Submitted:
24 July 2023
Posted:
25 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. CrNAKG expression is dependent on nitrogen source and growth phase
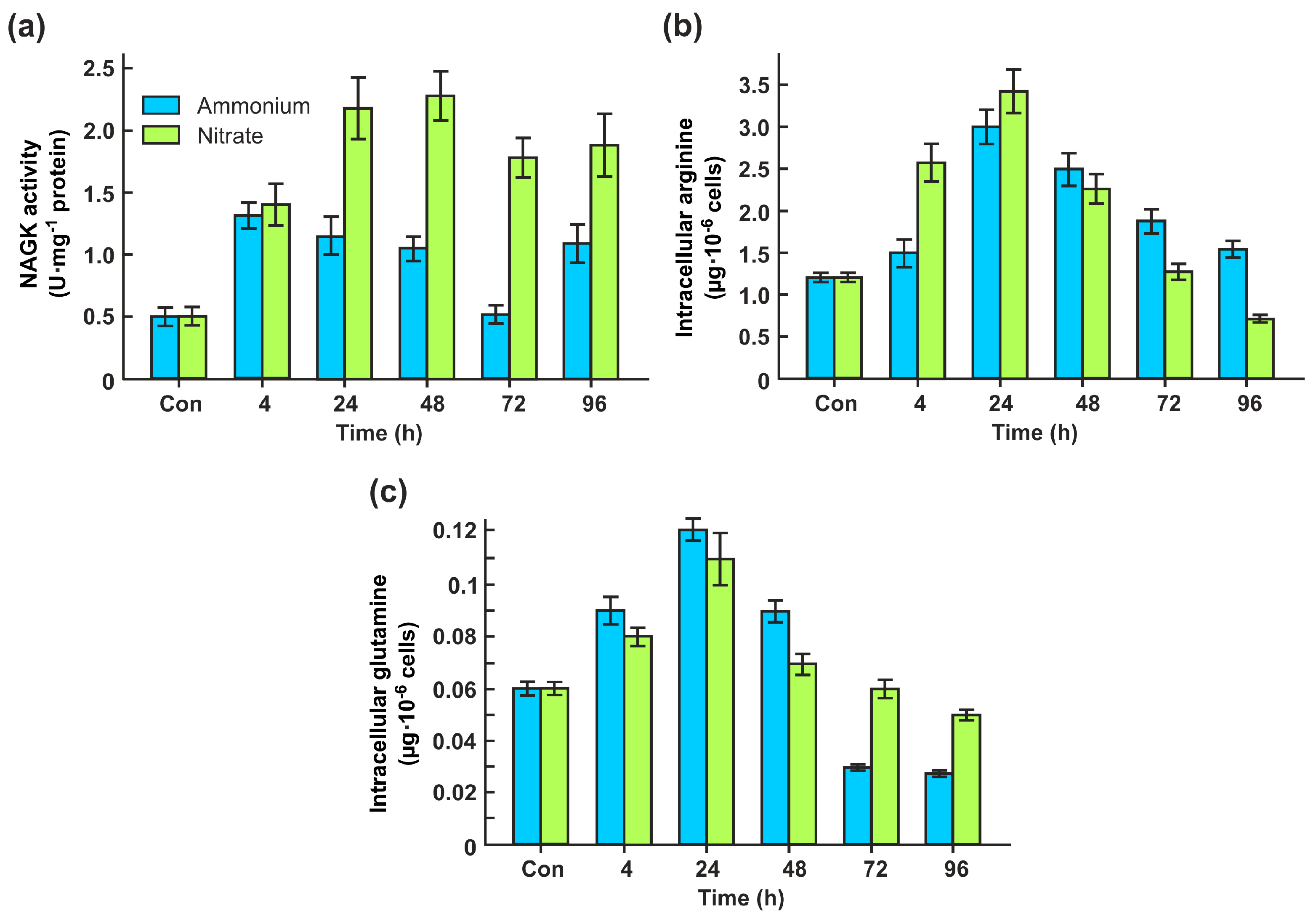
2.2. CrNAKG activity is dependent on nitrogen source
2.3. Nitrite promotes CrNAGK activity
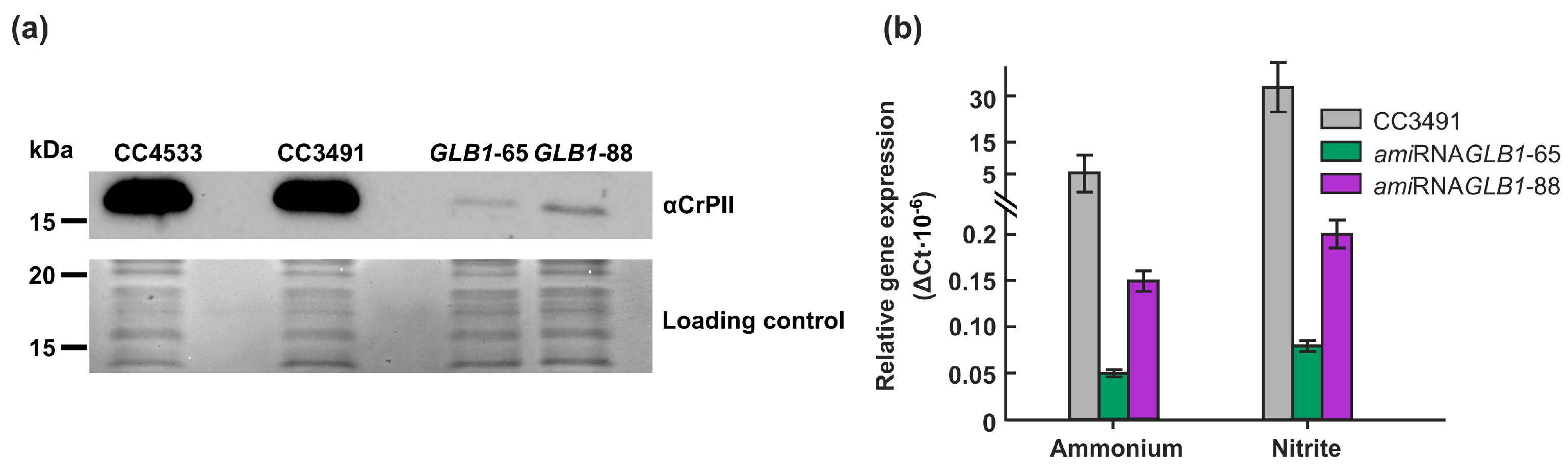
2.4. Underexpression of CrPII decreases the activity of CrNAGK but not the CrNAGK1 transcript level
2.5. CrNAGK activity is changed in N-starved cells
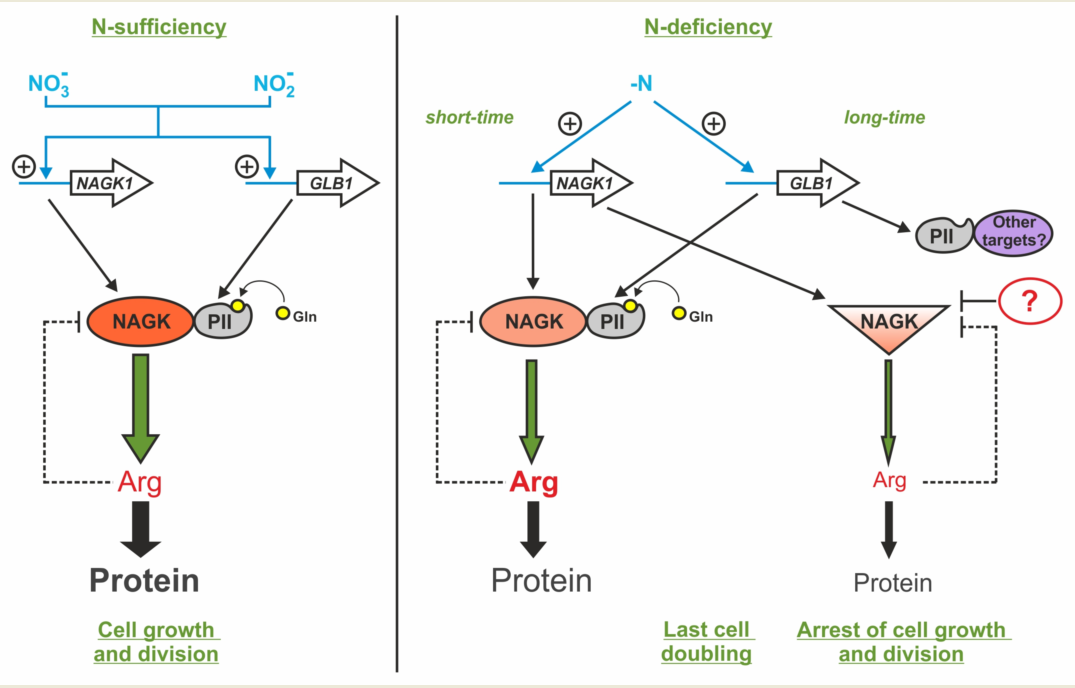
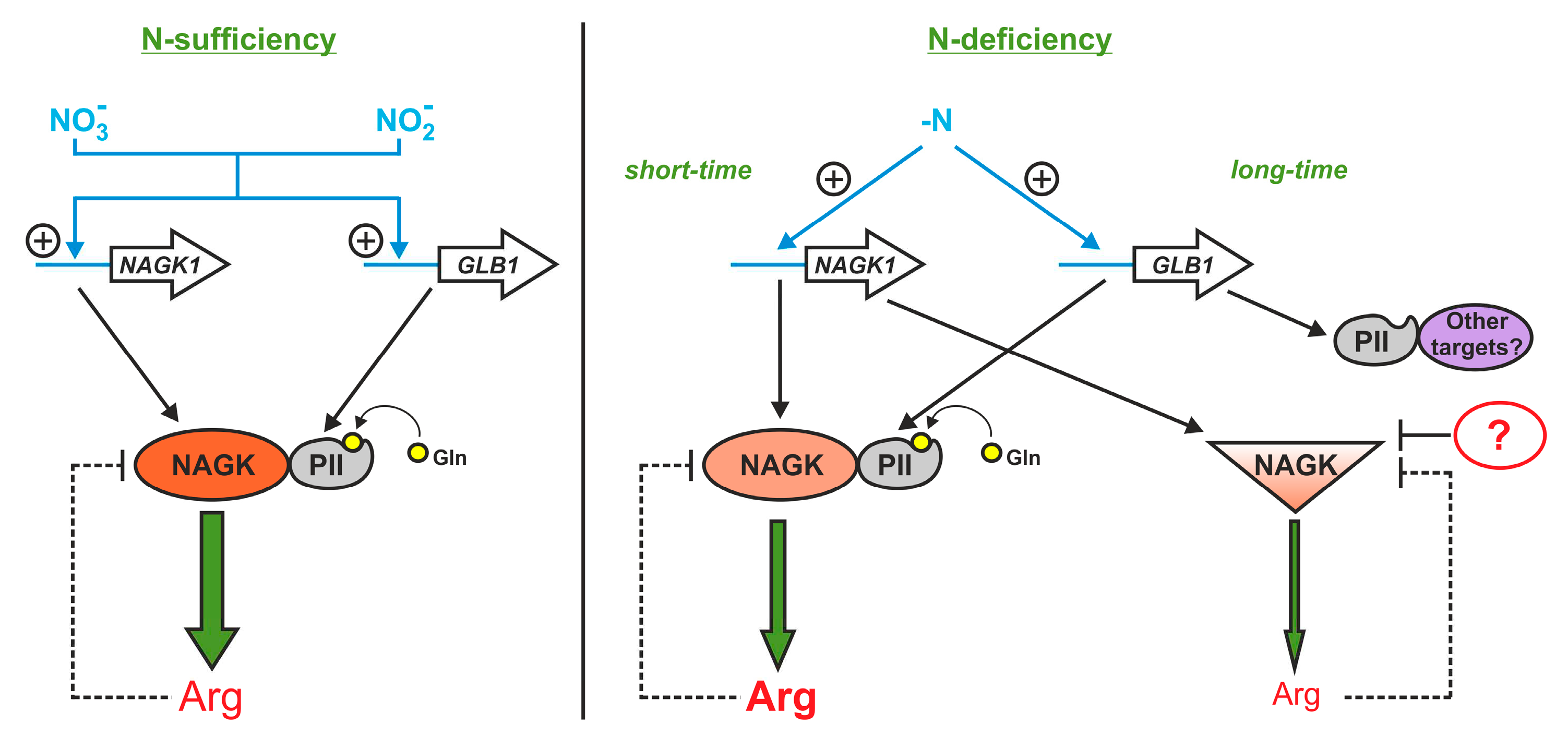
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Conditions
4.2. Generation of GLB1-underexpressing transformants
4.3. Quantitative real-time PCR
4.4. Protein isolation, SDS-PAGE, and Western blotting
4.5. NAGK activity assays
4.6. Determination of glutamine content
4.7. Determination of arginine content
4.8. Statistical analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Llácer, J.L.; Fita, I.; Rubio, V. Arginine and nitrogen storage. Curr. Opin. Struct. Biol. 2008, 18, 673–681. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef]
- Klimaszewska, K.; Morency, F.; Jones-Overton, C.; Cooke, J. Accumulation pattern and identification of seed storage proteins in zygotic embryos of Pinus strobus and in somatic embryos from different maturation treatments. Physiol. Plant 2004, 121, 682–690. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M.; del Río, L.A.; Barroso, J.B. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009, 184, 9–14. [Google Scholar] [CrossRef]
- Beez, S.; Fokina, O.; Herrmann, C.; Forchhammer, K. N-acetyl-L-glutamate kinase (NAGK) from oxygenic phototrophs: P(II) signal transduction across domains of life reveals novel insights in NAGK control. J. Mol. Biol. 2009, 389, 748–758. [Google Scholar] [CrossRef]
