Submitted:
27 July 2023
Posted:
28 July 2023
You are already at the latest version
Abstract
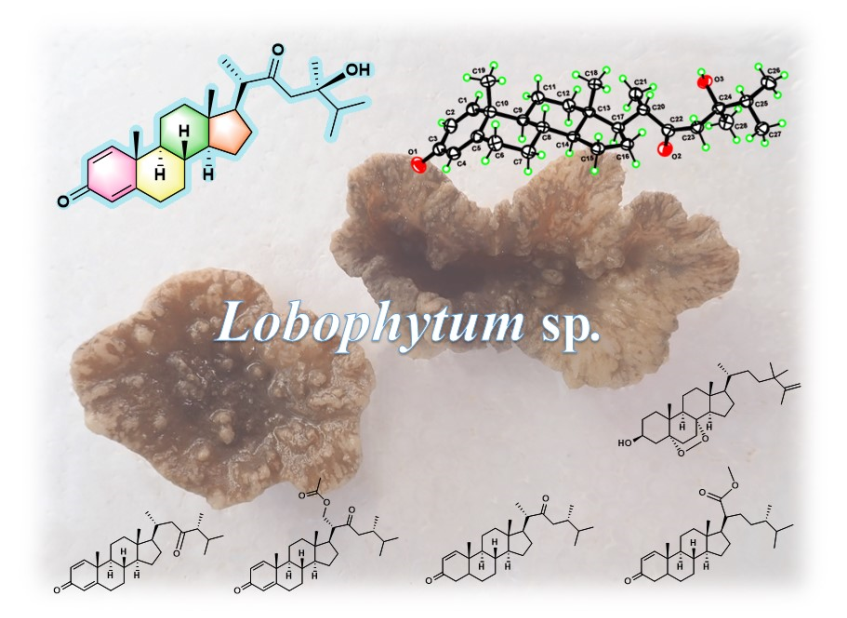
Keywords:
1. Introduction
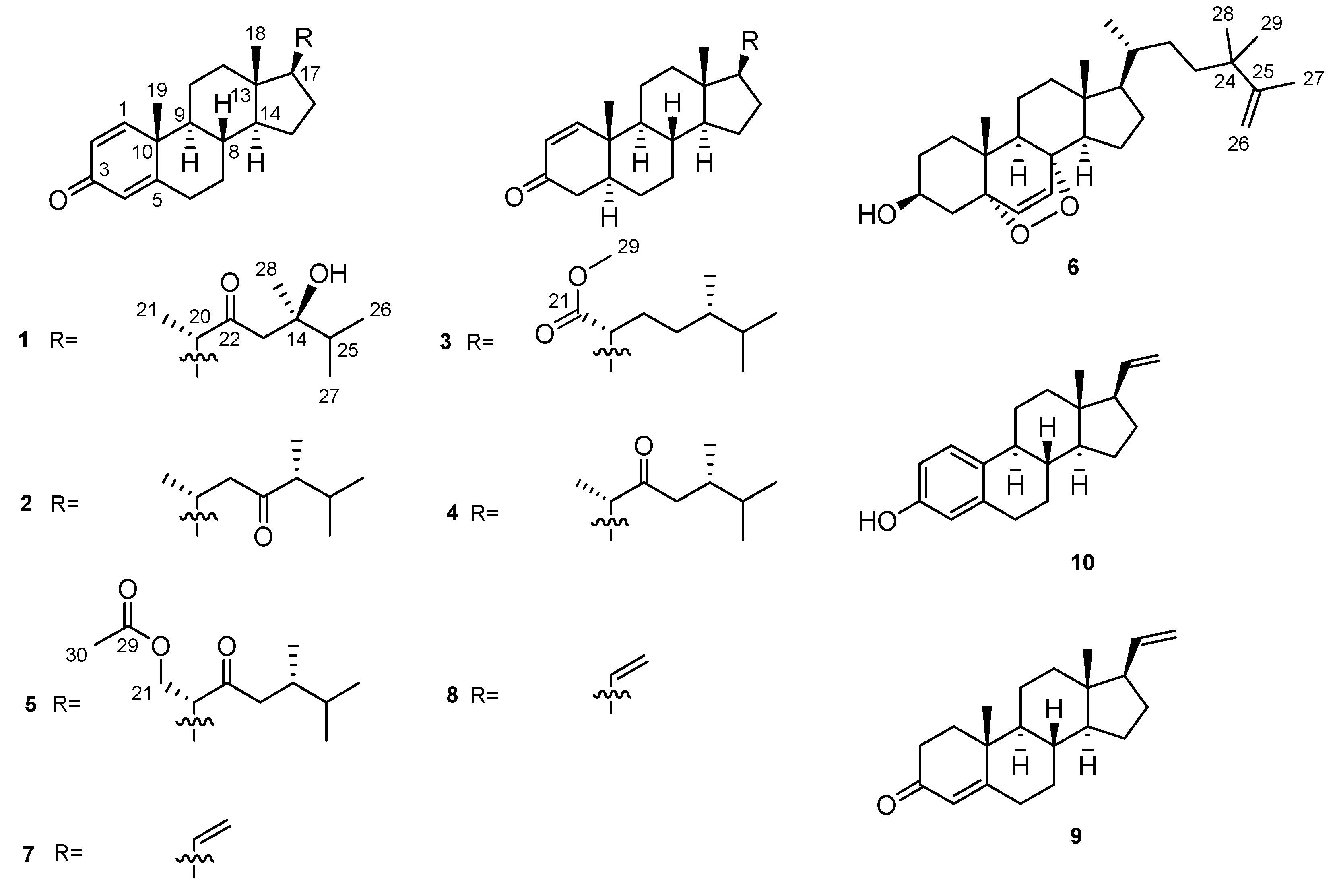
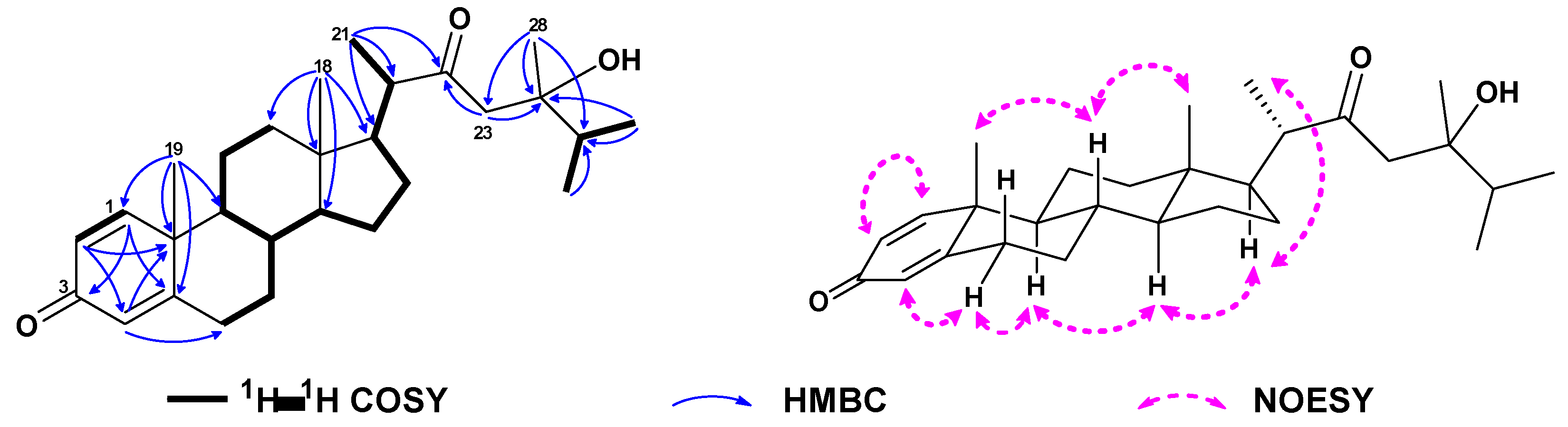
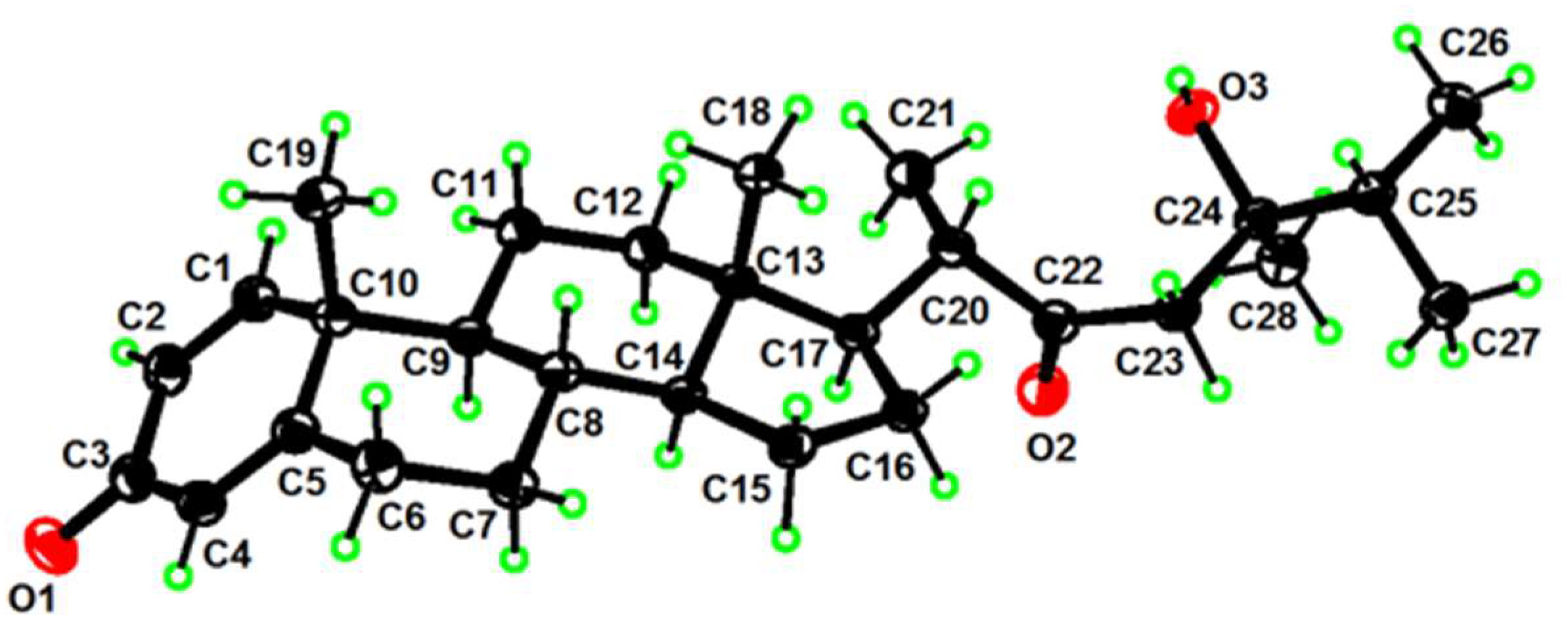
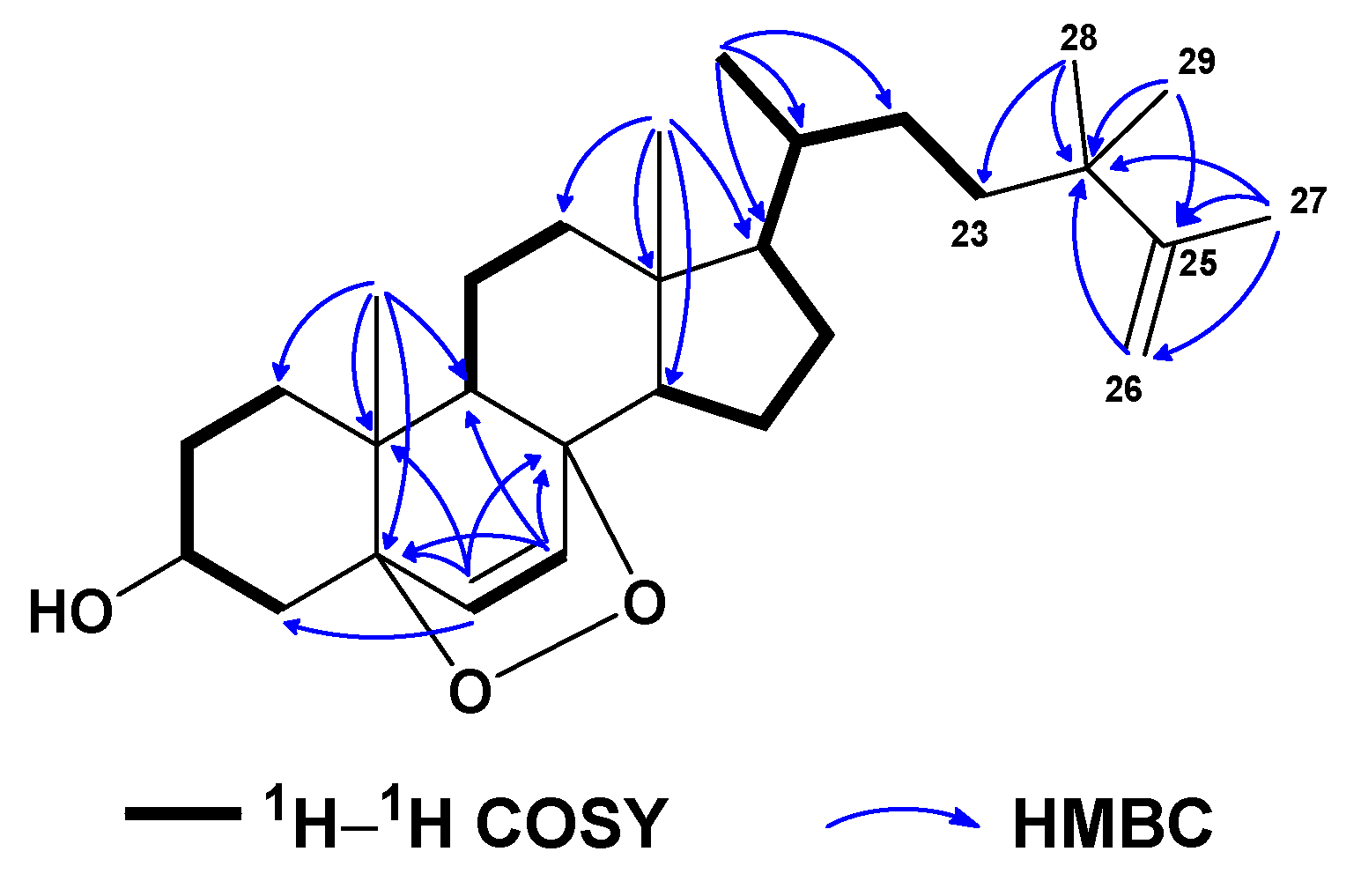
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Isolation
3.4. Spectroscopic Data of Compounds
3.5. X-ray Crystallographic Analysis for Compounds 1
4. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2023, 40, 275–325, and the previous reviews in this series. [Google Scholar] [CrossRef] [PubMed]
- Savić, M.P.; Sakač, M.N.; Kuzminac, I.Z.; Ajduković, J.J. Structural diversity of bioactive steroid compounds isolated from soft corals in the period 2015–2020. J. Steroid Biochem. 2022, 218, 106061. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, I.G.; Miguel, M.G.; Mnif, W. A brief review on new naturally occurring cembranoid diterpene derivatives from the soft corals of the genera Sarcophyton, Sinularia, and Lobophytum since 2016. Molecules 2019, 24, 781. [Google Scholar] [CrossRef]
