1. Introduction
Vibrio splendidus-related strains are ubiquitously spread in the marine environments including surface waters as well as sediments [
1], and these strains are clustered into a clade known as Splendidus which currently consists of 16 highly related species [
2]. In previous outbreaks of various aquatic animal epidemics,
V. splendidus has been proved to be an important strain in different mortality events, resulting in great losses to the worldwide aquaculture industry [
3].
V. splendidus has been shown to infect fish such as turbot
Scophthalmus maximus [
4], echinoderm such as sea cucumber
Apostichopus japonicus, and marine bivalves such as the scallop
Patinopecten yessoensis [
5], the pacific oyster
Crassostrea gigas [
6], the clam
Ruditapes decussatus [
7] and mussel
Mytilus sp. [
8].
It has been reported that the skin ulcer syndrome (SUS) of sea cucumber infected by
V. splendidus has caused over 80% mortality and over 30% economic losses in China and Japan [
3].
V. splendidus mainly affected immune signaling related pathways of
A. japonicus, such as endocytosis, lysosome, mitogen-activated protein kinase (MAPK) signaling pathway and chemokine [
9]. Till now, many virulence factors of
V. splendidus have been identified, which include the invasion porin OmpU [
10], metalloprotease Vsm [
11], invasive vesicular serine protease [
12], collagenase [
13], and type-six secretion system (T6SS) related proteins [
14]. As the major subunit of the flagellum, flagella C (FliC) has been shown to be associated with the virulence of
V. splendidus AJ01, mediating not only adhesion to coelomocytes of sea cucumber [
15] but also the host immune responses [
16]. With the further study of the pathogen-host interaction models, several amino acids (AAs) have been proved to mediate the virulence of pathogens that could infect their sensitive hosts [
17].
AAs are involved in different biochemical pathways and play metabolic and physiological roles [
18]. The host relies on AA metabolism to exert the defence response against the pathogen, while the pathogen uses AA metabolism to its own advantage [
18]. Therefore, some AAs are central points of competition between host and pathogen [
19,
20]. Pathogens require AAs to support their physiological functions and changes in AA availability have a significant impact on the growth of pathogens as well as the expression of their virulence factors [
21]. It has been generally acknowledged that L-glutamic acid (L-Glu) plays important roles in nutrient metabolism, energy supply, immune response, oxidative stress, and signal regulation in both the host cells and its corresponding pathogen [
22]. For example, L-Glu might serve as a nutrient source for the bacterium and enhanced the growth of
Pseudomonas syringae pv. tomato DC3000
in vitro culture [
23]. Plant pathogen
Ralstonia solanacearum utilized plant-derived L-Glu to promote the production of virulence factors, thus the virulence was increased [
24].
It has been previously reported that the higher temperatures elevated the levels of threonine, alanine, arginine, glutamic acid, tyrosine, histidine, glycine [
25], among which L-Glu was one of the best carbon sources for the growth of
V. splendidus AJ01 [
26]. Combined with these previous results lead us to wonder whether the increased L-Glu could elevate the virulence of
V. splendidus AJ01. In our present study, the effects of L-Glu on the cell viability and gene expression profile of coelomocyte from sea cucumber were determined. The effects of exogenous L-Glu on the virulence and expression of virulence related genes were determined both
in vivo and
in vitro. Our result showed that L-Glu could mediate the interaction between host and pathogen and influence the outcome of infection.
2. Materials and Methods
2.1. Animals
The sea cucumbers used in this study were commercially farmed animals purchased from Dalian Pacific Aquaculture Company (Dalian, China). Animal experiments were designed according to the recommendations in the National Institutes of Health's Guidelines for the Care and Use of Laboratory Animals. The experimental protocol was approved by the Laboratory Animal Ethics Committee of Ningbo University.
2.2. Coelomocyte Viability Assay
The culture of primary coelomocytes of sea cucumber was performed as described by Zhang et al [
27]. Briefly, the coelomic fluid was centrifuged at 800×g for 5 min and resuspended in Leiboviz's L-15 medium containing 0.39 M NaCl (Invitrogen, USA). One hundred microlitres of cell suspension were added to 96-well culture microplates and cultured at 16 °C overnight until the cells were in a healthy adherent state. Different concentrations of L-Glu were added into the wells, while Leiboviz's L-15 medium was added as a control. To determine the effects of
V. splendidus AJ01 on the viability of coelomocytes in the presence of L-Glu,
V. splendidus AJ01 at a concentration of 1×10
5 CFU/mL was added to coelomocytes and incubated overnight with or without L-Glu for 4 h. Cell counting kit-8 (CCK-8) assay was carried out to detect the cell viability. Ten microlitres of CCK-8 reagent (APExBIO, USA) were added into each well for 2 h at 16 °C, and absorbance at 450 nm was measured. The absorbance of
V. splendidus AJ01 incubated in Leiboviz's L-15 medium was subtracted to exclude the absorbance of
V. splendidus AJ01 itself. The viability of the coelomocytes was calculated according to manufacturer's instructions.
2.3. Histological Analysis
In order to explore the effects of different concentrations of L-Glu on the status of body wall, sea cucumbers incubated in seawater containing different concentrations of L-Glu for 72 h was taken for histological analysis [
28]. Sea cucumbers without any treatment was used as a control.
The tissue samples collected from each specimen were fixed with 10% neutral formaldehyde fixative for 24 h. Then, the fixed tissues were rinsed using 70% alcohol and dehydrated using gradient concentrations of 70, 85, 90, 95 and 100%, respectively. Tissue samples were clarified in xylene, embedded in paraffin wax at an average fusion temperature of 56 °C, and sectioned at 7 μm thicknesses with a microtome (KD-3358). The tissue sections were dewaxed, stained with hematoxylin for 10-20 min, and then stained with eosin for 3-5 min. Serial dilutions of ethyl alcohol were used for dehydration, and the sections were cleared in xylene and sealed using neutral glycerin. Then, the sections were observed under a microscope (ZEISS Axio Vert. A1).
