Submitted:
31 July 2023
Posted:
02 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of agronomic traits
2.3. Drought stress treatments
2.4. Histochemical and physiological analysis
2.5. Primer Design and Sequencing
2.6. RNA extraction and qRT-PCR
2.7. Sequence analysis
2.8. Subcellular localization
2.9. Statistical analysis
3. Results
3.1. Characterization of the dsm3 Mutant
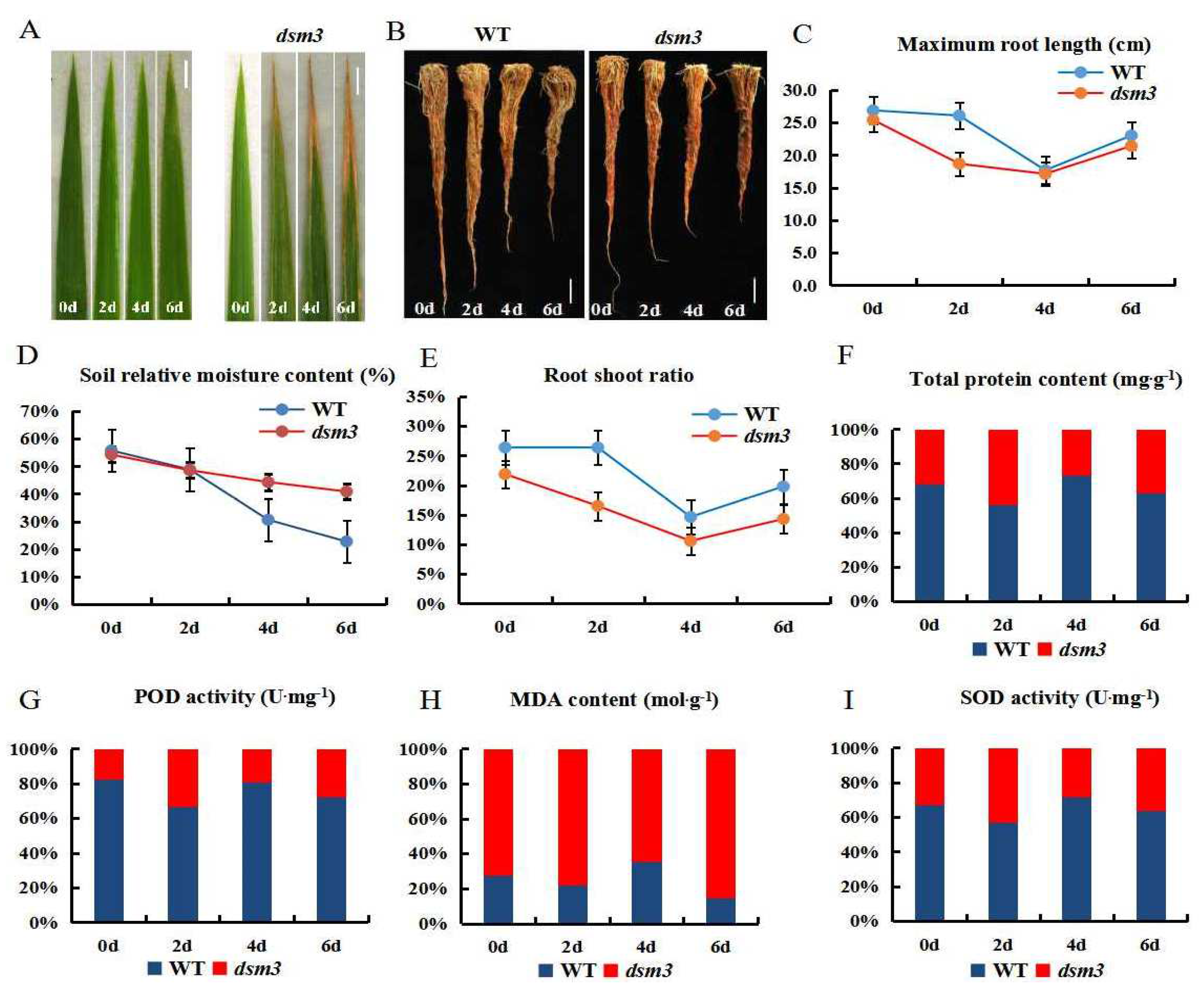
3.2. Sensitivity of dsm3 to Drought Stress
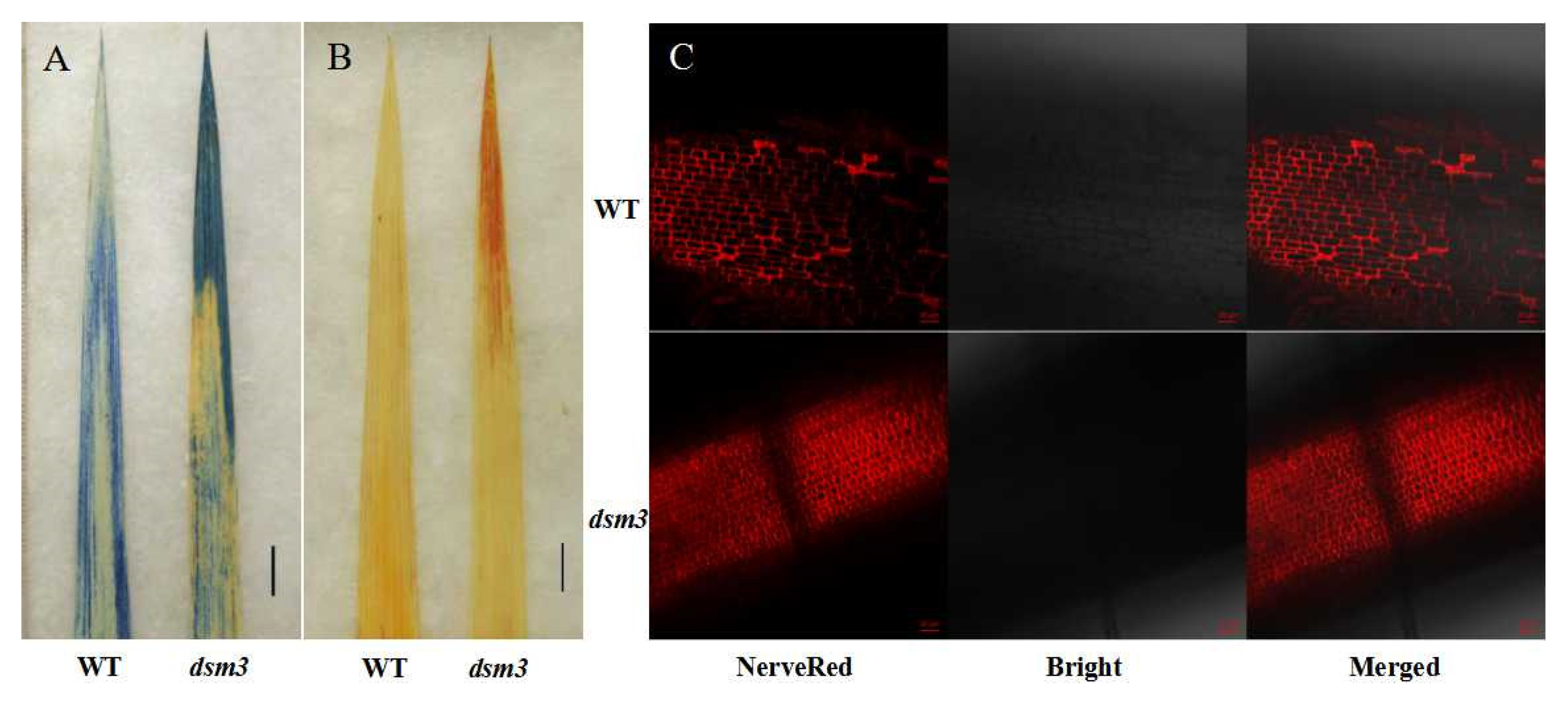
3.3. Abnormal H2O2 Accumulation, Cell Death, and Altered Cell Size in the Root Apical Meristem of the dsm3 Mutant
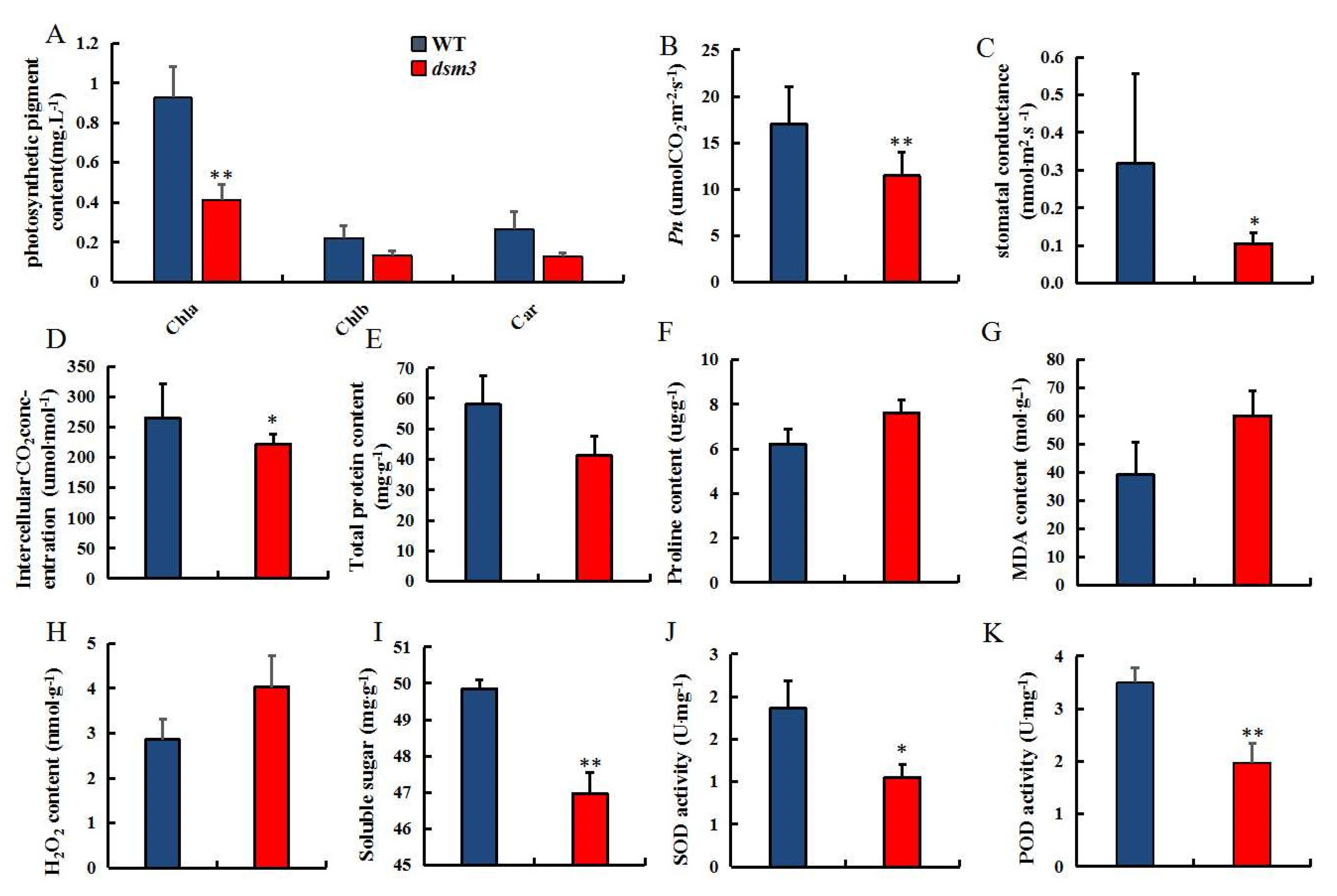
3.4. Evaluation of Photosynthetic and Physiological Indicators under Drought Stress
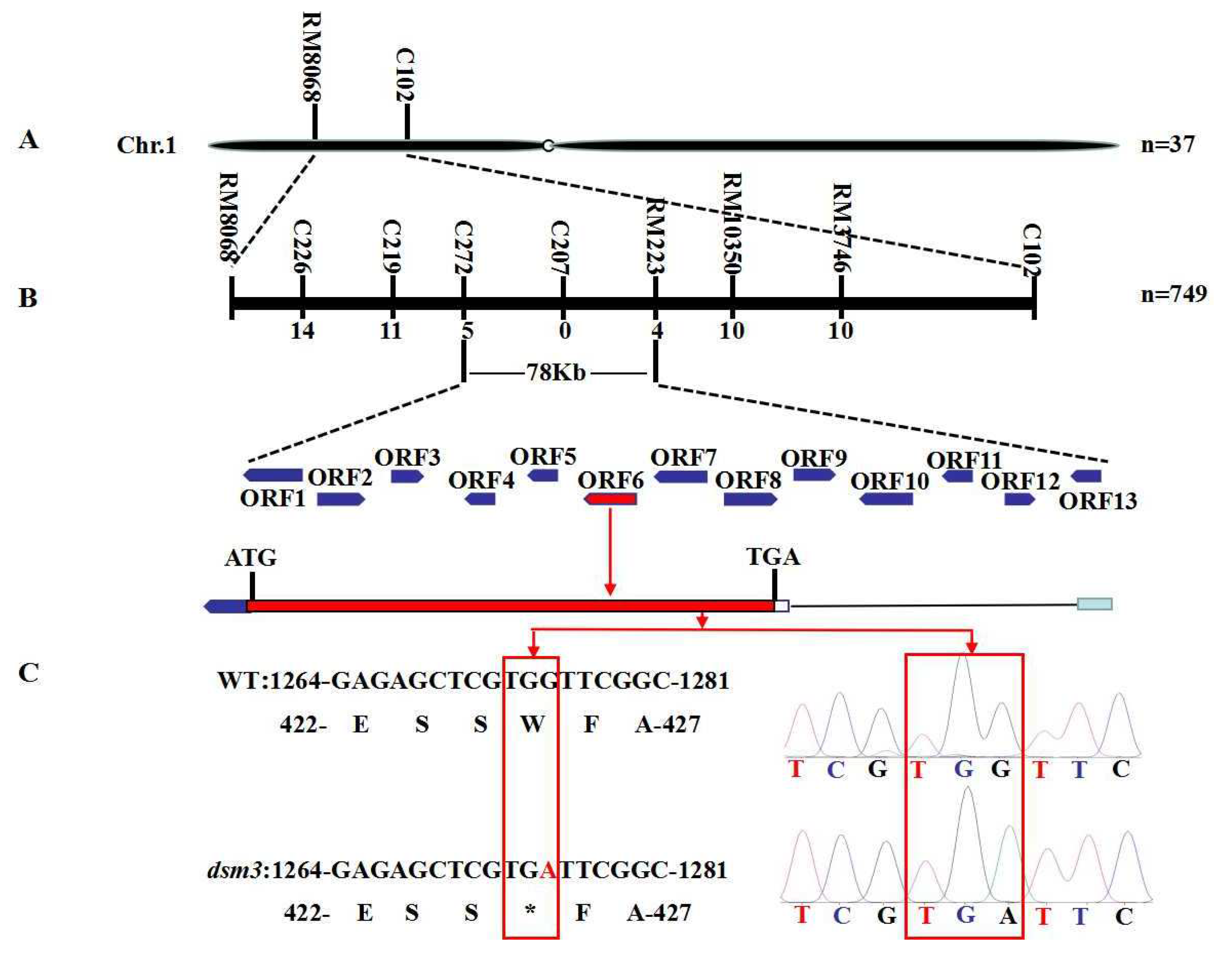
3.5. Genetic Analysis and Fine Mapping of Osdsm3
3.6. Analysis of Osdsm3 Expression Levels and Drought Stress-Related Genes
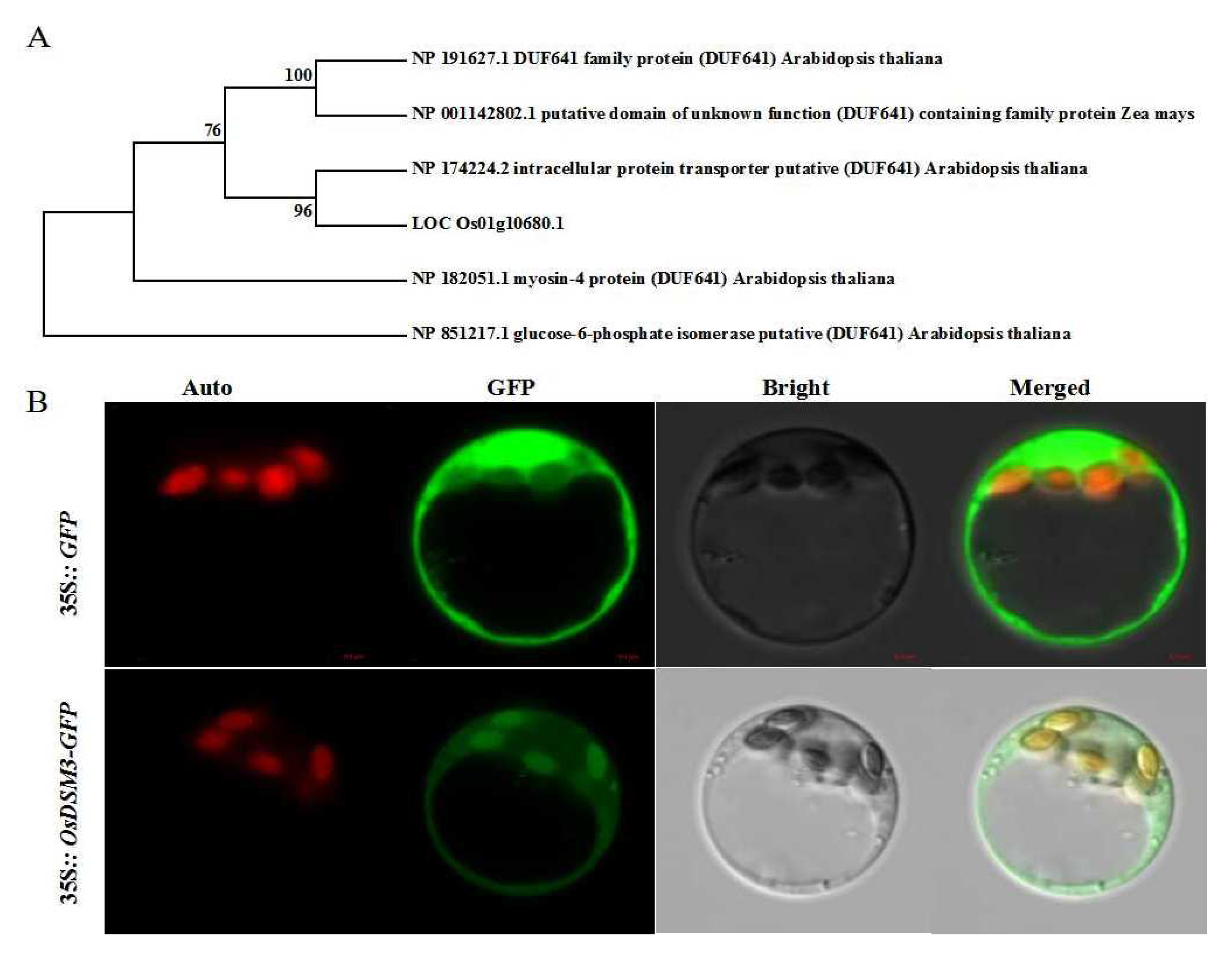
3.7. Phylogenetic Analysis and Subcellular Localization of OsDSM3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Upadhyaya, H.; Panda, S.K. Drought stress responses and its management in rice. In Advances in rice research for abiotic stress tolerance; Elsevier, 2019; pp. 177–200. [Google Scholar] [CrossRef]
- Yang, X.; Wang, B.; Chen, L.; Li, P.; Cao, C. The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, and quality. Scientific reports 2019, 9, 3742. [Google Scholar] [CrossRef]
- Kim, Y.; Chung, Y.S.; Lee, E.; Tripathi, P.; Heo, S.; Kim, K.H. Root Response to Drought Stress in Rice (Oryza sativa L.). Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef]