- Li, S.-W.; Cuadrado, C.; Yao, L.-G.; Daranas, A.H.; Guo, Y.-W. Quantum mechanical–NMR-aided configuration and conformation of two unreported macrocycles isolated from the soft coral Lobophytum sp.: Energy calculations versus coupling constants. Org. Lett. 2020, 22, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Z.; Han, X.; Li, X.-L.; Lu, Z.-Y.; Dou, B.-B.; Zhang, W.-Z.; Tang, X.-L.; Li, P.-L.; Li, G.-Q. Four bioactive new steroids from the soft coral Lobophytum pauciflorum collected in South China Sea. Beilstein J. Org. Chem. 2022, 18, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Ohno, O.; Mizuno, E.; Miyamoto, J.; Hoshina, T.; Sano, T.; Matsuno, K. Inhibition of lipopolysaccharide-induced inflammatory signaling by soft coral-derived prostaglandin A2 in RAW264.7 cells. Mar. Drugs 2022, 20, 316. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Li, C.-Y.; Lai, K.-H.; Liao, M.-Y.; Wang, W.-H.; Chung, H.-M. Chemical constituents from the octocoral Lobophytum sarcophytoides. Chem. Nat. Compd. 2022, 58, 1167–1169. [Google Scholar] [CrossRef]
- Peng, B.-R.; Lu, M.-C.; El-Shazly, M.; Wu, S.-L.; Lai, K.-H.; Su, J.-H. Aquaculture soft coral Lobophytum crassum as a producer of anti-proliferative cembranoids. Mar. Drugs 2018, 16, 15. [Google Scholar] [CrossRef]
- Ye, F.; Zhou, Y.-B.; Li, J.; Gu, Y.-C.; Guo, Y.-W.; Li, X.-W. New steroids from the South China Sea soft coral Lobophytum sp. Chem. Biodivers. 2020, 17, e2000214. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, L.