2.4. Immersion Infection Experiment
The survival percent of sea cucumber was calculated according to the description of Li et al. with minor modifications [
29]. To analyze the effect of L-Glu on the survival of sea cucumbers, L-Glu was added to seawater at concentrations of 0.1 mM, 1 mM and 10 mM, respectively. The status of sea cucumbers was observed daily and the number of deaths was recorded. To analyze the effect of L-Glu on
V. splendidus AJ01 infection, sea cucumbers were divided into three groups, one group was challenged with 1×10
7 CFU/mL
V. splendidus AJ01 cells in seawater without L-Glu and another group was challenged with the same volume of
V. splendidus AJ01 cells in seawater containing 0.1 mM L-Glu, whereas the group incubated with only 0.1 mM L-Glu was used as a control group.
2.5. Transcriptomic Library Construction
The sea cucumbers were evenly divided into two groups, one group was in the seawater containing 0.1 mM L-Glu, while another group was used as a control. After incubation for 72 h, the coelomic fluid was collected, and centrifuged at 800×g for 5 min to collect coelomocytes. The total RNA was isolated according to manufacturer's instructions (Vazyme, China). Library construction and high-throughput sequencing were carried out by Novogene Biotech (Beijing, China). The clean reads were subjected to the genomic database of
A. japonicus (ASM275485v1) using hisat2 [
30]. Gene counts were normalized using DESeq2 software [
31]. Differentially expressed genes (DEGs) were screened with a fold change > 2 as the criterion. Gene Ontology (GO) and KEGG were analyzed by Cluster Profile software, and significant enrichment was performed using padj<0.05 as the threshold.
2.6. Real Time Reverse Transcriptase PCR (RT‒PCR)
In order to analyze whether the expressions of virulence factors are regulated by L-Glu, mRNA levels of five virulence factors identified in
V. splendidus AJ01, namely
vspC [
13],
vsm [
32],
vshppd [
33],
hop [
34] and
fliC [
15] were detected using RT-PCR. The primers used in this study are listed in
Table 1. To collect the samples,
V. splendidus AJ01 was cultured to an OD
600 of approximately 0.5, washed twice using sterilized phosphate buffered saline (PBS) and divided into four aliquoats. The first aliquot was used as a control, the second aliquot was supplemented with 10 mM L-Glu, the third aliquot was supplemented with 100 μL of coelomic fluid from sea cucumbers, and the fourth aliquot was simultaneously added with 10 mM L-Glu and 100 μL of coelomic fluid. Then, all samples were incubated at 28 °C for 1 h, and cell pellet was collected after centrifugation. The extraction and reverse transcription of total RNA from bacterial cells was carried out according to the manufacturer's instructions (Vazyme, China). qRT‒PCR was performed in an ABI7500 instrument (Applied Biosystems, USA) using the SYBR ExScript RT‒PCR Kit (Vazyme, China). 16S rDNA was used as a reference gene to normalize the gene expression. Fold difference in mRNA levels was determined using the 2
−ΔΔCT method [
35].
2.7. Growth Measurement
V. splendidus AJ01 was cultured in 2216E medium containing 5 g/L tryptone, 1 g/L yeast extract and 0.01 g/L FePO
4 in filtered seawater at 28 °C. To analyze the effect of L-Glu on the growth of
V. splendidus AJ01, different concentrations of L-Glu were added to M9 minimal medium as the sole carbon source [
26], and then the absorbance of the culture was recorded at 600 nm. To determine the growth of
V. splendidus AJ01 in the coelomic fluid containing L-Glu, the filtrated and cell free coelomic fluid at a ratio of 1% or 10% (
v/v) was added into M9 medium containing 10 mM L-Glu, respectively. The absorbance at 600 nm was measured by Microplate Reader (FlexA-200, Allsheng, China) under the above incubation conditions. Each growth was repeated triplicates.
2.8. Swimming Motility Analysis
The swimming motility was measured as described previously [
29].
V. splendidus AJ01 was cultured in 2216E liquid medium to an OD
600 of approximately 0.5. After washed with PBS, the swimming motility was detected by needle inoculation on a soft M9 minimal medium plate containing 0.3% agar with 10 mM L-Glu as the solo carbon source. To analyze the effect of coelomic fluid on the bacterial swimming motility of
V. splendidus AJ01, the bacteria were inoculated into soft agar medium with 10% filtered coelomic fluid from sea cucumbers. Each group was repeated triplicates.
2.9. Data Accession Number
The raw data of the transcriptomic sequence have been deposited in the NCBI Short Read Archive (SRA) with accession number SRP446997.