- Salsinha, Y.C.F.; Maryani; Indradewa, D.; Purwestri, Y.A.; Rachmawati, D. Leaf physiological and anatomical characters contribute to drought tolerance of Nusa Tenggara Timur local rice cultivars. Journal of Crop Science and Biotechnology 2021, 24, 337–348. [Google Scholar] [CrossRef]
- Xu, W.; Cui, K.; Xu, A.; Nie, L.; Huang, J.; Peng, S. Drought stress condition increases root to shoot ratio via alteration of carbohydrate partitioning and enzymatic activity in rice seedlings. Acta physiologiae plantarum 2015, 37, 1–11. [Google Scholar] [CrossRef]
- Hazman, M.; Brown, K.M. Progressive drought alters architectural and anatomical traits of rice roots. Rice (N Y) 2018, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Sinha, R.; Singla-Pareek, S.L.; Pareek, A.; Singh, A.K. Shaping the root system architecture in plants for adaptation to drought stress. Physiol Plant 2022, 174, e13651. [Google Scholar] [CrossRef] [PubMed]
- Redillas, M.C.; Park, S.-H.; Lee, J.W.; Kim, Y.S.; Jeong, J.S.; Jung, H.; Bang, S.W.; Hahn, T.-R.; Kim, J.-K. Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant Biotechnology Reports 2012, 6, 89–96. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Yu, F.; Hu, B.; Jia, Y.; Sha, H.; Zhao, H. Differential activity of the antioxidant defence system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci Rep 2019, 9, 8543. [Google Scholar] [CrossRef]