-F.; Miao, Z.-H.; Wu, B.; Guo, Y.-W. Cytotoxic polyhydroxylated steroids from the South China Sea soft coral Lobophytum sp. Steroids 2019, 141, 76–80. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Y.-c.; Su, M.-z.; Guo, Y.-w. Chemistry and bioactivity of secondary metabolites from South China Sea marine fauna and flora: Recent research advances and perspective. Acta Pharmacol. Sin. 2022, 43, 3062–3079. [Google Scholar] [CrossRef]
- Song, Y.-T.; Yu, D.-D.; Su, M.Z.; Luo, H.; Cao, J.-G.; Yao, L.-G.; Liang, L.-F.; Guo, Y.-W.; Yang, F. Anti-tumor cembrane diterpenoids from the South China Sea soft coral Lobophytum sp. Chem. Biodivers. 2023, 20, e202300217. [Google Scholar] [CrossRef]
- Díaz-Marrero, A.R.; Porras, G.; Aragón, Z.; de la Rosa, J.M.; Dorta, E.; Cueto, M.; D’Croz, L.; Maté, J.; Darias, J. Carijodienone from the octocoral Carijoa multiflora. A spiropregnane-based steroid. J. Nat. Prod. 2011, 74, 292–295. [Google Scholar] [CrossRef]
- Seo, Y.; Jung, J.H.; Rho, J.-R.; Shin, J.; Song, J.-I. Isolation of novel bioactive steroids from the soft coral Alcyonium gracillimum. Tetrahedron 1995, 51, 2497–2506. [Google Scholar] [CrossRef]
- Wu, S.-L.; Wang, G.-H.; Dai, C.-F.; Sheu, J.-H. Pregnane-based steroids from a Formosan gorgonian Subergorgia mollis. J. Chin. Chem. Soc. 2004, 51, 205–208. [Google Scholar] [CrossRef]