3. Results
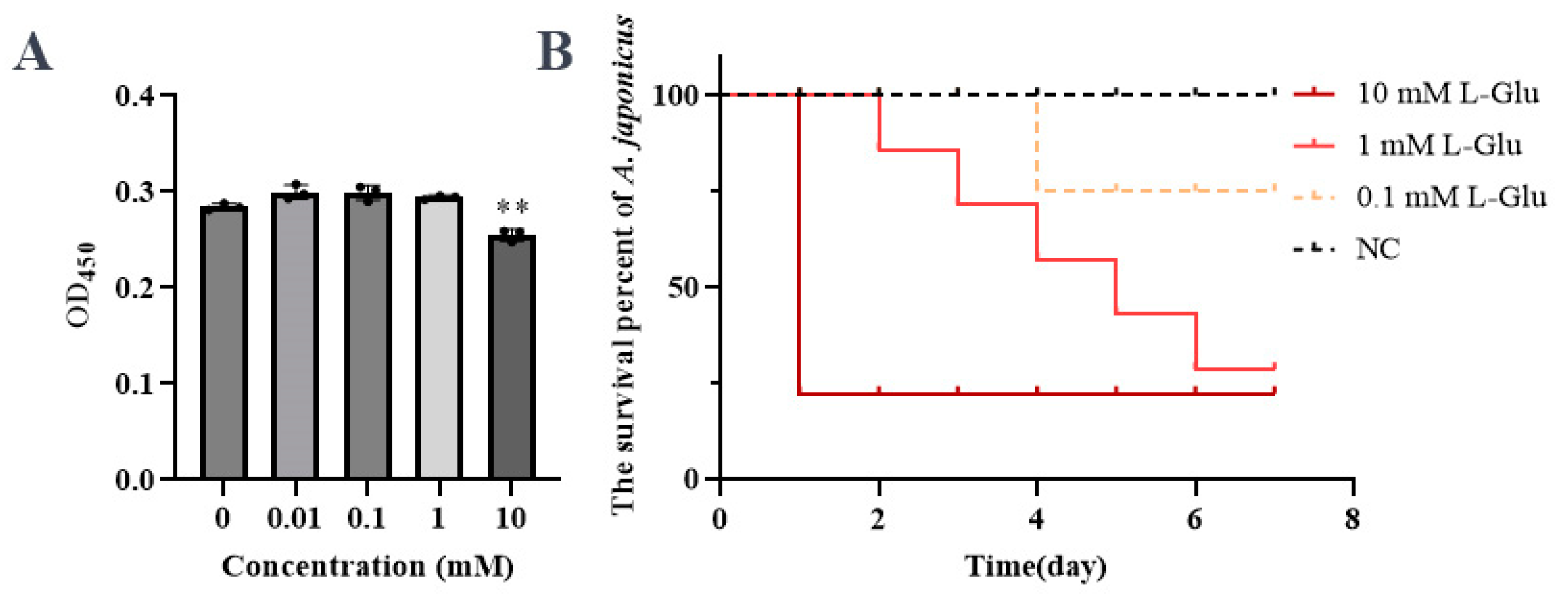
3.1. The Effect of L-Glu on Sea Cucumber Was at a Dose Dependent Manner
The effects of L-Glu on sea cucumbers were analyzed at both the cellular and individual levels. When 10 mM L-Glu was added to coelomocytes, the cell viability decreased significantly (
Figure 1A). At individual level, the survival percent decreased gradually as the concentration of L-Glu increased (
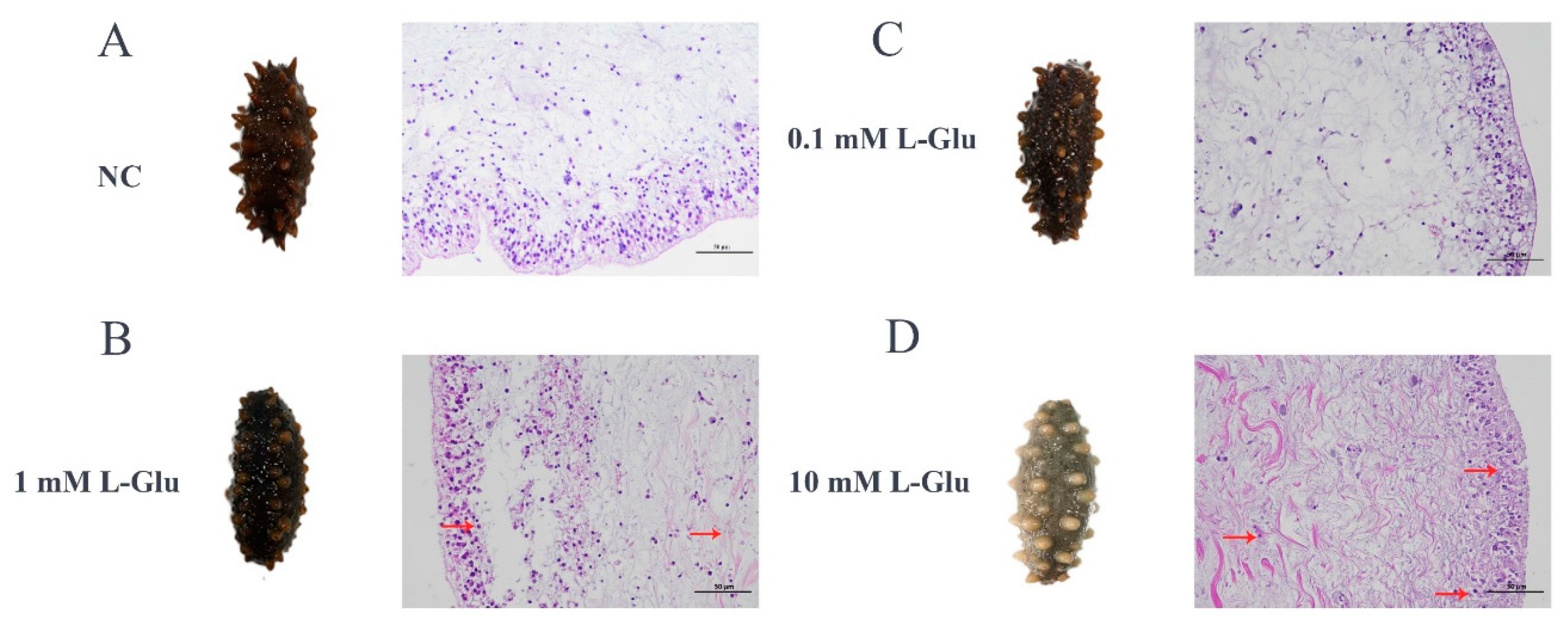
Figure 1B). Furthermore, histopathological observation of the body wall sections showed that L-Glu exhibited different degrees of tissue damage with tissue rupture and pyknosis at a dose dependent manner (
Figure 2). Combining the results of the above experiments, it could be concluded that 10 mM L-Glu showed obviously negative effects on coelomocyte, tissues and individual level, resulting in reduced cell viability, tissue damage and an extremely rapid decrease in survival. The damage to host gradually became unconspicuous as the concentration of exogenous L-Glu decreased. All these results indicated that L-Glu had a dose-dependent effect on sea cucumber, and excess L-Glu was detrimental to host survival.
