Submitted:
24 July 2023
Posted:
25 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
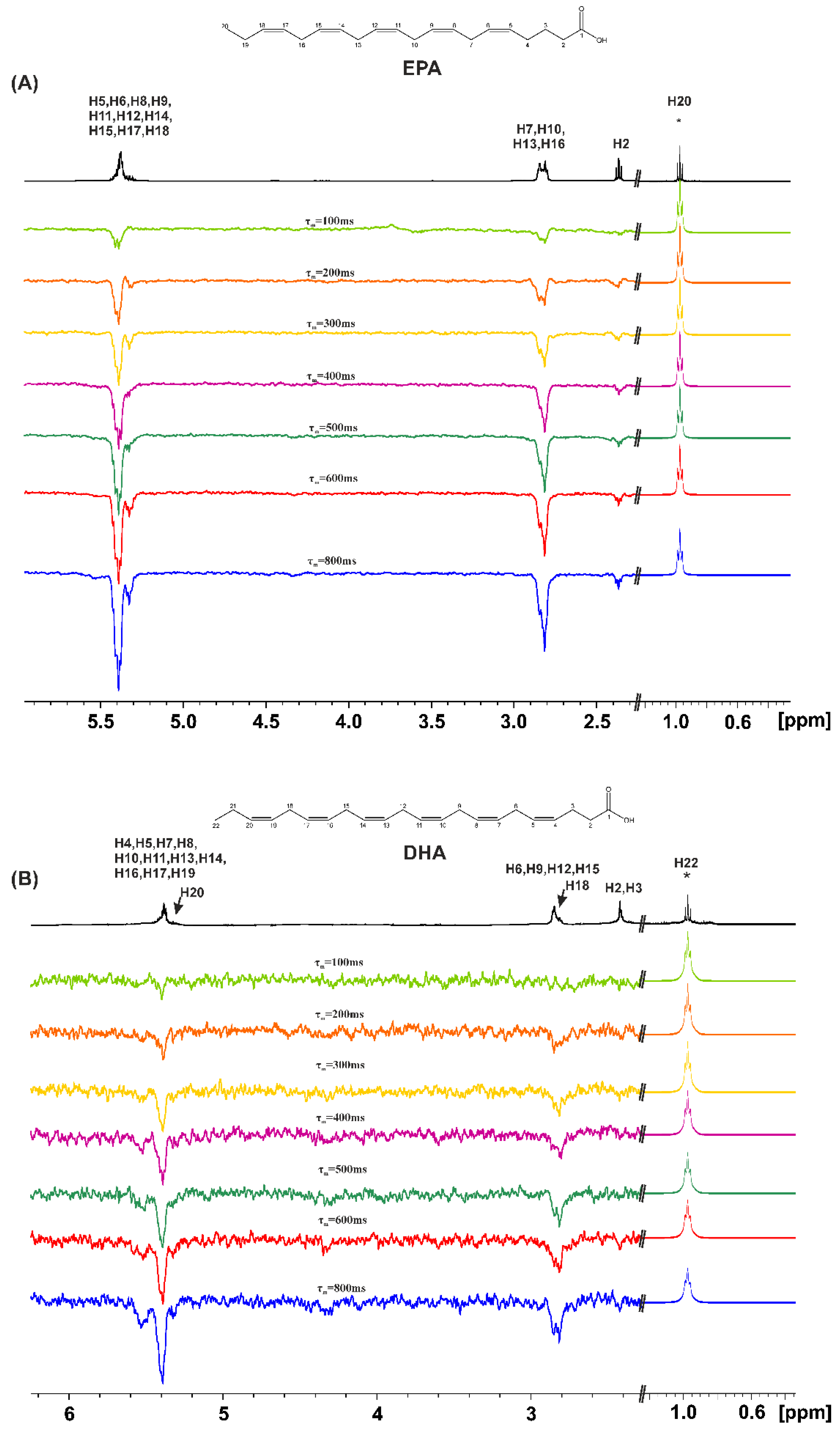
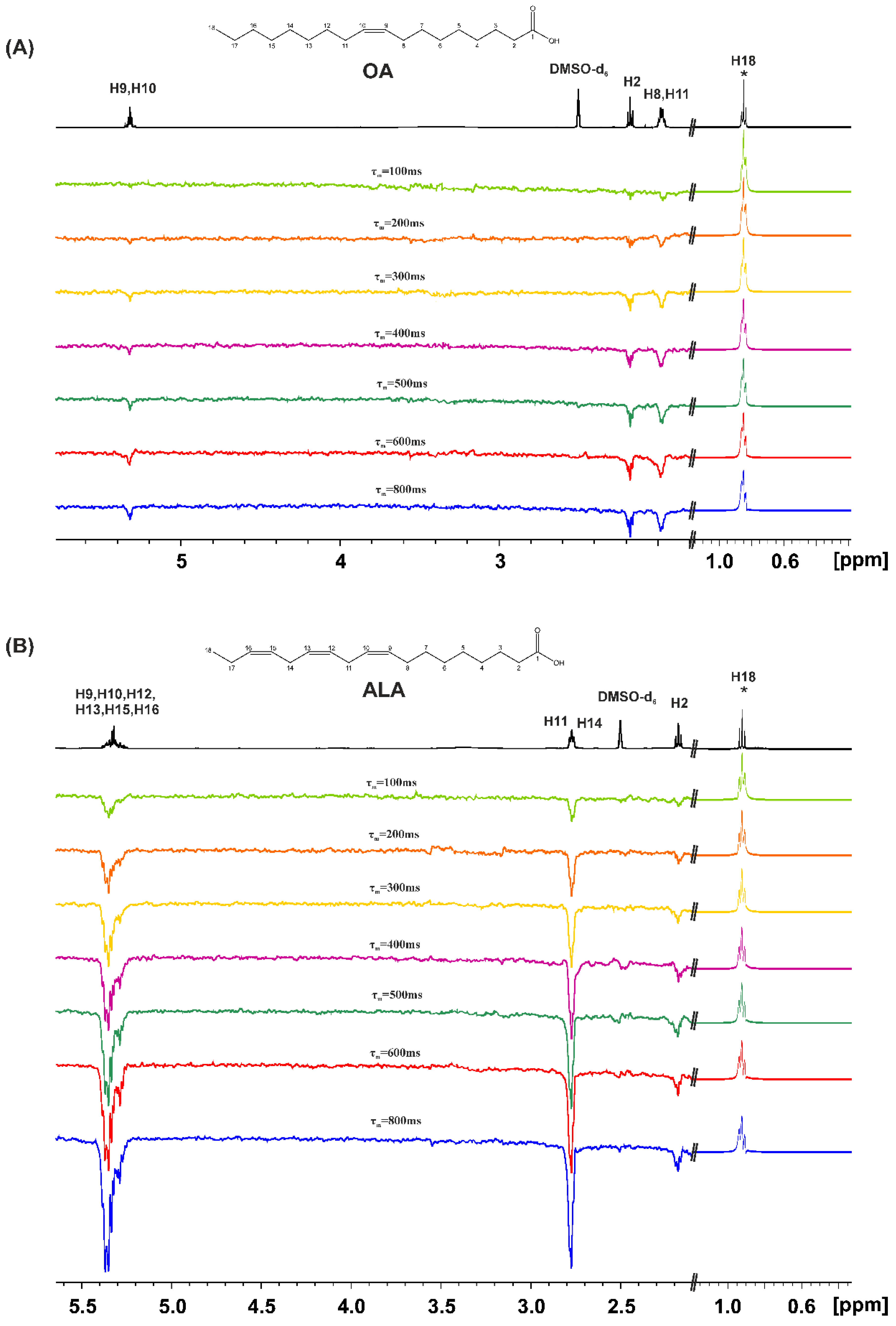
2.1. Variable Temperature and Concentration 1H NMR Chemical Shifts of Carboxylic Protons and 1D 1H NMR Transient NOE in CDCl3
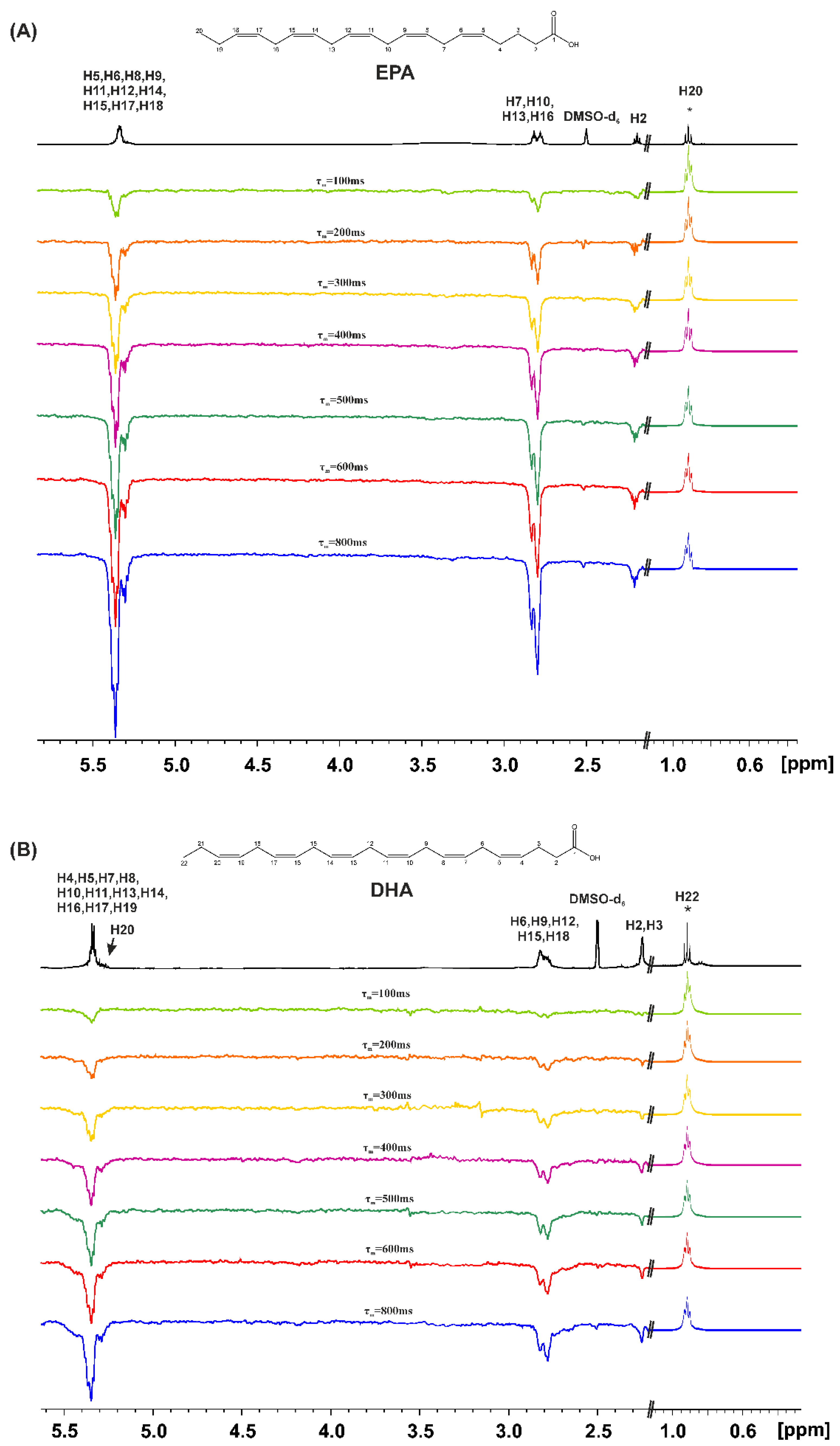
2.2. Variable Temperature 1H NMR Chemical Shifts of Carboxylic Protons and 1D 1H NMR Transient NOE in DMSO-d6
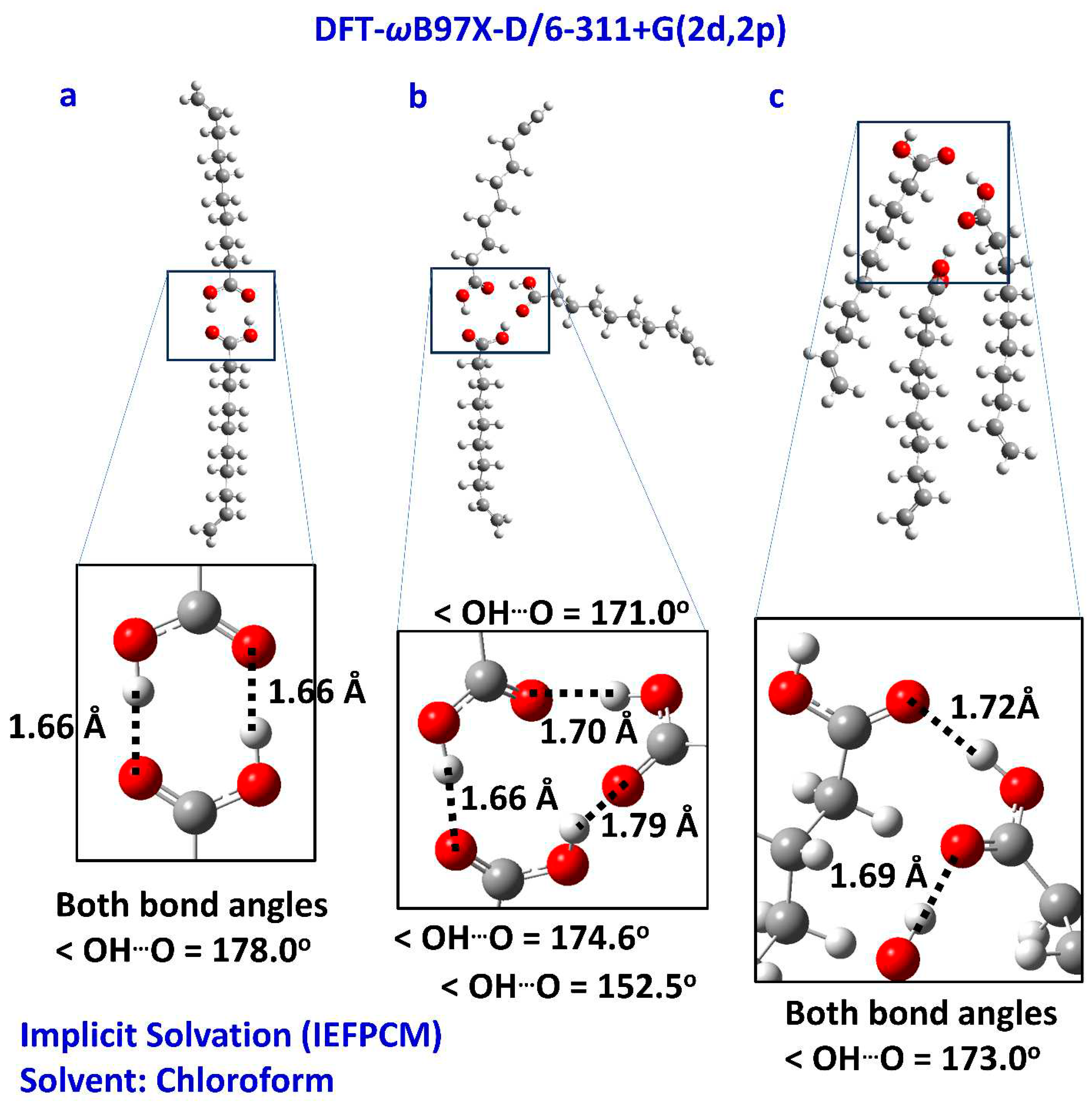
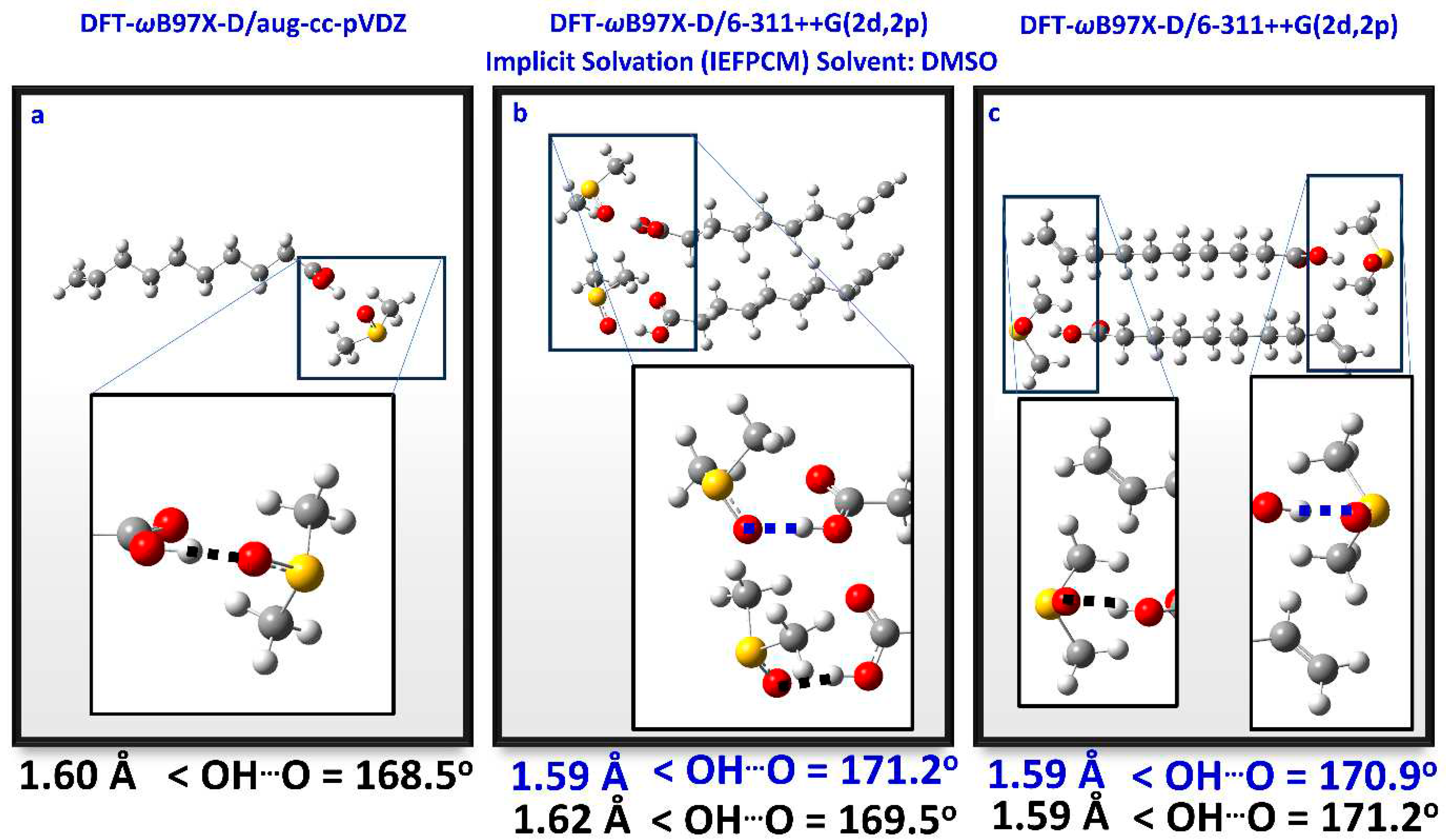
2.3. DFT Calculations in CHCl3 – Comparison with the Liquid State
2.4. DFT Calculations in DMSO
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Variable Temperature and Concentration 1H NMR Chemical Shifts and 1D 1H NMR Transient NOE
3.3. DFT Calculations of 1H NMR Chemical Shifts and Complexation Energies
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Gunstone, F.D. Fatty acid and lipid chemistry, 1st ed.; Springer: New York, NY, USA, 1996. [Google Scholar]
- Vance, D.E.; Vance, J.E. Biochemistry of lipids, lipoproteins and membranes (new comprehensive biochemistry), 5th ed.; Elsevier: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Akoh, C.C.; Min, D.B. Food lipids, chemistry, nutrition and biochemistry, 2nd ed.; Marcel Dekker Inc.: New York, NY, USA, 2002. [Google Scholar]
- Leray, C. Dietary lipids for healthy brain function; CRC Press: USA, 2021. [Google Scholar]
- Miles, E.A.; Calder, P.C. Modulation of immune function by dietary fatty acids. Proc. Nutr. Soc. 1998, 57, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y. Talukdar, S. Bae, E.J. Imamura, T. Morinaga, H. Fan, W. Li, P. Lu, W.J. Watkins, S.M. Olefsky, J.M. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell 2010, 142, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Simopoulos, A.P. The importance of the omega-6/omega-3 fatty acid ratio in cardiovascular disease and other chronic diseases. Exp. Biol. Med. 2008, 233, 674–688. [Google Scholar] [CrossRef]
- Gupta, R.; Lakshmy, R.; Abraham, R.A.; Reddy, K.S.; Jeemon, P.; Prabhakaran, D. Serum omega-6/omega-3 ratio and risk markers for cardiovascular disease in an industrial population of Delhi, Food Nutr. Sci. 2013, 4, 94–97. [Google Scholar] [CrossRef]
