>G | p.(Ser1235Arg) | rs34911792 |
| c.3964-28G>A | p.(?) | rs397508651 |
| c.4333G>A | p.(Asp1445Asn) | rs148783445 |
| HGVS cDNA change | Protein change | N° of carriers | db SNP | CFTR-Francea | CFTR2b | LOVDc | InterVard | Varsomee |
|---|---|---|---|---|---|---|---|---|
| c.125C>T | p.(Ser42Phe) | 3 | rs143456784 | VUS | n/a | P/VUS | VUS | VUS |
| c.902A>G | p.(Tyr301Cys) | 1 | rs150691494 | VUS | n/a | VUS | VUS | VUS |
| c.1495C>G | p.(Pro499Ala) | 1 | rs397508219 | n/a | n/a | n/a | VUS | LP |
| c.1582G>A | p.(Glu528Lys) | 1 | rs773018372 | n/a | n/a | n/a | VUS | VUS |
| c.2659A>C | p.(Thr887Pro) | 1 | rs770359007 | n/a | n/a | n/a | LB | VUS |
| c.2735C>T | p.(Ser912Leu) | 1 | rs121909034 | VUS | VUS | VUS | B | LB |
| c.2831T>C | p.(Val944Ala) | 1 | rs141747560 | n/a | n/a | n/a | VUS | LP |
| c.2876C>T | p.(Ala959Val) | 1 | rs397508448 | VUS | n/a | n/a | VUS | LP |
| c.3038C>T | p.(Pro1013Leu) | 1 | rs193922516 | VUS | n/a | VUS | VUS | LP |
| c.3389G>C | p.(Gly1130Ala) | 1 | rs397508550 | n/a | n/a | n/a | VUS | LP |
| c.3468+33A>G | p.(?) | 1 | rs1792459342 | n/a | n/a | n/a | n/a | VUS |
| c.3877G>A | p.(Val1293Ile) | 1 | rs769931559 | n/a | n/a | n/a | VUS | LP |
| c.4296C>G | p.(Asn1432Lys) | 1 | rs761669740 | n/a | n/a | n/a | LB | LP |
| c.53+56C>T (IVS1+56C>T) | p.(?) | 1 | rs140393487 | n/a | n/a | n/a | n/a | LB |
| c.2490+44A>C (IVS14+44A>C) | p.(?) | 1 | rs375692108 | n/a | n/a | n/a | n/a | LB |
| c.2909-93C>T (IVS17-93C>T) | p.(?) | 3 | rs144455881 | n/a | n/a | n/a | n/a | LB |
| c.3469-100C>G (IVS21-100C>G) | p.(?) | 2 | rs946757675 | n/a | n/a | n/a | n/a | LB |
| c.3964-86T>C (IVS24-86T>C) | p.(?) | 1 | rs1340773814 | n/a | n/a | n/a | n/a | LB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