- Lapina, T.; Selim, K.A.; Forchhammer, K.; Ermilova, E. The PII signaling protein from red algae represents an evolutionary link between cyanobacterial and Chloroplastida PII proteins. Sci. Rep. 2018, 8, 790. [Google Scholar] [CrossRef]
- Sugiyama, K.; Hayakawa, T.; Kudo, T.; Ito, T.; Yamaya, T. Interaction of N-acetylglutamate kinase with a PII-like protein in rice. Plant Cell Physiol. 2004, 45, 1768–1778. [Google Scholar] [CrossRef]
- Chen, Y.M.; Ferrar, T.S.; Lohmeier-Vogel, E.M.; Morrice, N.; Mizuno, Y.; Berenger, B.; Ng, K.K.; Muench, D.G.; Moorhead, G.B. The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetyl glutamate kinase. J. Biol. Chem. 2006, 281, 5726–5733. [Google Scholar] [CrossRef]
- Minaeva, E.; Forchhammer, K.; Ermilova, E. Glutamine assimilation and feedback regulation of L-acetyl-N-glutamate kinase activity in Chlorella variabilis NC64A results in changes in arginine pools. Protist 2015, 166, 493–505. [Google Scholar] [CrossRef]
- Chellamuthu, V.R.; Ermilova, E.; Lapina, T.; Lüddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A widespread glutamine-sensing mechanism in the plant kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef]
- Selim, K.; Ermilova, E.; Forchhammer, K. From cyanobacteria to Archaeplastida: new evolutionary insights into PII signalling in the plant kingdom. New Phytol. 2020, 227, 722–731. [Google Scholar] [CrossRef]
- Heinrich, A.; Maheswaran, M.; Ruppert, U.; Forchhammer, K. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol. Microbiol. 2004, 52, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.A.; Lapina, T.; Forchhammer, K.; Ermilova, E. Interaction of N-acetyl-L-glutamate kinase with the PII signal transducer in the non-photosynthetic alga Polytomella parva: co-evolution towards a hetero-oligomeric enzyme. FEBS J. 2020, 287, 465–482. [Google Scholar] [CrossRef]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.; Huergo, L. New views on PII signaling: from nitrogen sensing to global metabolic