- Blasbalg, T.L.; Hibbeln, J.R.; Ramsden, C.E.; Majchrzak, S.F.; Rawlings, R.R. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century, Am. J. Clin. Nutr. 2011, 93, 950–962. [Google Scholar] [CrossRef]
- Sacchi, Ρ.; Medina, Ι.; Paolillo, L.; Addeo, F. High-resolution 13C-NMR olefinic spectra of DHA and EPA acids, methyl esters and triacylglycerols, Chem. Phys. Lipids 1994, 69, 65–73. [Google Scholar] [CrossRef]
- Aursand, M.; Grasdalen, H. Interpretation of the 13C-NMR spectra of omega-3 fatty acids and lipid extracted from the white muscle of Atlantic salmon (Salmo salar), Chem. Phys. Lipids 1992, 62, 239–251. [Google Scholar] [CrossRef]
- Gunstone, F.D.; Seth, S.; Wolff, R.L. The distribution of Δ5 polyene acids in some pine seed oils between the α- and β-chains by 13C-NMR spectroscopy, Chem. Phys. Lipids 1995, 78, 89–96. [Google Scholar] [CrossRef]
- Gunstone, F.D.; Seth, S. A study of the distribution of eicosapentaenoic acid and docosahexaenoic acid between the α and β glycerol chains in fish oils by 13C-NMR spectroscopy, Chem. Phys. Lipids 1994, 72, 119–126. [Google Scholar] [CrossRef]
- Alexandri, E.; Ahmed, R.; Sidiqui, H.; Choudhary, M.I.; Tsiafoulis, C.G.; Gerothanassis, I.P. High resolution NMR spectroscopy as a structural and analytical tool of unsaturated lipids in solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef]
- Applegate, K.R.; Glomset, J.A. Computer-based modeling of the conformation and packing properties of docosahexanoic acid, J. Lipid Res. 1986, 27, 658–680. [Google Scholar] [CrossRef]
- Smith, P.; Lorenz, C.D. LiPyphylic: A python toolkit for the analysis of lipid membrane simulations, J. Chem. Theory Comput. 2021, 17, 5907–5919. [Google Scholar] [CrossRef] [PubMed]
- Manna, M.; Nieminen, T.; Vattulainen, I. Understanding the role of lipids in signaling through atomistic and multiscale simulations of cell membranes, Annu. Rev. Biophys. 2019, 48, 421–439. [Google Scholar] [CrossRef] [PubMed]