- Blackman, A.J.; Heaton, A.; Skelton, B.W.; White, A.H. Pregnane derivatives from two soft corals of the genus Capnella. Aust. J. Chem. 1985, 38, 565–573. [Google Scholar] [CrossRef]
- Yan, X.-H.; Liu, H.-L.; Huang, H.; Li, X.-B.; Guo, Y.-W. Steroids with aromatic A-rings from the Hainan soft coral Dendronephthya studeri Ridley. J. Nat. Prod. 2010, 74, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Marinozzi, M.; Castro Navas, F.F.; Maggioni, D.; Carosati, E.; Bocci, G.; Carloncelli, M.; Giorgi, G.; Cruciani, G.; Fontana, R.; Russo, V. Side-chain modified ergosterol and stigmasterol derivatives as liver X receptor agonists. J. Med. Chem. 2017, 60, 6548–6562. [Google Scholar] [CrossRef]
- Yan, X.-H.; Lin, L.-P.; Ding, J.; Guo, Y.-W. Methyl spongoate, a cytotoxic steroid from the Sanya soft coral Spongodes sp. Bioorg. Med. Chem. Lett. 2007, 17, 2661–2663. [Google Scholar] [CrossRef]
- Ahmed, A.F.; Wu, M.-H.; Wu, Y.-C.; Dai, C.-F.; Sheu, J.-H. Metabolites with cytotoxic activity from the Formosan soft coral Cladiella australis. J. Chin. Chem. Soc. 2006, 53, 489–494. [Google Scholar] [CrossRef]