control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Chellamuthu, V.R.; Alva, V.; Forchhammer, K. From cyanobacteria to plants: conservation of PII functions during plastid evolution. Planta 2013, 237, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, W.; Sun, L.P.; Zhou, Z.G. Evidence for PII with NAGK interaction that regulates Arg synthesis in the microalga Myrmecia incisa in response to nitrogen starvation. Sci. Rep. 2017, 7, 16291. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.; Galván, A. Inorganic nitrogen assimilation in Chlamydomonas. J. Exp. Bot. 2007, 58, 2279–2287. [Google Scholar] [CrossRef]
- Sanz-Luque, E.; Chamizo-Ampudia, A.; Llamas, A.; Galván, A.; Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 2015, 6, 899. [Google Scholar] [CrossRef]
- Tejada-Jimenez, M.; Llamas, A.; Galván, A.; Fernández, E. Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plants 2019, 8, 56. [Google Scholar] [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Sanz-Luque, E.; Tejada-Jiménez, M.; Llamas, Á; Plouviez, M. ; Guieysse, B.; Fernández, E.; Galván, A. Chlamydomonas reinhardtii, an algal model in the nitrogen cycle. Plants 2020, 9, 903. [Google Scholar] [PubMed]
- Ermilova, E.; Lapina, T.; Zalutskaya, Z.; Minaeva, E.; Fokina, O.; Forchhammer, K. PII signal transduction protein in Chlamydomonas reinhardtii: localization and expression pattern. Protist 2013, 164, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-J.; Wang, H.; Gargouri, M.; Deshpande, R.R.; Skepper, J.N.; Holguin, F.O.; Juergens, M.T.; Shachar-Hill, Y.; Hicks, L.M.; Gang, D.R. The response of Chlamydomonas reinhardtii to nitrogen deprivation: A systems biology analysis. Plant J. 2015, 81, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Wase, N.; Black, P.N.; Stanley, B.A.; DiRusso, C.C. Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J. Proteome Res. 2014, 13, 1373–1396. [Google Scholar] [CrossRef]