- Alexandri, E.; Primikyri, A.; Papamokos, G.; Venianakis, T.; Gkalpinos, V.G.; Tzakos, A.G.; Karydis-Messinis, A.; Moschovas, D.; Avgeropoulos, A.; Gerothanassis, I.P. NMR and computational studies reveal novel aspects in molecular recognition of unsaturated fatty acids with non-labeled serum albumin. FEBS J. 2022, 289, 5617–5636. [Google Scholar] [CrossRef]
- Alexandri, E.; Venianakis, T.; Primikyri, A.; Papamokos, G.; Gerothanassis, I.P. Molecular basis for the selectivity of DHA and EPA in Sudlow’s drug binding sites in human serum albumin with the combined use of NMR and docking calculations. Molecules 2023, 28, 3724. [Google Scholar] [CrossRef] [PubMed]
- Eldho, N.V.; Feller, S.E.; Tristram-Nagle, S.; Polozov, I.V.; Gawrisch, K. Polyunsaturated docosahexaenoic vs docosapentaenoic acids differences in lipid matrix properties from the loss of one double bond, J. Am. Chem. Soc. 2003, 125, 6409–6421. [Google Scholar] [CrossRef]
- Law, J.M.S.; Szori, M.; Izsak, R.; Pekne, B.; Csizmadia, I.G.; Viskolcz, B. Folded and unfolded conformations of the ω-3 polyunsaturated fatty acid family: CH3CH2[CH=CHCH2]B[CH2]MCOOH. First principles study, J. Phys. Chem. A 2006, 110, 6100–6111. [Google Scholar] [CrossRef]
- Iwahashi, M.; Kasahara, Y.; Matsuzawa, H.; Yagi, K.; Nomura, K.; Terauchi, H.; Ozaki, Y.; Susuki, M. Self-diffusion, dynamical molecular conformation and liquid structures of n-saturated and unsaturated fatty acids, J. Phys. Chem. B 2000, 104, 6186–6194. [Google Scholar] [CrossRef]
- Iwahashi, M.; Kasahara, Y. Dynamic molecular movements and aggregation structures of lipids in a liquid state, Curr. Opin. Colloid Interface Sci. 2011, 16, 359–366. [Google Scholar] [CrossRef]
- Broadhurst, C.L.; Schmidt, W.F.; Nguyen, J.K.; Qin, J.; Chao, K.; Aubuchon, S.R.; Kim, M.S. Continuous gradient temperature Raman spectroscopy and differential scanning calorimetry of N-3DPA and DHA from -100 to 10oC, Chem. Phys. Lipids 2017, 204, 94–104. [Google Scholar] [CrossRef]
- Schmidt, W.F.; Chen, F.; Broadhurst, C.L.; Nguyen, J.K.; Qin, J.; Chao, K.; Kim, M.S. GTRS and 2D-NMR studies of alpha and gamma linolenic acids each containing the same H2C14-(H–C=C–H)–C11H2–(H–C=C–H)–C8H2 moiety. J. Mol. Struct. 2019, 1196, 258–270. [Google Scholar] [CrossRef]