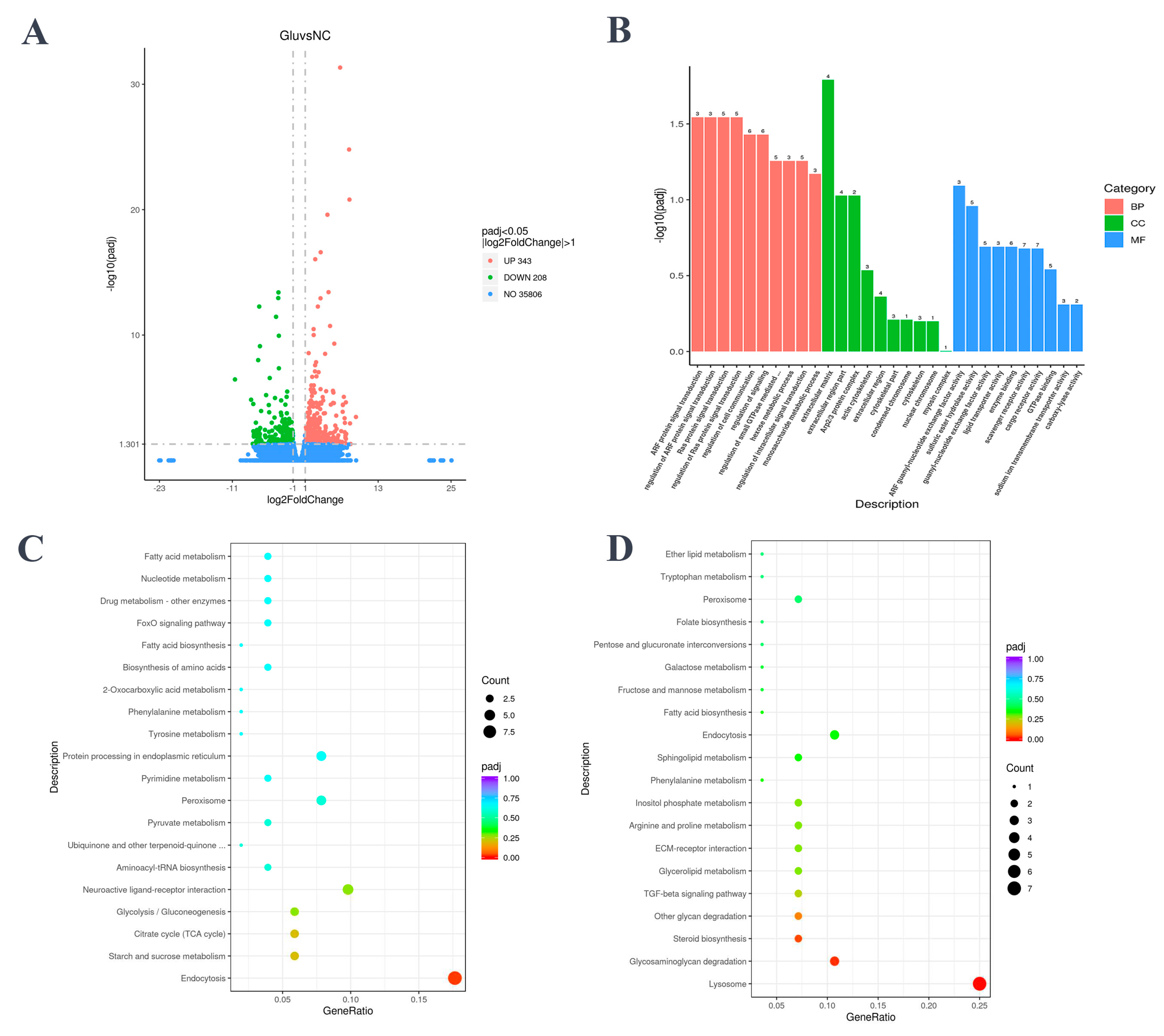
3.2. L-Glu Affected the Immune-Related Pathways in the Sea Cucumber
In the coelomocytes collected from individuals treated with L-Glu, 343 genes were significantly upregulated and 206 genes were significantly downregulated (
Figure 3A). The DEGs were enriched in three parts of the GO database, namely, cellular component (CC), molecular function (MF), and biological process (BP). In BP part, the terms with the most enriched DEGs was regulation of cell communication, and the most significant enriched terms were ADP-ribosylation factors (ARF) protein signal transduction and Ras protein signal transduction. In CC part, extracellular matrix (ECM) term was enriched for the most enriched DEGs and was the most significantly different CC. And in MF part, scavenger receptor activity, cargo receptor activity and enzyme binding were the top three entries (
Figure 3B).
In KEGG analysis, two pathways were significantly enriched. There were 9 upregulated DEGs significantly enriched in endocytosis pathway, while 7 downregulated GEGs were significantly enriched in lysosome pathway (
Figure 3C,D).