- Monteiro, L.d.F.R.; Giraldi, L.A.; Winck, F.V. From Feasting to Fasting: The Arginine Pathway as a Metabolic Switch in Nitrogen-Deprived Chlamydomonas reinhardtii. Cells 2023, 12, 1379. [Google Scholar] [CrossRef]
- Zalutskaya, Z.; Kochemasova, L.; Ermilova, E. Dual positive and negative control of Chlamydomonas PII signal transduction protein expression by nitrate/nitrite and NO via the components of nitric oxide cycle. BMC Plant Biol. 2018, 18, 305. [Google Scholar] [CrossRef]
- Forchhammer, K.; Tandeau de Marsac, N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 1994, 176, 84–91. [Google Scholar] [CrossRef]
- Bellido-Pedraza, C.M.; Calatrava, V.; Llamas, A.; Fernandez, E.; Sanz-Luque, E.; Galván, A. Nitrous oxide emissions from nitrite are highly dependent on nitrate reductase in the microalga Chlamydomonas reinhardtii. Int. J. Mol. Sci. 2022, 23, 9412. [Google Scholar] [CrossRef]
- Forchhammer, K.; Tandeau de Marsac, N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1995, 177, 2033–40. [Google Scholar] [CrossRef]
- Lácer, J.L.; Contreras, A.; Forchhammer, K.; Marco-Marín, C.; Gil-Ortiz, F.; Maldonado, R.; Fita, I.; Rubio, V. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 17644–17649. [Google Scholar] [CrossRef]
- Forcada-Nadal, A.; Llácer, J.L.; Contreras, A.; Marco-Marín, C.; Rubio, V. The PII-NAGK-PipX-NtcA regulatory axis of cyanobacteria: a tale of changing partners, allosteric effectors and non-covalent interactions. Front. Mol. Biosci. 2018, 5, 91. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2019, 44, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Zalutskaya, Z.; Kharatyan, N.; Forchhammer, K.; Ermilova, E. Reduction of PII signaling protein enhances lipid body production in Chlamydomonas reinhardtii. Plant Sci. 2015, 240, 1–9. [Google Scholar] [CrossRef]
- Takeuchi, T.; Benning, C. Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas. Biotechnol. Biofuels 2019, 12, 292. [Google Scholar] [CrossRef] [PubMed]