- Bagheri Novir, S.; Tirandaz, A.; Lotfipour, H. Quantum study of DHA, DPA and EPA anticancer fatty acids for microscopic explanation of their biological functions, J. Mol. Liq. 2021, 325, 114646. [Google Scholar] [CrossRef]
- Venianakis, T.; Primikyri, A.; Alexandri, E.; Papamokos, G.; Gerothanassis, I.P. Molecular models of three ω-3 fatty acids based on NMR and DFT calculations of 1H NMR chemical shifts, J. Mol. Liq. 2021, 342, 117460. [Google Scholar] [CrossRef]
- Venianakis, T.; Siskos, M.; Papamokos, G.; Gerothanassis, I.P. NMR and DFT studies of monounsaturated and ω-3 polyunsaturated free fatty acids in the liquid state reveal a novel atomistic structural model of DHA, J. Mol. Liq. 2023, 376, 121459. [Google Scholar] [CrossRef]
- Neratzaki, A.A.; Tsiafoulis, C.G.; Charisiadis, P.; Kontogianni, V.G.; Gerothanassis, I.P. Novel determination of the total phenolic contents in crude plant extracts by the use of 1H NMR of the ‒OH spectral region, Anal. Chim. Acta 2011, 688, 54–60. [Google Scholar] [CrossRef]
- Siskos, M.G.; Tzakos, A.G.; Gerothanassis, I.P. Accurate ab initio calculations of O–H⋯O and O–H⋯-O proton chemical shifts: towards elucidation of the nature of the hydrogen bond and prediction of hydrogen bond distances, Org. Biomol. Chem. 2015, 13, 8852–8868. [Google Scholar] [CrossRef]
- Siskos, M.G.; Iqbal Choudhary, M.; Gerothanassis, I.P. Hydrogen atomic positions of O–H···O hydrogen bonds in solution and in the solid state: the synergy of quantum chemical calculations with 1H-NMR chemical shifts and X-ray diffraction methods, Molecules 2017, 22, 415. [Google Scholar] [CrossRef]
- Yamamoto, S.; Matsuda, H.; Kasahara, Y.; Iwahashi, M.; Takagi, T.; Bada, T.; Kanamori, T. Dynamic molecular behavior of semi-fluorinated oleic, elaidic and stearic acids in the liquid state, J. Oleo Sci. 2012, 61, 649–657. [Google Scholar] [CrossRef]
- Goldman, M.A.; Emerson, M.T. Hydrogen-bonded species of acetic acid in inert solvents, J. Phys. Chem. 1973, 77, 2295–2299. [Google Scholar] [CrossRef]
- Marechal, Y. H-bonded open and cyclic dimers in the gas phase, J. Mol. Struct. 1988, 189, 55–63. [Google Scholar] [CrossRef]
