Submitted:
02 August 2023
Posted:
04 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
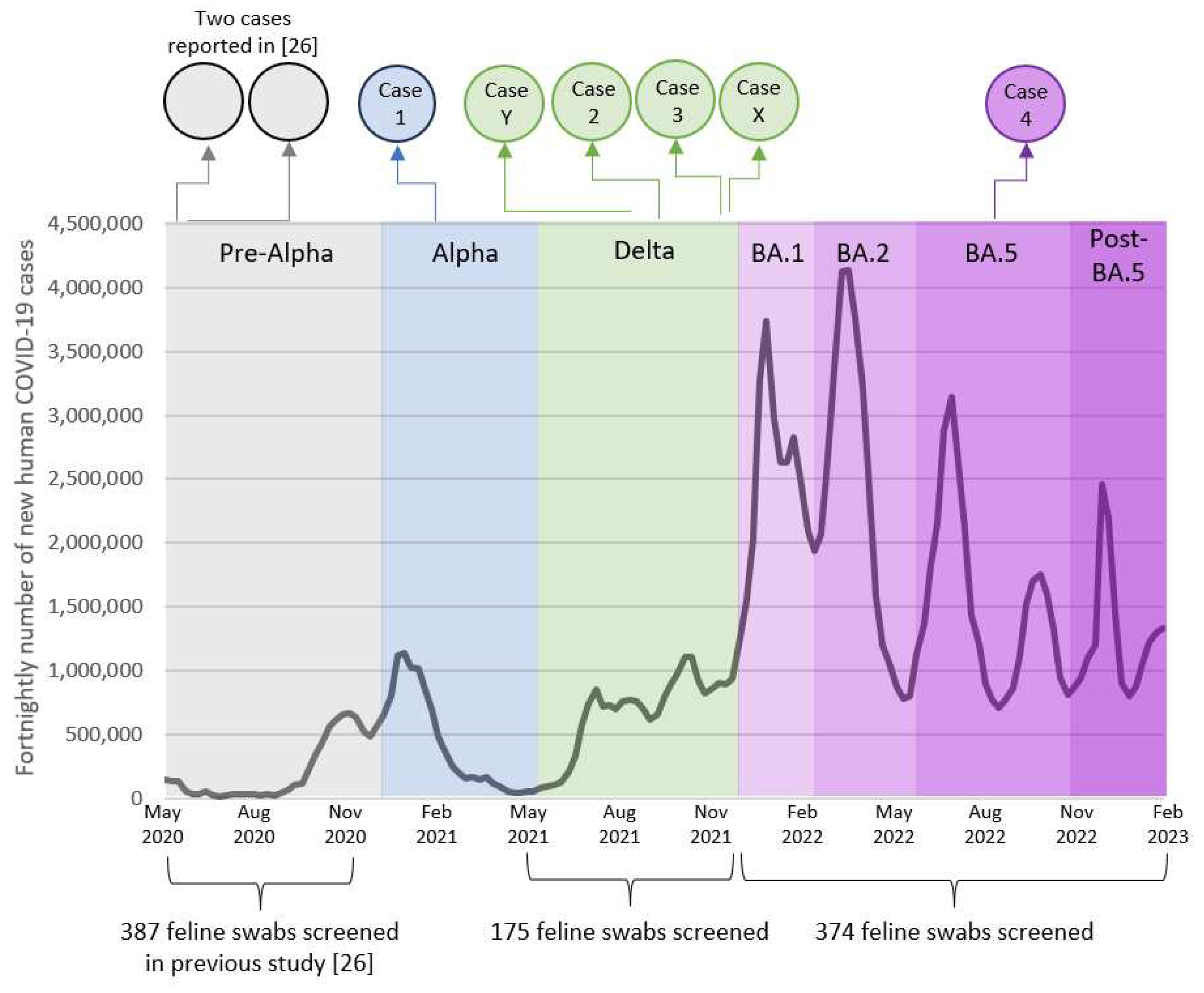
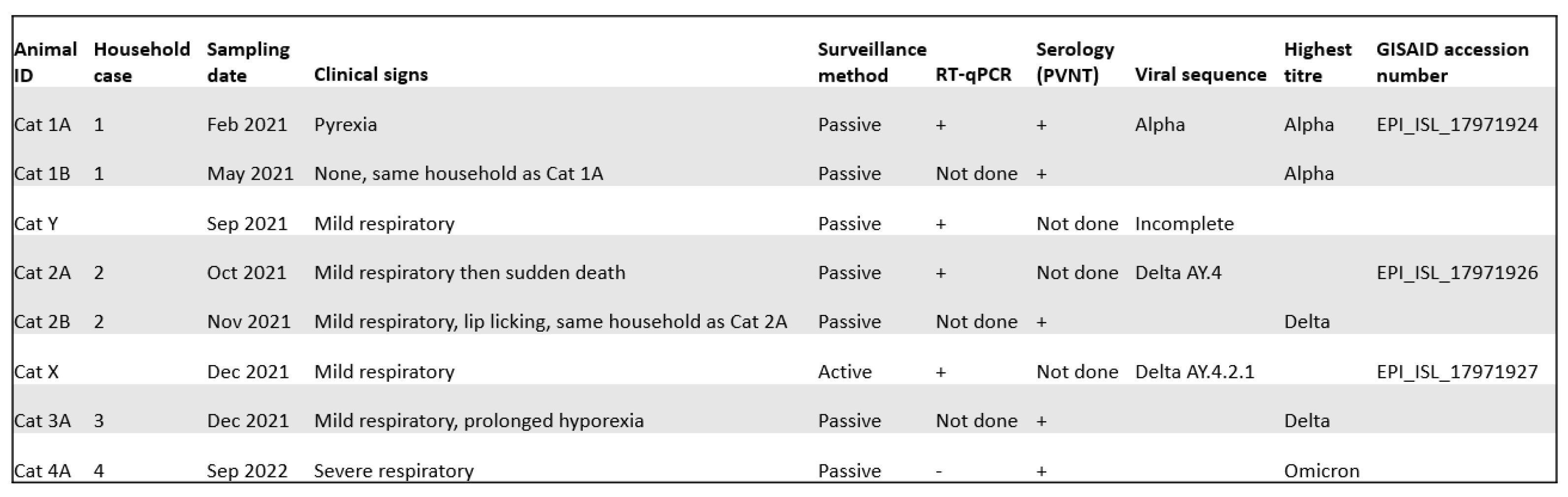
3. Results
3.1. Active surveillance
3.2. Passive surveillance
3.3. Seropositive cases identified previously