- Yang, M.; Liang, L.-F.; Li, H.; Tang, W.; Guo, Y.-W. A new 5α,8α-epidioxysterol with immunosuppressive activity from the South China Sea soft coral Sinularia sp. Nat. Prod. Res. 2020, 34, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. SHELXT - Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. 2015, C71, 3–8. [Google Scholar]
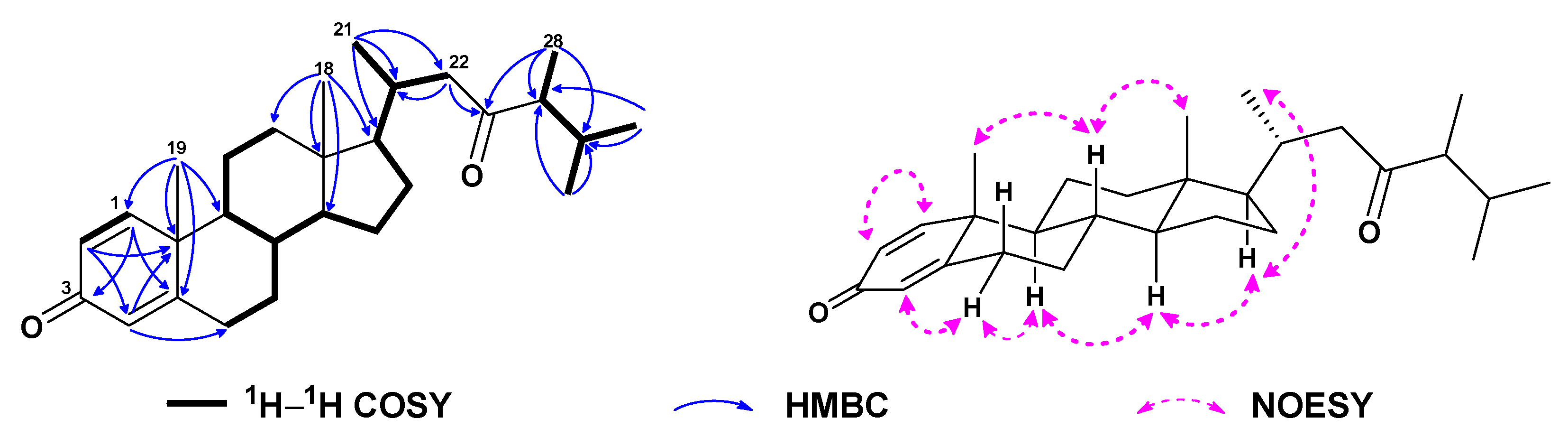
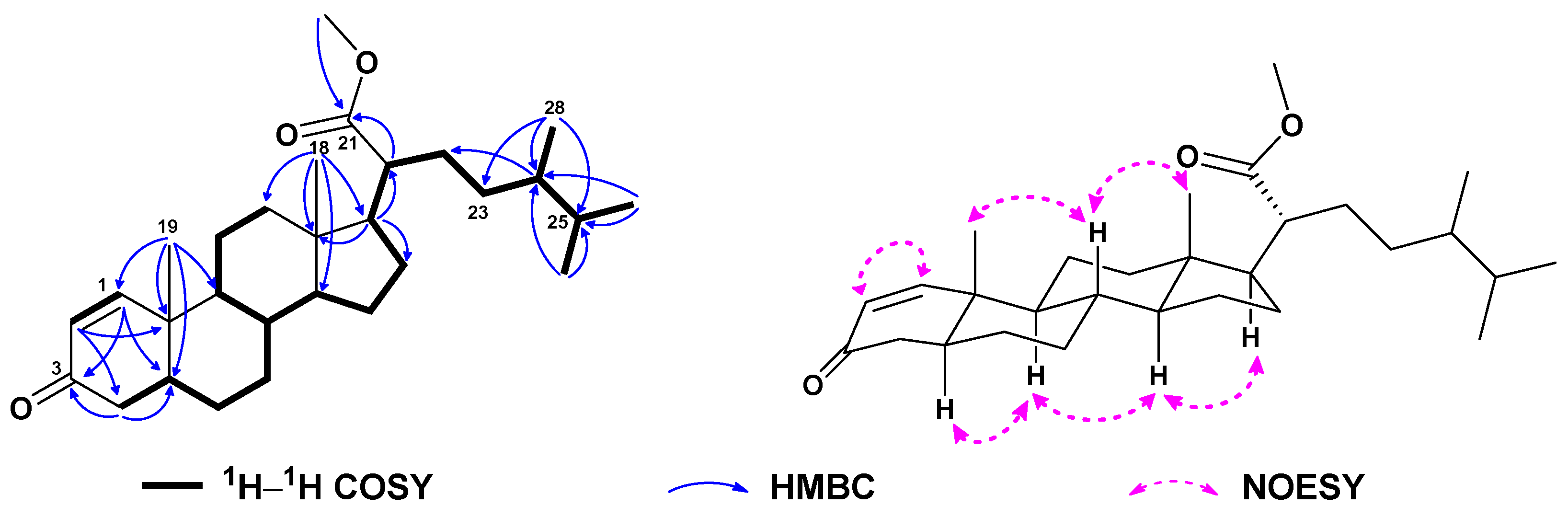
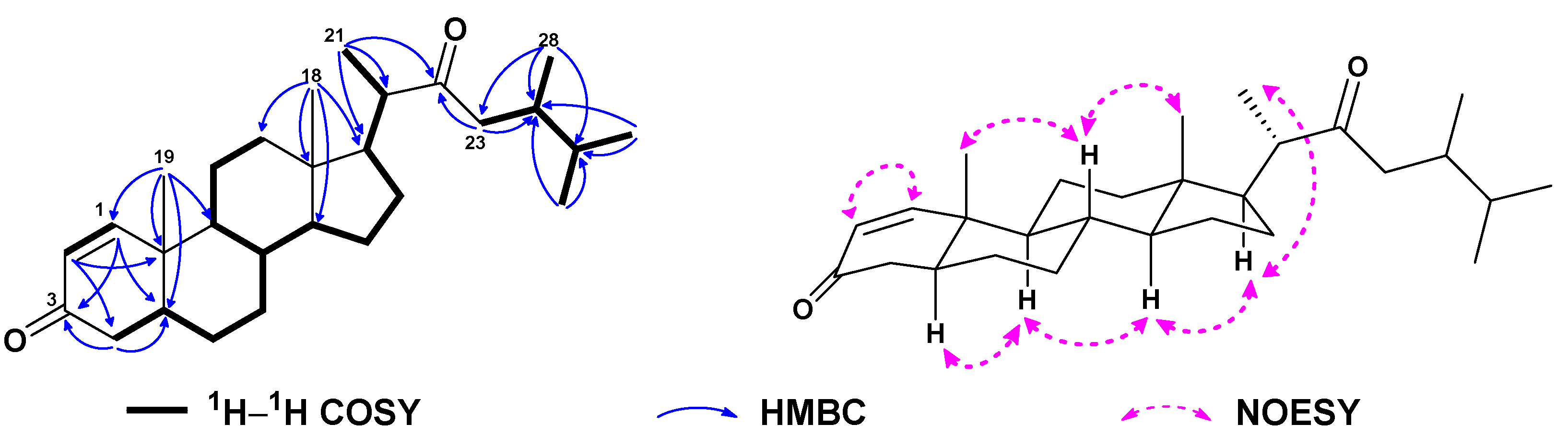
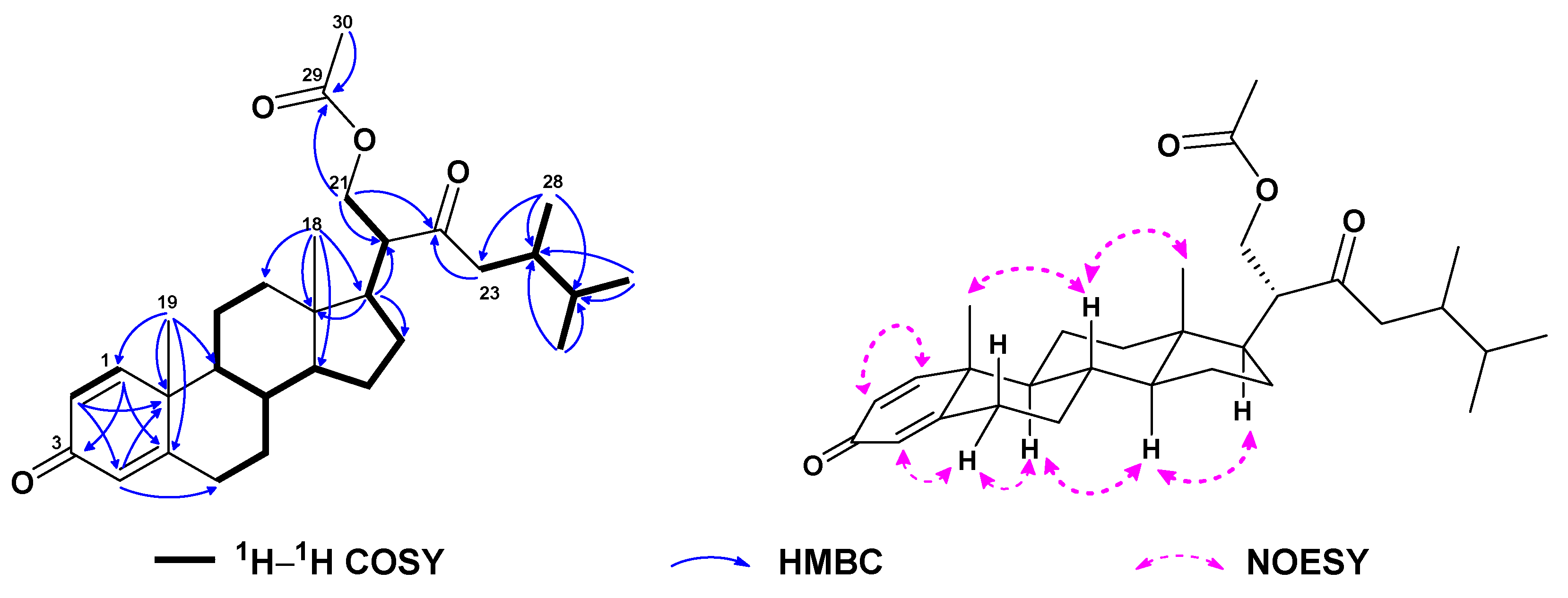
| No. | 1a | 2b | 3a | 4a | 5a | 6c |
| δH mult. (J in Hz) | δH mult. (J in Hz) | δH mult. (J in Hz) | δH mult. (J in Hz) | δH mult. (J in Hz) | δH mult. (J in Hz) | |
| 1 | 7.04 d (10.2) | 7.05 d (10.1) | 7.11 d (10.2) | 7.13 d (10.2) | 7.03 d (10.1) | 1.70 m |
| 1.94 m | ||||||
| 2 | 6.23 dd (10.2, 2.0) | 6.22 dd (10.1, 1.9) | 5.84 dd (10.2, 1.0) | 5.85 d (10.2) | 6.23 dd (10.1, 1.9) | 1.54 m |
| 1.84 m | ||||||
| 3 | 3.97 m | |||||
| 4 | 6.07 t (1.7) | 6.06 br s | 2.21 m | 2.23 m | 6.07 br s | 1.91 m |
| 2.35 dd (17.7, 14.2) | 2.36 dd (17.8, 14.2) | 2.11 dd (13.7, 5.2) | ||||
| 5 | 1.90 m | 1.91 m | ||||
| 6 | 2.35 m | 2.36 m | 1.37 m | 1.31 m | 2.35 m | 6.24 d (8.5) |
| 2.45 m | 2.45 m | 1.42 m | 1.42 m | 2.46 m | ||
| 7 | 1.93 m | 1.03 m | 0.96 m | 0.96 m | 1.06 m | 6.50 d (8.6) |
| 2.48 m | 1.96 m | 1.70 m | 1.71 m | 1.94 m | ||
| 8 | 1.61 m | 1.62 m | 1.45 m | 1.45 m | 1.61 m | |
| 9 | 1.59 m | 1.04 m | 0.96 m | 0.99 m | 1.06 m | 1.49 m |