- Tjahjono, M.; Cheng, S.; Li, C.; Garland, M. Self-association of acetic acid in dilute deuterated chloroform. wide-range spectral reconstructions and analysis using FTIR spectroscopy, BTEM, and DFT, J. Phys. Chem. A 2010, 114, 12168–12175. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.I. Competing intramolecular vs. intermolecular hydrogen bonds in solution. Int. J. Mol. Sci. 2014, 15, 19562–19633. [Google Scholar] [CrossRef] [PubMed]
- Issaoui, N.; Ghalla, H.; Brandan, S.A.; Bardak, F.; Flakus, H.I.; Atac, A.; Oujia, B. Experimental FTIR and FT-Raman and theoretical studies on the molecular structures of monomer and dimer of 3-thiopheneacrylic acid, J. Mol. Struct. 2017, 1135, 209–221. [Google Scholar] [CrossRef]
- Lengvinaitė, D.; Aidas, K.; Kimtys, L. Molecular aggregation in liquid acetic acid: insight from molecular dynamics/quantum mechanics modelling of structural and NMR properties, Phys. Chem. Chem. Phys. 2019, 21, 14811–14820. [Google Scholar] [CrossRef]
- Jozwiak, K.; Jezierska, A.; Panek, J.J.; Goremychkin, E.A.; Tolstoy, P.M.; Shenderovich, I.G.; Filarowski, A. Inter- vs. intramolecular hydrogen bond patterns and proton dynamics in nitrophthalic acid associates, Molecules 2020, 25, 4720. [Google Scholar] [CrossRef]
- Ernst, J.; Sheldrick, W.S.; Fuhrhop, J.-H. The structures of the essential unsaturated fatty acids. Crystal structures of linoleic and evidence for the crystal structures of α-linolenic acid and arachidonic acid, Z. Naturforsch. 1979, 34b, 706–711. [Google Scholar] [CrossRef]
- Iwahashi, M.; Kasahara, Y.; Minami, H.; Matsazawa, H.; Susuki, M.; Ozaki, Y. Molecular behaviors of n-fatty acids in liquid state, J. Oleo Sci. 2002, 51, 157–164. [Google Scholar] [CrossRef]
- Takahashi, O.; Kohno, Y.; Nishio, M. Relevance of weak hydrogen bonds in the conformation of organic compounds and bioconjugates: evidence from recent experimental data and high-level ab initio MO calculations, Chem. Rev. 2010, 110, 6049–6076. [Google Scholar] [CrossRef]
- Maier, J.M.; Li, P.; Vik, E.C.; Yehl, C.J.; Strickland, S.M.S.; Shimizu, K.D. Measurement of solvent OH-pi interactions using a molecular balance, J. Am. Chem. Soc. 2017, 139, 6550–6553. [Google Scholar] [CrossRef]