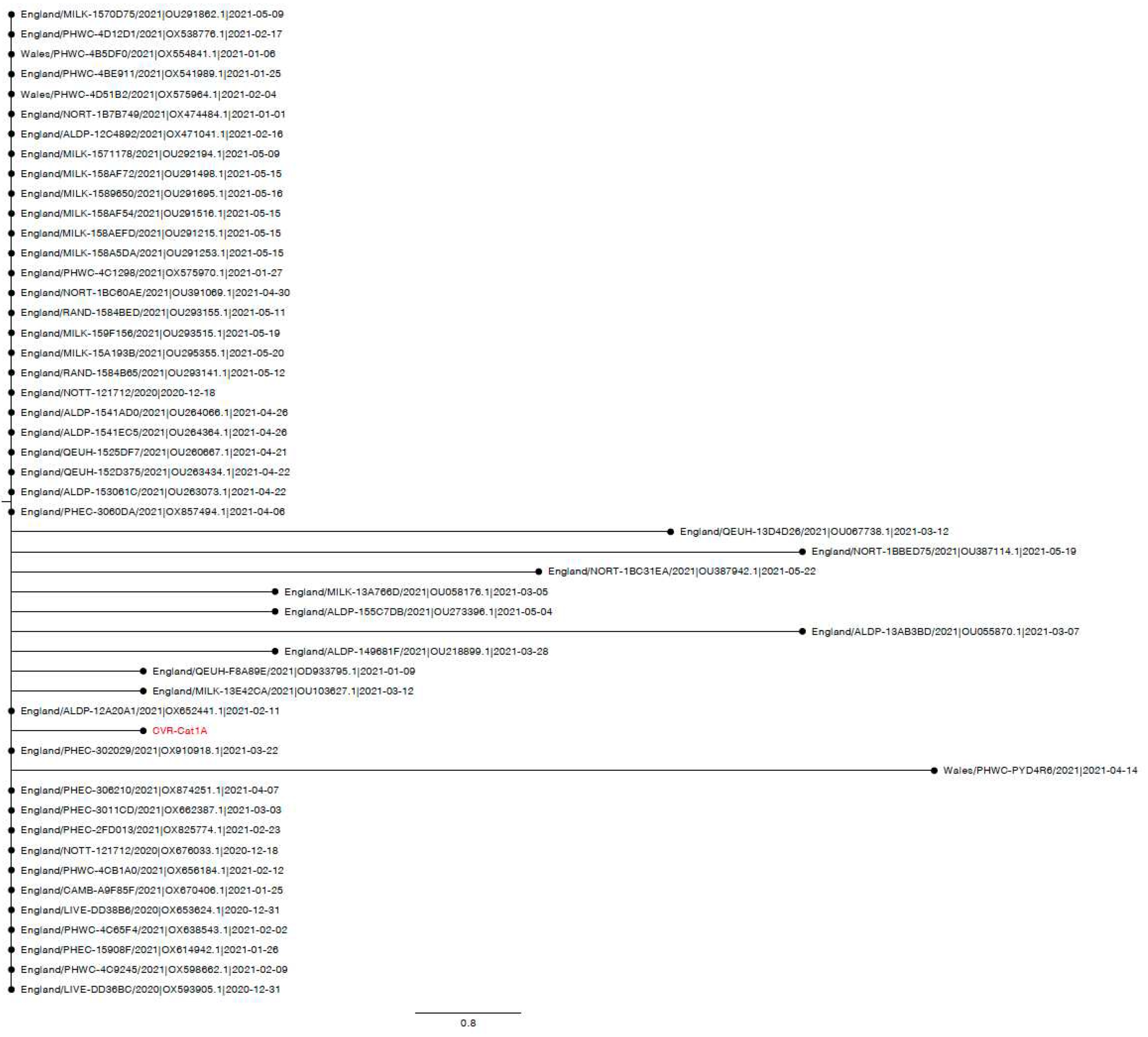
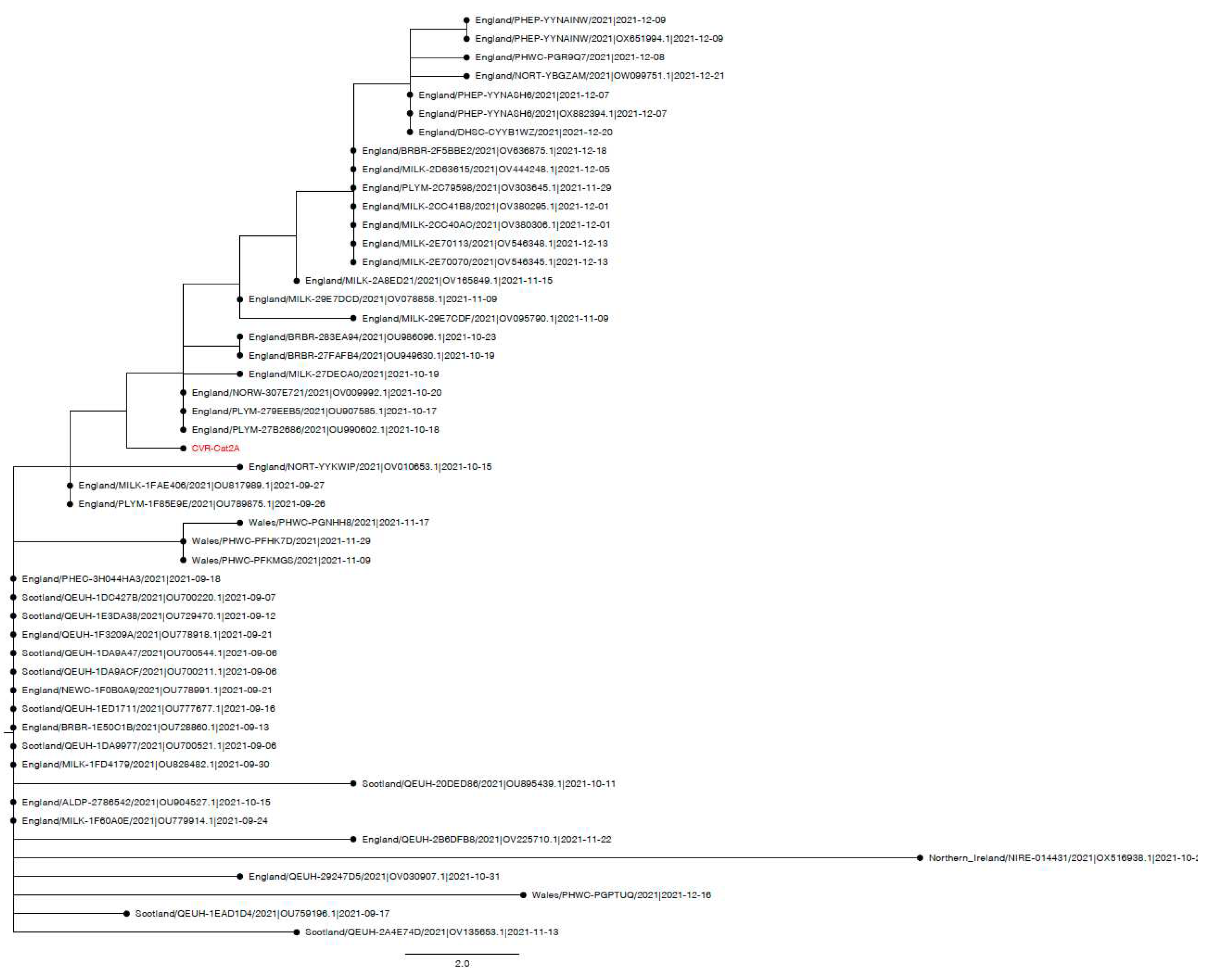
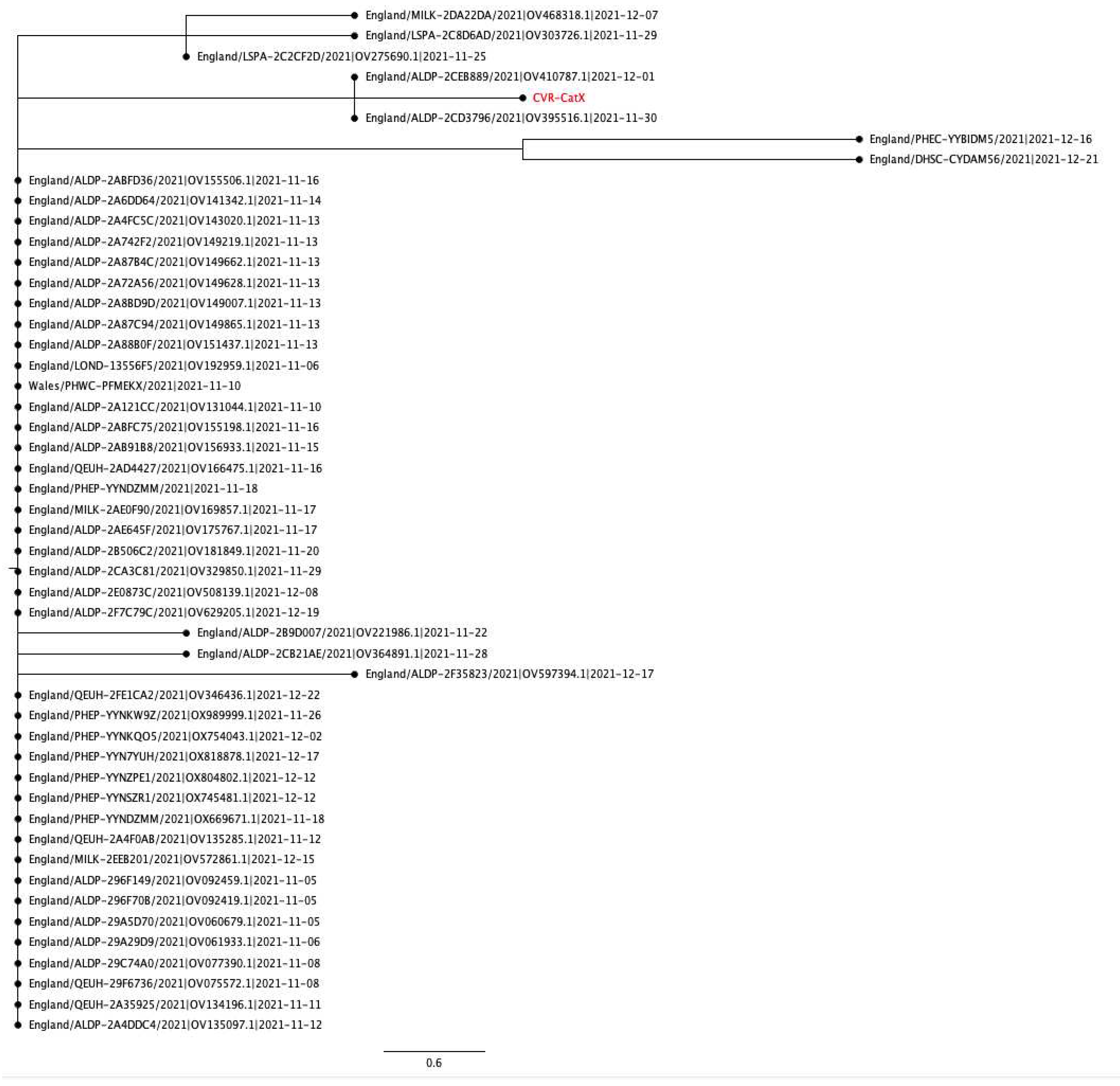
3.4. Case series
3.4.1. Case one: Acute pyrexia
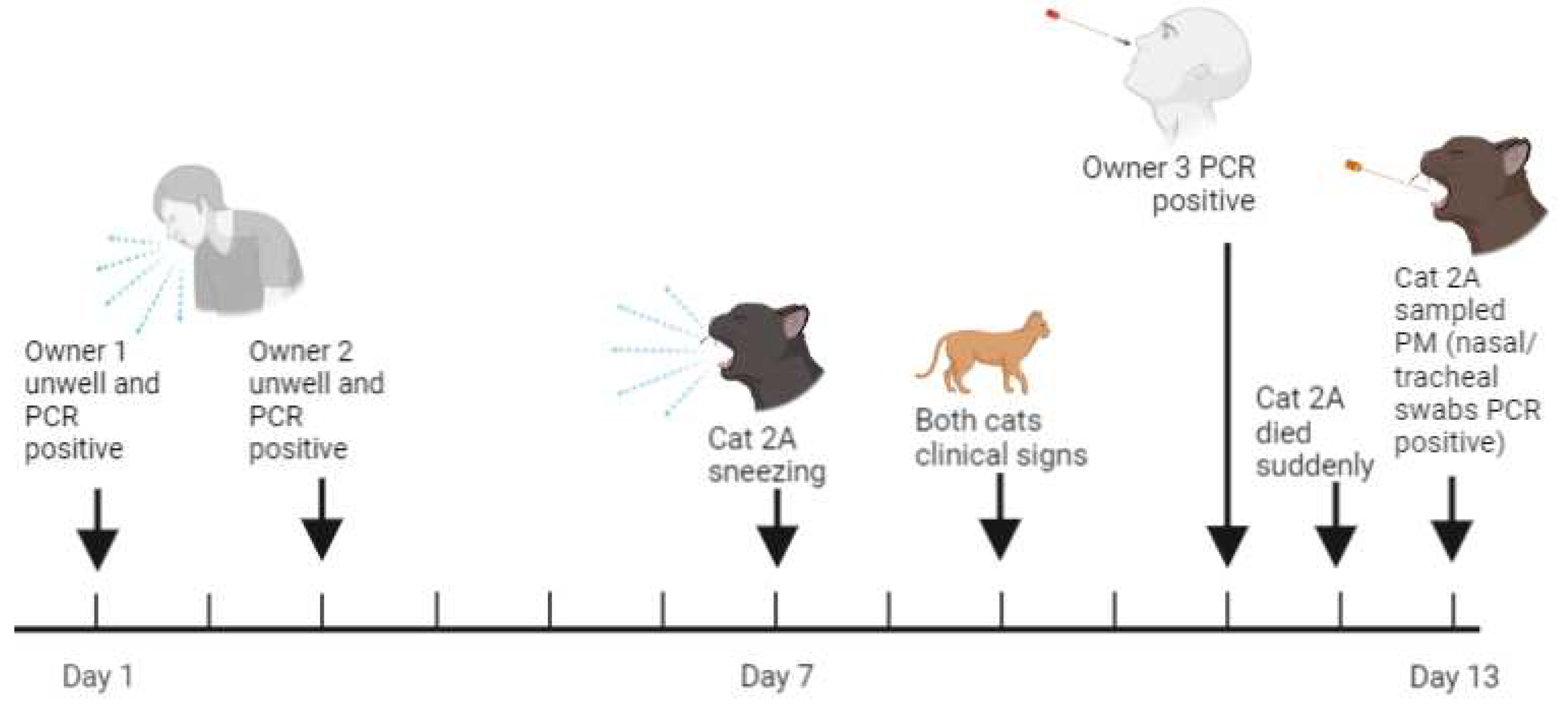
3.4.2. Case two: Sudden death
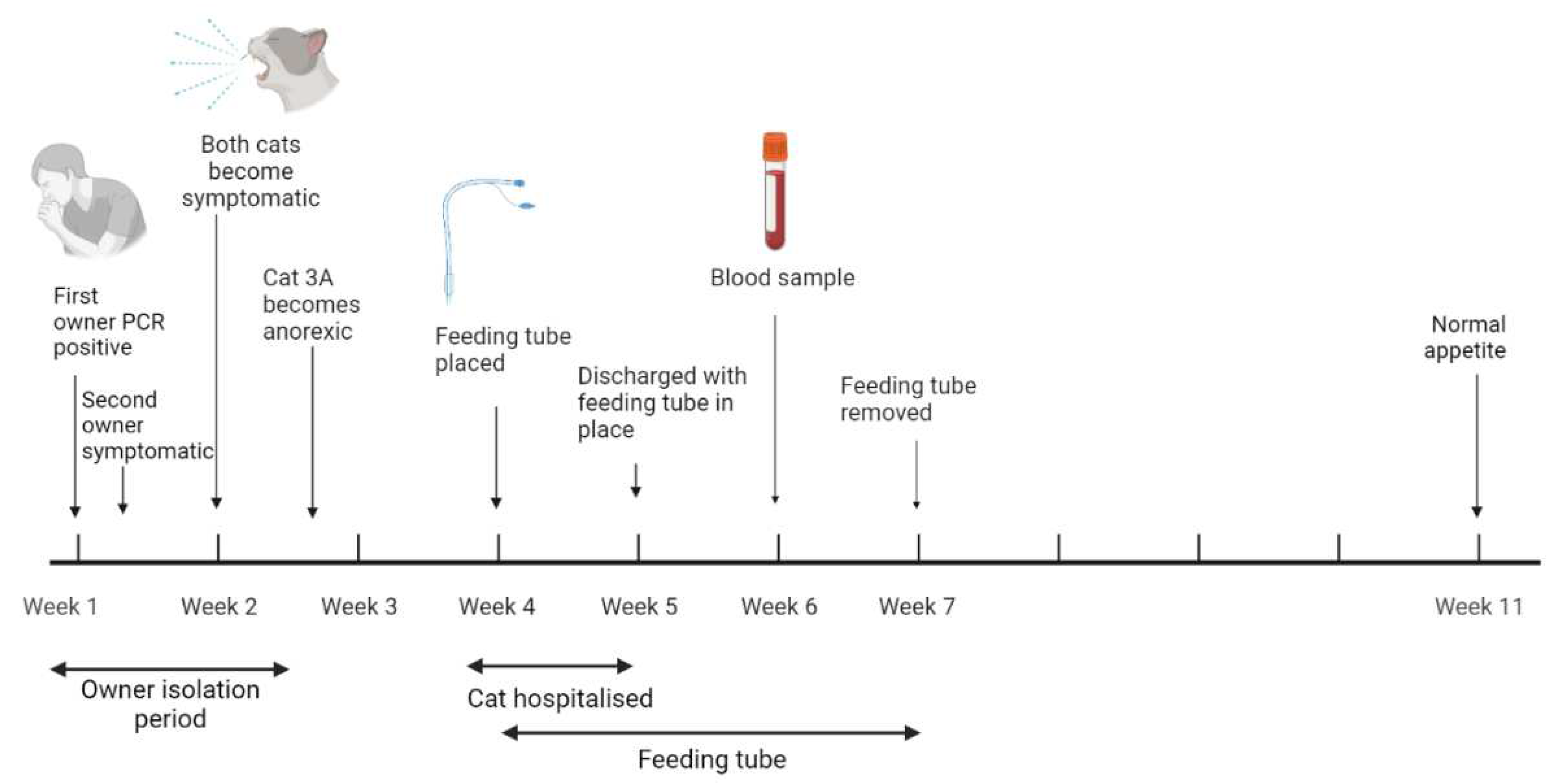
3.4.3. Case three: Anorexia