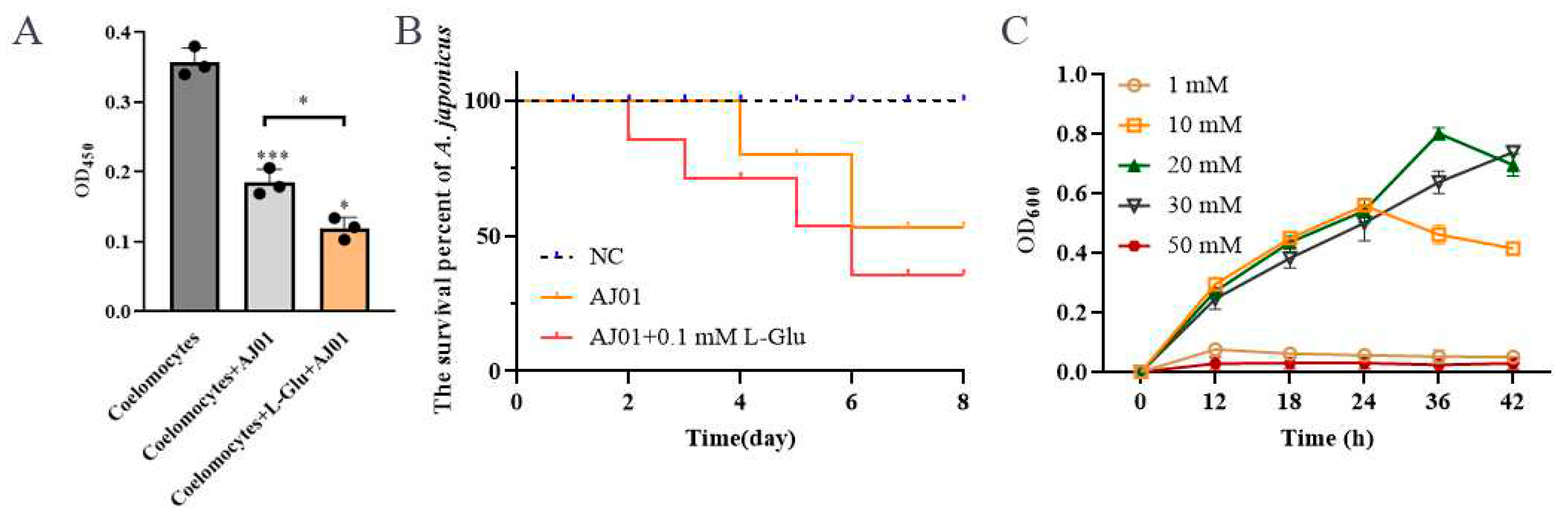
3.3. L-Glu Promoted the Virulence of V. splendidus AJ01
To determine the role of L-Glu in pathogen-host interactions, viability of coelomocyte and bacterial infection experiment were analyzed.
V. splendidus AJ01 significantly reduced the viability of cultured primary coelomocytes and led to a decrease in survival percent of sea cucumber. In the presence of L-Glu, cell viability and survival percent were further reduced (
Figure 4A,B). The viability of the coelomocytes reduced to 79% after the addition of
V. splendidus AJ01, and the viability further decreased 20% with the addition of L-Glu. Therefore, L-Glu significantly increased the virulence of
V. splendidus AJ01 on coelomocytes. These results suggested that L-Glu contributed to the virulence of
V. splendidus AJ01 to its host sea cucumber. To show whether L-Glu could promote the propagation of
V. splendidus, the bacterial growth was determined in the presence of L-Glu. It was shown that
V. splendidus AJ01 did not show obvious growth when L-Glu was added at a low concentration of 1 mM. As the concentration of L-Glu increased,
V. splendidus AJ01 showed obvious growth. But when the concentration of L-Glu increased to 50 mM, the growth of
V. splendidus AJ01 was completely inhibited (
Figure 4C).
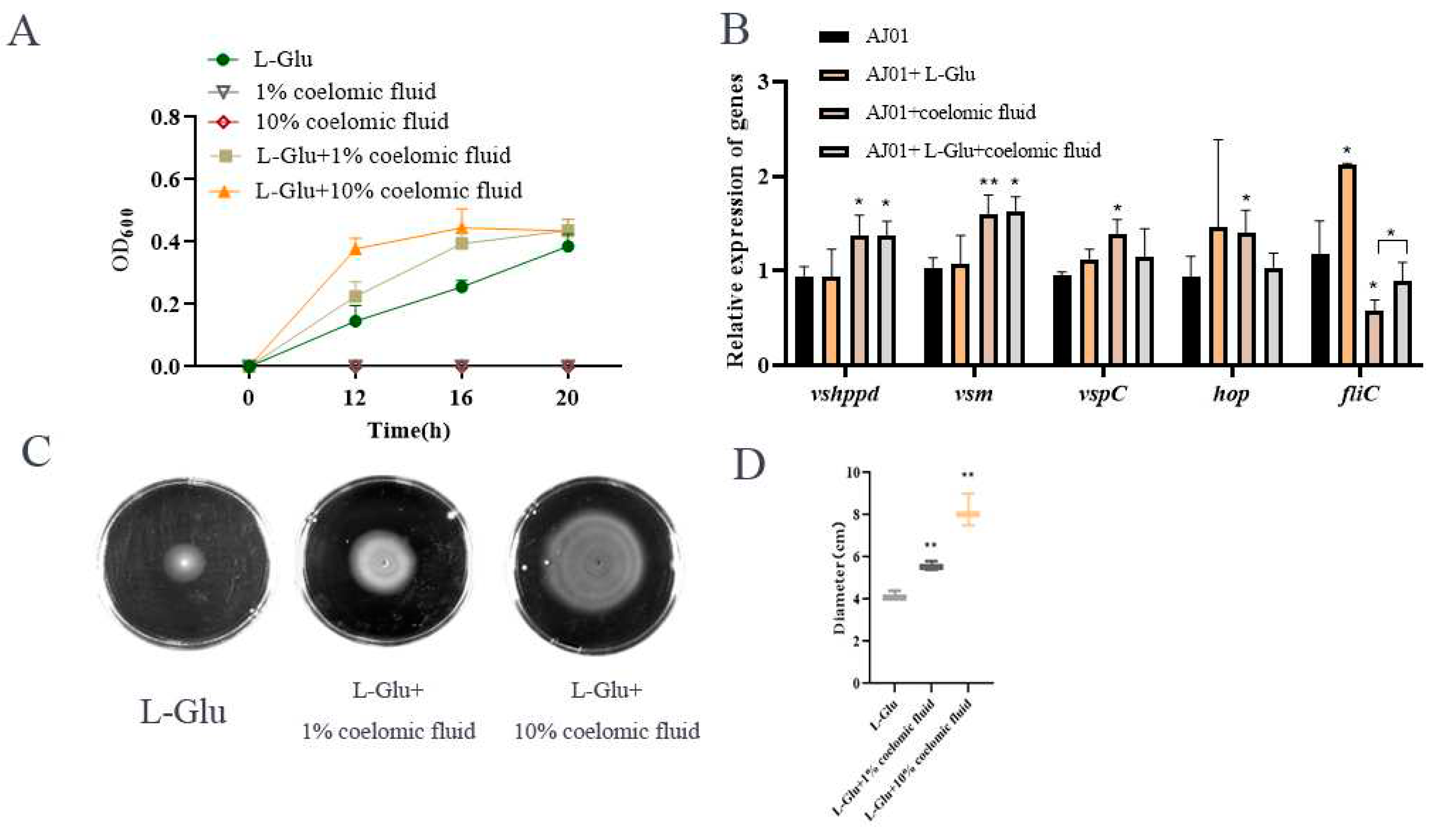
3.4. L-Glu Enhanced the Swimming Motility of V. splendidus AJ01
V. splendidus AJ01 could not grow in media supplemented with only 1% (
v/v) or 10% (
v/v) of coelomic fluid (
Figure 5A). But when
V. splendidus AJ01 was inoculated into the M9 minimal medium containing different volumes of coelomic fluid and L-Glu, the growths of
V. splendidus AJ01 were obvious with the amount of coelomic fluid increased, which was totally different from the growth in the coelomic fluid or L-Glu (
Figure 5A). Based on
Figure 4C, 10 mM L-Glu which was just sufficient for the growth of
V. splendidus AJ01, was selected to study the expressions of the virulence factor related genes. The virulence factors were mainly upregulated in the presence of coelomic fluid, but surprisingly, however, among the tested genes, only the mRNA level of the
fliC gene was significantly upregulated in the presence of L-Glu and then recovered, when the coelomic fluid and L-Glu were simultaneously presented (
Figure 5B).