- Kalra, K.; Gorle, S.; Cavallo, L.; Oliva, R.; Chawla, M. Occurrence and stability of lone pair-pi and OH-pi interactions between water and nucleobases in functional RNAs. Nucleic Acids Res. 2020, 48, 5825–5838. [Google Scholar] [CrossRef]
- Oku, K.; Watanabe, H.; Kubota, M.; Fukuda, S.; Kurimoto, M.; Tsujisaka, Y.; Komori, M.; Inoue, Y.; Sakurai, M. NMR and quantum chemical study on the OH···π and CH···O interactions between trehalose and unsaturated fatty acids: implication for the mechanism of antioxidant function of trehalose, J. Am. Chem. Soc 2003, 125, 12739–12748. [Google Scholar] [CrossRef] [PubMed]
- Wudarczyk, J.; Papamokos, G.; Marszalek, T.; Nevolianis, T.; Schollmeyer, D.; Pisula, W.; Floudas, G.; Baumgarten, M.; Mullen, K. Dicyanobenzothiadiazole derivatives possessing switchable dielectric permittivities, ACS Appl. Mater. Interf. 2017, 9, 20527–20535. [Google Scholar] [CrossRef] [PubMed]
- Pashkovskaya, A.A.; Vazdar, M.; Zimmermann, L.; Jovanovic, O.; Pohl, P.; Pohl, E.E. Mechanism of long-chain free fatty acid protonation at the membrane-water interface, Biophys. J. 2018, 114, 2142–2151. [Google Scholar] [CrossRef]
- Cross, B.P.; Schleich, T. Temperature dependence of the chemical shifts of commonly employed proton n. m.r. reference compounds, Org. Magn. Reson. 1977, 10, 82–85. [Google Scholar] [CrossRef]
- Hoffman, R.E. Standardization of chemical shifts of TMS and solvent signals in NMR solvents, Magn. Reson. Chem. 2006, 44, 606–616. [Google Scholar] [CrossRef]
- Kessler, H.; Oschkinat, H.; Griesinger, C.; Bermel, W. Transformation of homonuclear two-dimensional NMR techniques into one-dimensional techniques using Gaussian pulses, J. Magn. Reason. 1986, 70, 106–133. [Google Scholar] [CrossRef]
- Stott, K.; Stonehouse, J.; Keeler, J.; Hwang, T.-L.; Shaka, A. J. Excitation sculpting in high-resolution nuclear magnetic resonance spectroscopy: Application to selective NOE experiments, J. Am. Chem. Soc. 1995, 117, 4199–4200. [Google Scholar] [CrossRef]
- Neuhaus, D.; Williamson, P.M. The nuclear Overhauser effect in structural and conformational analysis; VHC Publishers: New York, NY, USA, 1989. [Google Scholar]