| 11 | 1.07 m | 1.15 m | 0.85 m | 0.88 m | 1.66 m | 1.20 m |
| 1.69 m | 1.61 m | 1.72 m | 1.76 m | 1.71 m | 1.50 m | |
| 12 | 1.28 m | 1.17 m | 1.07 m | 1.31 m | 1.23 m | 1.21 m |
| 1.97 m | 2.03 m | 1.50 m | 1.97 m | 1.90 m | 1.96 m | |
| 14 | 1.04 m | 1.00 m | 1.08 m | 1.09 m | 1.01 m | 1.53 m |
| 15 | 1.18 m | 1.62 m | 1.09 m | 1.11 m | 1.20 m | 0.89 m |
| 1.63 m | 1.68 m | 1.64 m | 1.60 m | 1.64 m | 1.64 m | |
| 16 | 1.18 m | 1.25 m | 1.30 m | 1.32 m | 1.30 m | 1.34 m |
| 1.73 m | 1.79 m | 1.90 m | 1.69 m | 1.65 m | 1.90 m | |
| 17 | 2.47 m | 1.13 m | 1.65 m | 1.63 m | 1.60 m | 1.18 m |
| 18 | 0.76 s | 0.78 s | 0.72 s | 0.72 s | 0.81 s | 0.78 s |
| 19 | 1.23 s | 1.23 s | 0.99 s | 1.01 s | 1.23 s | 0.89 s |
| 20 | 2.48 m | 2.04 m | 2.20 m | 2.50 m | 2.84 td (10.4, 4.4) | 1.32 m |
| 21 | 1.11 d (7.6) | 0.90 d (7.0) | 1.09 d (6.9) | 3.96 t (10.7) | 0.89 d (6.9) | |
| 4.48 dd (10.7, 4.4) | ||||||
| 22 | 2.20 m | 1.27 m | 1.17 m | |||
| 2.45 m | 1.40 m | 1.26 m | ||||
| 23 | 2.52 d (17.7) | 1.39 m | 2.17 m | 2.22 dd (17.7, 9.1) | 1.17 m | |
| 2.66 d (17.7) | 1.51 m | 2.45 dd (17.0, 4.3) | 2.47 dd (17.5, 3.8) | 1.37 m | ||
| 24 | 2.29 m | 1.24 m | 1.93 m | 1.94 m | ||
| 25 | 1.75 m | 1.92 m | 1.55 m | 1.55 m | 1.55 m | |
| 26 | 0.88 d (6.9) | 0.84 d (6.8) | 0.75 d (6.9) | 0.82 d (6.8) | 0.83 d (7.1) | 4.65 br s |