- Sales, C.R.; Ribeiro, R.V.; Silveira, J.A.; Machado, E.C.; Martins, M.O.; Lagoa, A.M. Superoxide dismutase and ascorbate peroxidase improve the recovery of photosynthesis in sugarcane plants subjected to water deficit and low substrate temperature. Plant Physiol Biochem 2013, 73, 326–336. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kusaba, M.; Tanaka, A.; Sakamoto, W. Protection of Chloroplast Membranes by VIPP1 Rescues Aberrant Seedling Development in Arabidopsis nyc1 Mutant. Front Plant Sci 2016, 7, 533. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R. Plant responses to drought stress. From Morphological to Molecular Features; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–5. [Google Scholar] [CrossRef]
- Ahmad, I.; Devonshire, J.; Mohamed, R.; Schultze, M.; Maathuis, F.J. Overexpression of the potassium channel TPKb in small vacuoles confers osmotic and drought tolerance to rice. New Phytol 2016, 209, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, L.; Xue, Y.; Zhang, Q.; Wang, L.; Shou, H. Overexpression of OsVP1 and OsNHX1 increases tolerance to drought and salinity in rice. Journal of Plant Biology 2010, 53, 444–452. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiology 2017, 173, 944–955. [Google Scholar] [CrossRef]
- Du, H.; Liu, L.; You, L.; Yang, M.; He, Y.; Li, X.; Xiong, L. Characterization of an inositol 1, 3, 4-trisphosphate 5/6-kinase gene that is essential for drought and salt stress responses in rice. Plant molecular biology 2011, 77, 547–563. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wu, J.; Zheng, X.; Zheng, S.; Sun, X.; Qiu, Q.; Lu, T. Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PloS one 2013, 8, e57472. [Google Scholar] [CrossRef]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem Biophys Res Commun 2016, 478, 703–709. [Google Scholar] [CrossRef]
- Sharma, E.; Bhatnagar, A.; Bhaskar, A.; Majee, S.M.; Kieffer, M.; Kepinski, S.; Khurana, P.; Khurana, J.P. Stress-induced F-Box protein-coding gene OsFBX257 modulates drought stress adaptations and ABA responses in rice. Plant Cell Environ 2023, 46, 1207–1231. [Google Scholar] [CrossRef]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef]
- Pa, V.; Vijayaraghavareddy, P.; Uttarkar, A.; Dawane, A.; D, S.; V, A.; Kc, B.; Niranjan, V.; Ms, S.; Cv, A.; et al. Novel small molecules targeting bZIP23 TF improve stomatal conductance and photosynthesis under mild drought stress by regulating ABA. FEBS J 2022, 289, 6058–6077. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Chai, C.; Qian, Q.; Li, C.; Tang, J.; Sun, L.; Huang, Z.; Guo, X.; Sun, C.; Liu, M.; et al. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 2008, 54, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.G.; Lee, C.Y.; Tseng, C.S. Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot Stud 2018, 59, 2. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 2006, 103, 12987–12992. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 2004, 9, 236–243. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Min, M.K.; Choi, E.H.; Kim, N.; Moon, S.J.; Yoon, I.; Kwon, T.; Jung, K.H.; Kim, B.G. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol Biol 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, K.; Niu, X.; Wang, Q.; Wan, Y.; Yang, F.; Li, G.; Wang, Y.; Wang, R. Genome-wide Identification of PP2C Genes and Their Expression Profiling in Response to Drought and Cold Stresses in Medicago truncatula. Sci Rep 2018, 8, 12841. [Google Scholar] [CrossRef]
- Zong, W.; Tang, N.; Yang, J.; Peng, L.; Ma, S.; Xu, Y.; Li, G.; Xiong, L. Feedback Regulation of ABA Signaling and Biosynthesis by a bZIP Transcription Factor Targets Drought-Resistance-Related Genes. Plant Physiol 2016, 171, 2810–2825. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Peng, Z.; Sprague, S.A.; Wang, W.; Park, J.; Akhunov, E.; Jagadish, K.S.V.; Nakata, P.A.; Cheng, N.; et al. Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci Rep 2017, 7, 15950. [Google Scholar] [CrossRef]
- Li, D.; Liu, P.; Yu, J.; Wang, L.; Dossa, K.; Zhang, Y.; Zhou, R.; Wei, X.; Zhang, X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol 2017, 17, 152. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Meng, H.; Wen, H.; Fan, Y.; Zhao, J. Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol Biol 2011, 75, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ahn, J.W.; Jin, U.H.; Choi, D.; Paek, K.H.; Pai, H.S. Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem 2003, 278, 19406–19415. [Google Scholar] [CrossRef] [PubMed]