- Harris, E.H. Culture and storage methods. In The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use; Harris, E., Ed.; Academic Press: San Diego, CA, USA, 1989; pp. 25–63. [Google Scholar]
- Kropat, J.; Hong-Hermesdorf, A.; Casero, D.; Ent, P.; Castruita, M.; Pellegrini, M.; Merchant, S.S.; Malasarn, D. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011, 66, 770–80. [Google Scholar] [CrossRef]
- Baker, C.J.; Mock, N.M. An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell, Tissue Organ Cult. 1994, 39, 7–12. [Google Scholar] [CrossRef]
- Molnar, A.; Bassett, A.; Thuenemann, E.; Schwach, F.; Karkare, S.; Ossowski, S.; Weigel, D. Baulcombe, D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009, 58, 165–174. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 1228–1232. [Google Scholar] [CrossRef]
- Zalutskaya, Z.; Ostroukhova, M.; Ermilova, E. The Chlamydomonas alternative oxidase 1 is regulated by cadmium stress: new insights into control of expression. Environ. Exp. Bot. 2016, 130, 133–140. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Popov, N.; Schmitt, M.; Schulzeck, S.; Matthies, H. Reliable micromethod for determination of the protein content in tissue homogenates. Acta Biol. Med. Germ. 1975, 34, 1441–1446. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Maikova, A.; Zalutskaya, Z.; Lapina, T.; Ermilova, E. The HSP70 chaperone machines of Chlamydomonas are induced by cold stress. J. Plant Physiol. 2016, 204, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Lund, P. L-Glutamine and L-Glutamate: UV-method with glutaminase and glutamate dehydrogenase. In Methods of Enzymatic Analysis; Bergmeyer, H.U., Ed.; VCH, Verlagsgesellschaft: Weinheim, Germany, 1986; Volume 8, pp. 357–363. [Google Scholar]
- Sakaguchi, S. A new method for the colorimetric determination of arginine. J. Biochem. 1950, 37, 231–236. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