- Kontogianni, V.G.; Gerothanassis, I.P. Analytical and structural tools of lipid hydroperoxides: Present state and future perspectives, 2022, 27, 2139. Molecules 2022, 27, 2139. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Siskos, M.G.; Siddiqui, H.; Gerothanassis, I.P. DFT calculations of δ(13C) and (1H) chemical shifts and 3J(13C-O-O-1H) coupling constants as structural and analytical tools in hydroperoxides: prospects and limitations of 1H-13C HMBC experiments, Magn. Reson. Chem. 2022, 60, 970–984. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Systematic optimization of long-range corrected hybrid density functionals, J. Chem. Phys. 2008, 128, 084106. [Google Scholar] [CrossRef]
- Chai, J.D.; Head-Gordon, M. Long-range corrected hybrid density functionals with damped atom–atom dispersion corrections, Phys. Chem. Chem. Phys. 2008, 10, 6615–6620. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, J.R.; Trucks, G.W.; Keith, T.A.; Frisch, M.J. A Comparison of models for calculating nuclear magnetic resonance shielding tensors, J. Chem. Phys. 1996, 104, 5497–5509. [Google Scholar] [CrossRef]
- Boys, S.F.; Bernardi, F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors, Mol. Phys. 1970, 19, 553–566. [Google Scholar] [CrossRef]
- Tomasi, J.; Mennucci, B.; Cammi, R. Quantum mechanical continuum solvation models, Chem. Rev. 2005, 105, 2999–3094. [Google Scholar] [CrossRef]
- Bursch, M.; Mewes, J.-M.; Hansen, A.; Grimme, S. Best-practice DFT protocols for basic molecular computational chemistry, Angew. Chem. Int. Ed. 2022, 61, e202205735. [Google Scholar] [CrossRef]
- Papamokos, G.; Dimitriadis, T.; Bikiaris, D.N.; Papageorgiou, G.Z.; Floudas, G. Chain conformation, molecular dynamics, and thermal properties of poly(n-methylene 2,5-furanoates) as a function of methylene unit sequence length. Macromolecules 2019, 52, 6533–6546. [Google Scholar] [CrossRef]
- Gerothanassis, I.P. Ligand-observed in-tube NMR in natural products research: A review on enzymatic biotransformations, protein–ligand interactions, and in-cell NMR spectroscopy, Arab. J. Chem. 2023, 16, 104536. [Google Scholar] [CrossRef]