- Allen, G.C.; Flores-Vergara, M.A.; Krasynanski, S.; Kumar, S.; Thompson, W.F. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Ning, J.; Li, X.; Hicks, L.M.; Xiong, L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol 2010, 152, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.S.; Mawlong, I.; Ali, K.; Tyagi, A. Regulation of phytosterol biosynthetic pathway during drought stress in rice. Plant Physiol Biochem 2018, 129, 11–20. [Google Scholar] [CrossRef]
- Du, H.; Wang, N.; Cui, F.; Li, X.; Xiao, J.; Xiong, L. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol 2010, 154, 1304–1318. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjarvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J Integr Plant Biol 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Kalsoom, U.; Bhatti, H.N.; Asgher, M. Characterization of Plant Peroxidases and Their Potential for Degradation of Dyes: a Review. Appl Biochem Biotechnol 2015, 176, 1529–1550. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr Opin Plant Biol 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv Exp Med Biol 2018, 1081, 189–214. [Google Scholar] [CrossRef]
- Hirota, K.; Yoshikiyo, K.; Guilbaud, G.; Tsurimoto, T.; Murai, J.; Tsuda, M.; Phillips, L.G.; Narita, T.; Nishihara, K.; Kobayashi, K.; et al. The POLD3 subunit of DNA polymerase delta can promote translesion synthesis independently of DNA polymerase zeta. Nucleic Acids Res 2015, 43, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Palaniyar, N.; Nadesalingam, J.; Clark, H.; Shih, M.J.; Dodds, A.W.; Reid, K.B. Nucleic acid is a novel ligand for innate, immune pattern recognition collectins surfactant proteins A and D and mannose-binding lectin. J Biol Chem 2004, 279, 32728–32736. [Google Scholar] [CrossRef]