In addition, the motility of
V. splendidus AJ01 was also promoted in the simultaneous presence of coelomic fluid and L-Glu, as the volume of coelomic fluid increased (
Figure 5C,D). The growth and motility of
V. splendidus AJ01 under the above conditions indicated that that L-Glu promoted the response of
V. splendidus AJ01 to coelomic fluid by increasing proliferation ability and the bacterial cell motility.
4. Discussion
L-Glu is a multi-functional AA that occupies a central position in amino acid metabolism. In the studies of the effects of L-Glu on aquaculture animals, it is mainly used as one of the feed additives and has been found to improve the growth performance and development of animals [
36]. For example, dietary supplementation with 2% Amino Gut (a mixture of feed-grade Glu and Gln) enhanced the survival of juvenile Nile tilapia [
37], while less study was focused on its effects on immunity [
38]. Our present study first proposed the inhibitory effect of exogenously redundant L-Glu on the cell viability of coelomocytes from sea cucumber, which is confirmed by the results of tissue damage and reduced survival rate. This result was similar to that of L-Glu-treated A1 astrocytes, where excess L-Glu could induces neuronal hyperexcitability and excitotoxicity [
39]. In this study, DEGs of sea cucumbers immersed in seawater containing L-Glu were analyzed to determine the direct effect of L-Glu on the immunity of sea cucumber.
Endocytosis is the process of translocating extracellular substances into the cell [
40], and
V. splendidus has been shown to enter the coelomocytes via endocytosis [
41]. Therefore, the upregulation of endocytosis pathway suggested that more
V. splendidus could internalized into the coelomocyte of sea cucumber. While the lysosomal pathway mainly uses a variety of proteases to digest a variety of macromolecules [
42], through which
V. splendidus can be recognized by the host and cleared through the lysosomal pathway [
43]. Thus, the downregulation of lysosomal related proteases will lead to impaired lysosomal function and increasing the burden on the host, which in turn might bring a burden on the host. In all, the upregulation of endocytosis pathways combined with the downregulation of lysosomal pathway in sea cucumber with exogenous L-Glu suggested an attenuated immune related ability to lyse bacteria during infection, thus would promote bacterial invasion. In our transcriptomic analysis, AFRs-related proteins were significantly upregulated and were significantly enriched in endocytosis pathway. It has been shown that ARFs related proteins can affect signaling pathways and participate in the regulation of endocytosis [
44]. In addition, studies have shown that ARF protein mediates susceptibility to a variety of pathogens [
45], which was consistent with our hypothesis that L-Glu may promote the infection of
V. splendidus AJ01 to sea cucumber.
Both host and pathogen can influence AA availability to their respective advantage [
21]. Certain amino acids such as arginine influence competition between host and pathogen, because pathogens compete with the host for these amino acids through certain strategies [
46]. There have been some sporadic reports of L-Glu mediated interactions between host and pathogen. L-Glu upregulated the expressions of pathogen-associated molecular patterns in the host, while it increased the colonization and the expressions of virulence related genes of extracellular polysaccharide production, cellulase activity, swimming ability and biofilm formation of
R. solanacearum [
47]. In our study, L-Glu could also promote
V. splendidus AJ01 infection, however, contrary to the upregulated cholera toxin production in
Vibrio cholerae [
48], L-Glu only upregulated the expression of the
fliC gene in
V. splendidus to increase bacterial swimming ability in the host environment, which has been well defined as a virulence factor mediating pathogenicity [
16].
5. Conclusions
In the present study, the effects of exogenous L-Glu on the aspects of both host and pathogen were analyzed. On one hand, L-Glu could affect host immunity at a dose dependent manner, leading to upregulation of endocytosis pathway and downregulation of lysosomal pathway at higher additive concentration. On the other hand, L-Glu could not only be used by V. splendidus AJ01 as the carbon source, but also increase the bacterial swimming motility, both of which promoted the infection of sea cucumber by V. splendidus. The results of this study demonstrated that L-Glu affected the outcome of infection by regulating the immune responses of the host and the growth and motility of the pathogens.