| 4.72 br s | ||||||
| 27 | 0.93 d (6.8) | 0.90 d (7.0) | 0.84 d (6.8) | 0.87 d (6.8) | 0.87 d (6.8) | 1.67 s |
| 28 | 1.12 s | 0.98 d (6.9) | 0.76 d (6.9) | 0.81 d (6.8) | 0.81 d (7.6) | 1.00 s |
| 29 | 3.65 s | 1.00 s | ||||
| 30 | 2.00 s | |||||
| OH | 4.09 s |
| No. | 1a | 2b | 3a | 4a | 5a | 6c |
| δC (mult.) | δC (mult.) | δC (mult.) | δC (mult.) | δC (mult.) | δC (mult.) | |
| 1 | 155.8 (d) | 156.2 (d) | 158.7 (d) | 158.6 (d) | 155.7 (d) | 34.8 (t) |
| 2 | 127.7 (d) | 127.6 (d) | 127.5 (d) | 127.6 (d) | 127.8 (d) | 30.3 (t) |
| 3 | 186.5 (s) | 186.6 (s) | 200.4 (s) | 200.4 (s) | 186.5 (s) | 66.6 (d) |
| 4 | 124.1 (d) | 123.9 (d) | 41.1 (t) | 41.1 (t) | 124.1 (d) | 37.1 (d) |
| 5 | 169.1 (s) | 169.5 (s) | 44.4 (d) | 44.4 (d) | 169.0 (s) | 82.3 (s) |
| 6 | 33.0 (t) | 33.0 (t) | 27.7 (t) | 27.7 (t) | 32.9 (t) | 135.6 (d) |
| 7 | 33.7 (t) | 33.8 (t) | 32.1 (t) | 31.4 (t) | 33.6 (t) | 130.9 (d) |
| 8 | 35.6 (d) | 35.6 (d) | 35.8 (d) | 35.8 (d) | 35.6 (d) | 79.6 (s) |
| 9 | 52.3 (d) | 52.4 (d) | 50.1 (d) | 49.9 (d) | 52.2 (d) | 51.2 (d) |
| 10 | 43.6 (s) | 43.8 (s) | 39.1 (s) | 39.1 (s) | 43.6 (s) | 37.1 (s) |
| 11 | 22.9 (t) | 24.5 (t) | 30.1 (t) | 21.3 (t) | 22.8 (t) | 23.5 (t) |
| 12 | 39.5 (t) | 39.5 (t) | 37.5 (t) | 39.8 (t) | 38.8 (t) | 39.5 (t) |