- Primikyri, A.; Papamokos, G.; Venianakis, T.; Sakka, M.; Kontogianni, V.; Gerothanassis, I.P. Structural basis of artemisinin binding sites in serum albumin with the combined use of nmr and docking calculations. Molecules 2022, 27, 5912. [Google Scholar] [CrossRef]
- Bhattacharya, A.A.; Grüne, Τ.; Curry, S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin, J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef]
- Petitpas, I.; Grüne, T.; Bhattacharya, A.A.; Curry, S. Crystal structures of human serum albumin complexed with monounsaturated and polyunsaturated fatty acids, J. Mol. Biol. 2001, 314, 955–960. [Google Scholar] [CrossRef]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin, J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]


| CDCl3 | DMSO-d6 | Liquid statea | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FFA | δ (ppm) |
R2 | Δδ/ΔΤ (ppb K-1) | Inter. | δ (ppm) |
R2 | Δδ/ΔΤ (ppb K-1) | Inter. | δ (ppm) |
R2 | Δδ/ΔΤ (ppb K-1) | Inter. |
| CA | 11.08 | 0.999 | -32.69 | 20.81 | 11.94 | 0.999 | -6.62 | 13.92 | 12.25 | 0.999 | -11.31 | 15.98 |
| OA | 11.17 | 0.999 | -31.50 | 20.54 | 11.94 | 0.999 | -6.88 | 13.99 | 12.13 | 0.998 | -10.32 | 15.21 |
| ALA | 10.39 | 0.998 | -42.74 | 23.13 | 11.95 | 0.999 | -6.79 | 13.97 | 10.88 | 0.998 | -13.06 | 14.76 |
| EPA | 10.77 | 0.997 | -35.41 | 21.31 | 12.01 | 0.997 | -7.27 | 14.18 | 10.91 | 0.999 | -14.38 | 14.19 |
| DHA | 9.07 | 0.992 | -29.52 | 17.90 | 12.08 | 0.993 | -6.45 | 14.00 | 8.60 | 0.986 | -16.43 | 13.51 |
| FFA | Intermolecular interaction | δ(COOH) (ppm) |
|---|---|---|
| CA dimer | COO-H…O=COH | 13.6 |
| CA cyclic trimer | COO-H…O=COH | 12.9/11.2/10.7 |
| CA linear trimer | COO-H…O=COH COOH (free) |
12.2/12.2 6.8 |
| CA tetramer parallel | COO-H…O=COH | 14.3, 14.0, 13.7, 13.0 |
| CA tetramer antiparallel | COO-H…O=COH | 13.8, 13.8, 13.8, 13.2 |
| FFA | Intermolecular interaction |
δ(COOH) (ppm) |
|---|---|---|
| CA | COO-H…DMSO | 13.6 |
| CA dimer parallel | COO-H… DMSO | 14.3, 14.0, 13.7, 13.0 |
| CA dimer antiparallel | COO-H… DMSO | 13.8, 13.8, 13.8, 13.2 |
| ALA | COO-H… DMSO | 13.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