Author Contributions
Conceptualization, W.Z. and Y.L.; methodology, Y.L. and W.S.; software, Y.L. and W.S.; validation, Y.L., W.S. and W.Z.; formal analysis, Y.L.; investigation, Y.L.; resources, Y.L. and W.S.; data curation, Y.L. and W.S.; writing—original draft preparation, Y.L.; writing—review and editing, W.Z.; visualization, Y.L.; supervision, W.Z.; project administration, Y.L. and W.Z.; funding acquisition, W.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholar (LR20C190001), the National Natural Science Foundation of China (31972833), and the K.C. Wong Magna Fund at Ningbo University.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from one of the corresponding authors, Weiwei Zhang, on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vezzulli, L.; Pezzati, E.; Stauder, M.; Stagnaro, L.; Venier, P.; Pruzzo, C. Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus. Environ Microbiol 2015, 17, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Kita-Tsukamoto, K.; Thompson, F.L. Inferring the evolutionary history of Vibrios by means of multilocus sequence analysis. J Bacteriol 2007, 189, 7932–7936. [Google Scholar] [CrossRef]
- Zhang, W.; Li, C.H. Virulence mechanisms of Splendidus clade strains, emerging aquaculture pathogens, from case studies and the genome database. Rev Aquac 2021, 13, 2004–2026. [Google Scholar] [CrossRef]
- Gatesoupe, F.J.; Lambert, C.; Nicolas, J.L. Pathogenicity of Vibrio splendidus strains associated with turbot larvae, Scophthalmus maximus. J Appl Microbiol 1999, 87, 757–763. [Google Scholar] [CrossRef]
- Liu, R.; Qiu, L.; Yu, Z.; Zi, J.; Yue, F.; Wang, L.; Song, L. Identifification and characterisation of pathogenic Vibrio splendidus from Yesso scallop (Patinopecten yessoensis) cultured in a low temperature environment. J Invertebr Pathol 2013, 114, 144–150. [Google Scholar] [CrossRef]
- Garnier, M.; Labreuche, Y.; Garcia, C.; Robert, M.; Nicolas, J.L. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacifific oyster Crassostrea gigas. Microb. Ecol 2007, 53, 187–196. [Google Scholar] [CrossRef]
- Gevers, D.; Cohan, F.M.; Lawrence, J.G.; Spratt, B.G.; Coenye, T.; Feil, E.J.; Stackebrandt, E.; Van de Peer, Y.; Vandamme, P.; Thompson, F.L.; Swings, J. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol 2005, 3, 733–739. [Google Scholar] [CrossRef]
- Charles, M.; Trancart, S.; Oden, E.; Houssin, M. Experimental infection of Mytilus edulis by two Vibrio splendidus-related strains: Determination of pathogenicity level of strains and influence of the origin and annual cycle of mussels on their sensitivity. J Fish Dis 2020, 43, 9–21. [Google Scholar] [CrossRef]
- Gao, Q.; Liao, M.; Wang, Y.; Li, B.; Zhang, Z.; Rong, X.; Chen, G.; Wang, L. Transcriptome analysis and discovery of genes involved in immune pathways from coelomocytes of sea cucumber (Apostichopus japonicus) after Vibrio splendidus challenge. Int J Mol Sci 2015, 16, 16347–16377. [Google Scholar] [CrossRef]
- Duperthuy, M.; Binesse, J.; Le Roux, F.; Romestand, B.; Caro, A.; Got, P.; Givaudan, A.; Mazel, D.; Bachère, E.; Destoumieux-Garzón, D. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol 2010, 12, 951–963. [Google Scholar] [CrossRef]
- Binesse, J.; Delsert, C.; Saulnier, D.; Champomier-Verge`s, M.C.; Zagorec, M.; Munier-Lehmann, H.; Le Roux, F. Metalloprotease vsm is the major determinant of toxicity for extracellular products of Vibrio splendidus. Appl Environ Microbiol 2008, 74, 7108–7117. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, A,S. ; Duperthuy, M.; Charrie`re, G.M.; Le Roux, F.; Goudene`ge, D.; Gourbal, B.; Destoumieux-Garzo´n, D. Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ Microbiol 2015, 17, 1152–1165. [Google Scholar] [CrossRef]
- Li, Y.; Yang, H.R.; Zhang, J.X.; Shi, W.B.; Li, W.S.; Zhang, W.W. VspC from Vibrio splendidus is responsible for collagen degradation in Apostichopus japonicus. Aquaculture 2023, 571. [Google Scholar] [CrossRef]
- Oyanedel, D.; Labreuche, Y.; Bruto, M.; Amraoui, H.; Robino, E.; Haffner, P.; Rubio, T.; Charrière, G.M.; Le Roux, F. Destoumieux-Garzón, D. Vibrio splendidus O-antigen structure a trade-off between virulence to oysters and resistance to grazers. Environ Microbiol 2020, 22, 4264–4278. [Google Scholar]
- Dai, F.; Li, Y.; Shao, Y.N.; Li, C.H.; Zhang, W.W. FliC of Vibrio splendidus-related strain involved in adhesion to Apostichopus japonicus. Microb Pathogenesis 2020, 149, 104503. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Guo, M.; Shao, Y.; Li, C. Vibrio splendidus flagellin C binds tropomodulin to induce p38 MAPK-mediated p53-dependent coelomocyte apoptosis in Echinodermata. J Biol Chem 2022, 298, 102091. [Google Scholar] [CrossRef] [PubMed]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol Biol 2016, 90, 549–559. [Google Scholar] [CrossRef]
- Pashaei, S.; Yarani, R.; Mohammadi, P.; Emami Aleagha, M.S. The potential roles of amino acids and their major derivatives in the management of multiple sclerosis. Amino Acids 2022, 54, 841–858. [Google Scholar] [CrossRef]
- Olive, A.J.; Sassetti, C.M. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat Rev Microbiol 2016, 14, 221–234. [Google Scholar] [CrossRef]
- Yang, J.; Sun, C.; Fu, D.; Yu, T. Test for l-glutamate inhibition of growth of Alternaria alternata by inducing resistance in tomato fruit. Food Chem 2017, 230, 145–153. [Google Scholar] [CrossRef]