| 13 | 43.1 (s) | 42.9 (s) | 42.4 (s) | 43.1 (s) | 43.0 (s) | 44.8 (s) |
| 14 | 54.9 (d) | 55.6 (d) | 55.8 (d) | 55.8 (d) | 54.6 (d) | 51.7 (d) |
| 15 | 24.7 (t) | 23.0 (t) | 23.8 (t) | 24.5 (t) | 24.5 (t) | 20.8 (t) |
| 16 | 27.5 (t) | 28.4 (t) | 27.3 (t) | 27.7 (t) | 26.9 (t) | 28.3 (t) |
| 17 | 52.0 (d) | 56.0 (d) | 52.9 (d) | 52.4 (d) | 49.6 (d) | 56.2 (d) |
| 18 | 12.4 (q) | 12.2 (q) | 12.4 (q) | 12.6 (q) | 12.5 (q) | 12.7 (q) |
| 19 | 18.8 (q) | 18.8 (q) | 13.1 (q) | 13.1 (q) | 18.8 (q) | 18.3 (q) |
| 20 | 50.9 (d) | 32.0 (d) | 48.0 (d) | 50.0 (d) | 53.6 (d) | 35.8 (d) |
| 21 | 16.4 (q) | 20.0 (q) | 176.9 (s) | 16.7 (q) | 64.6 (t) | 18.9 (q) |
| 22 | 217.6 (s) | 49.2 (t) | 29.8 (t) | 214.8 (s) | 211.8 (s) | 30.4 (t) |
| 23 | 48.2 (t) | 215.0 (s) | 21.2 (t) | 46.8 (t) | 49.6 (t) | 37.1 (t) |
| 24 | 74.0 (s) | 53.0 (d) | 38.7 (d) | 33.8 (d) | 33.3 (d) | 38.8 (s) |
| 25 | 37.4 (d) | 30.2 (d) | 31.4 (d) | 32.1 (d) | 32.1 (d) | 152.3 (s) |
| 26 | 17.0 (q) | 18.8 (q) | 17.5 (q) | 18.3 (q) | 18.6 (q) | 109.6 (t) |
| 27 | 17.9 (q) | 21.6 (q) | 20.6 (q) | 20.0 (q) | 19.9 (q) | 19.5 (q) |
| 28 | 23.0 (q) | 12.7 (q) | 15.3 (q) | 16.1 (q) | 16.2 (q) | 27.3 (q) |
| 29 | 51.2 (q) | 170.7 (s) | 27.7 (q) | |||
| 30 | 21.0 (q) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