- Zhu, W.; Ma, T.; Zhang, M.; Sha, W. Research progress of receptor-like protein kinases in plants. Genomics and Applied Biology 2018, 37, 451–458. [Google Scholar]
- Allen, T.; Ingles, P.J.; Praekelt, U.; Smith, H.; Whitelam, G.C. Phytochrome-mediated agravitropism in Arabidopsis hypocotyls requires GIL1 and confers a fitness advantage. Plant J 2006, 46, 641–648. [Google Scholar] [CrossRef] [PubMed]


| Trait | ZH8015 | dsm3 |
|---|---|---|
| Plant height (cm) | 101.90 ± 2.70 | 71.25 ± 3.16 ** |
| No.of effective tillerings | 6.30 ± 1.80 | 6.70 ± 1.40 |
| Grain length (cm) | 1.01 ± 0.10 | 0.97 ± 0.13 * |
| Grain width (cm) | 0.33 ± 0.04 | 0.29 ± 0.05 * |
| Grain length-width ratio | 0.31 ± 0.05 | 0.33 ± 0.07 ** |
| Thousand grain weight (g) | 38.33 ± 0.99 | 26.91 ± 1.53 ** |
| Panicle length (cm) | 24.40 ± 1.40 | 18.80 ± 1.14 ** |
| Flag leaf length (cm) | 30.38 ± 4.11 | 22.33 ± 3.11 ** |
| Flag leaf width (cm) | 1.58 ± 0.09 | 1.26 ± 0.13 * |
| Second upper leaf length (cm) | 41.66 ± 4.64 | 33.87 ± 4.00 * |
| Second upper leaf width (cm) | 1.48 ± 0.12 | 1.25 ± 0.07 ** |
| Third upper leaf length (cm) | 45.68 ± 4.60 | 41.33 ± 3.59 * |
| Third upper leaf width (cm) | 1.52 ± 0.12 | 1.25 ± 0.08 ** |
| F2 population | F1 generation | N0. of normal plants | No. of mutant plants | Theoretic ratio | Chi-square value |
|---|---|---|---|---|---|
| dsm3×02428 | Normal | 2591 | 809 | 3:1 | 2.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