- Ren, W.; Rajendran, R.; Zhao, Y.; Tan, B.; Wu, G.; Bazer, F.W.; Zhu, G.; Peng, Y.; Huang, X.; Deng, J.; Yin, Y. Amino acids as mediators of metabolic cross talk between host and pathogen. Front Immunol 2018, 9, 319. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Brosnan, M.E. Glutamate: a truly functional amino acid. Amino Acids 2013, 45, 413–418. [Google Scholar] [CrossRef] [PubMed]
- O'Malley, M.R.; Kpenu, E.; Peck, S.C.; Anderson, J.C. Plant-exuded chemical signals induce surface attachment of the bacterial pathogen Pseudomonas syringae. PeerJ 2023, 11, e14862. [Google Scholar] [CrossRef]
- Shen, F.; Yin, W.; Song, S.; Zhang, Z.; Ye, P.; Zhang, Y.; Zhou, J.; He, F.; Li, P.; Deng, Y. Ralstonia solanacearum promotes pathogenicity by utilizing l-glutamic acid from host plants. Mol Plant Pathol 2020, 21, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Li, C.; Chen, X. Metabolomic responses of sea cucumber Apostichopus japonicus to thermal stresses. Aquaculture 2015, 435, 390–397. [Google Scholar] [CrossRef]
- Jiang, G.; Li, Y.; Li, Y.; Zhang, W.; Li, C. Selection of the amino acid and saccharide that increase the tetracycline susceptibility of Vibrio splendidus. Front Vet Sci 2022, 8, 823332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.J.; Li, C.H.; Zhang, P.; Jin, C.H.; Pan, D.D.; Bao, Y.B. iTRAQ-based proteomics reveals novel members involved in pathogen challenge in sea cucumber Apostichopus japonicus. PloS One 2014, 9, e100492. [Google Scholar] [CrossRef]
- Lv, Z.; Guo, M.; Zhao, X.; Shao, Y.; Zhang, W.; Li, C. IL-17/IL-17 Receptor pathway-mediated inflammatory response in apostichopus japonicus supports the conserved functions of cytokines in invertebrates. J Immunol 2022, 208, 464–479. [Google Scholar] [CrossRef]
- Li, Y.; Dai, F.; Li, Y.N.; Liang, W.K.; Li, C.H.; Zhang, W.W. Hfq, a global regulator contributes to the virulence of Vibrio splendidus AJ01. Aquaculture 2022, 546, 737416. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. ; HISAT, a fast spliced aligner with low memory requirements. Nat Methods 2015, 12, 357. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhang, C.; Liang, W.; Zhang, W.; Li, C. Characterization of a metalloprotease involved in Vibrio splendidus infection in the sea cucumber. Apostichopus japonicus 2016, 101, 96–103. [Google Scholar]
- Liang, W.K.; Zhang, C.; Liu, N.N.; Zhang, W.W.; Han, Q.X.; Li, C.H. Cloning and characterization of Vshppd, a gene inducing haemolysis and immune response of Apostichopus japonicus. Aquaculture 2016, 464, 246–252. [Google Scholar] [CrossRef]
- Zhuang, Q.T.; Dai, F.; Zhao, X.L.; Shao, Y.N.; Guo, M.; Lv, Z.M.; Li, C.H.; Zhang, W.W. Cloning and characterization of the virulence factor Hop from Vibrio splendidus. Microb Pathog. 2020, 139, 103900. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Valdebenito, I.; Moreno, C.; Lozano, C.; Ubilla, A. Effect of L-glutamate and glycine incorporated in activation media, on sperm motility and fertilization rate of rainbow trout (Oncorhynchus mykiss) spermatozoa. J Appl Ichthyol 2010, 26, 702–706. [Google Scholar] [CrossRef]
- Li, X.; Zheng, S.; Wu, G. Nutrition and metabolism of glutamate and glutamine in fish. Amino Acids 2020, 52, 671–691. [Google Scholar] [CrossRef]
- Cheng, Z.; Buentello, A.; Gatlin, D.M., III. Effects of dietary arginine and glutamine on growth performance, immune responses and intestinal structure of red drum, Sciaenops ocellatus. Aquaculture 2011, 319, 247–252. [Google Scholar] [CrossRef]
- Schousboe, A.; Scafidi, S.; Bak, L.K.; Waagepetersen, H.S.; McKenna, M. C. Glutamate metabolism in the brain focusing on astrocytes. Adv Neurobiol 2014, 11, 13–30. [Google Scholar]
- Cossart, P.; Helenius, A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol 2014, 6, a016972. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, S.; Shao, Y.; Guo, M.; Zhang, W.; Li, C. A unique NLRC4 receptor from echinoderms mediates Vibrio phagocytosis via rearrangement of the cytoskeleton and polymerization of F-actin. PLoS Pathog 2021, 17, e1010145. [Google Scholar] [CrossRef]
- Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Chiaradia, E.; Urbanelli, L.; Emiliani, C. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int J Mol Sci 2020, 21, 2576. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.; Guo, M.; Shao, Y.; Li, C. Novel secreted STPKLRR from Vibrio splendidus AJ01 promotes pathogen internalization via mediating tropomodulin phosphorylation dependent cytoskeleton rearrangement. PLoS Pathog 2023, 19, e1011419. [Google Scholar] [CrossRef] [PubMed]
- D'Souza-Schorey, C.; Chavrier, P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 2006, 7, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Reiling, J.H.; Olive, A.J.; Sanyal, S.; Carette, J.E.; Brummelkamp, T.R.; Ploegh, H.L.; Starnbach, M.N.; Sabatini, D.M. A CREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat Cell Biol 2013, 15, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Tattoli, I.; Sorbara, M.T.; Vuckovic, D.; Ling, A.; Soares, F.; Carneiro, L.A.; Yang, C.; Emili, A.; Philpott, D.J.; Girardin, S.E. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 2012, 11, 563–575. [Google Scholar] [CrossRef]
- Goto, Y.; Maki, N.; Ichihashi, Y.; Kitazawa, D.; Igarashi, D.; Kadota, Y.; Shirasu, K. Exogenous treatment with glutamate induces immune responses in arabidopsis. Mol Plant Microbe Interact 2020, 33, 474–487. [Google Scholar] [CrossRef]
- Nishiyama, S.; Suzuki, D.; Itoh, Y.; Suzuki, K.; Tajima, H.; Hyakutake, A.; Homma, M.; Butler-Wu, S.M.; Camilli, A.; Kawagishi, I. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun 2012, 80, 3170–3178. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).