3.4.4. Case four: Respiratory distress and renal disease
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Supplementary methods
Appendix A.1. SARS-CoV-2 RT-qPCR
-
Reagents:PCR kit: NEB Luna® Universal Probe One-Step RT-qPCR Kit E3006Reaction mix: IDT 2019-nCoV RUO Kit #10006713PCR positive control plasmid: IDT 2019-nCoV_N_Positive Control #10006625Primers/probes: IDT CDC panel #10006713
-
Primer/probe sequences:SARS-CoV-2 N1 forward: GACCCCAAAATCAGCGAAATSARS-CoV-2 N1 reverse: TCTGGTTACTGCCAGTTGAATCTGSARS-CoV-2 N1 probe: ACCCCGCATTACGTTTGGTGGACC (label: FAM/BHQ1)SARS-CoV-2 N2 forward: TTACAAACATTGGCCGCAAASARS-CoV-2 N2 reverse: GCGCGACATTCCGAAGAASARS-CoV-2 N2 probe: ACAATTTGCCCCCAGCGCTTCAG (label: FAM/BHQ1)
- PCR machine: Applied Biosystems 7500 Real time PCR System
| Temp (oC) | Time | Cycles |
|---|---|---|
| 55 | 10 minutes | 1 |
| 95 | 1 minute | 1 |
| 95 | 10 seconds | 45 |
| 58 | 1 minute |
Appendix A.2. SARS-CoV-2 Viral Full Genome Sequencing
Appendix A.2.1. Cats 1A, 2A and Y
Appendix A.2.2. Cat X
Appendix A.3. Phylogenetic and Mutation analysis of Cat sequences
Appendix B. Supplementary information
| Swab | N1 assay Ct | N2 assay Ct |
|---|---|---|
| Nasal | 18 | 17.1 |
| Oral | Undet. | Undet. |
| Tracheal | 18.9 | 18.1 |
| Rectal | Undet. | 37.5 |
| Cat X (Delta AY.4.2.1) | Cat 2A (Delta AY.4) | Cat 1A (Alpha B.1.1.7) |
|---|---|---|
| T19R | T19R | |
| V36F | ||
| T95I | T95I | |
| I68I | ||
| H69del | ||
| V70del | ||
| G142D | G142D | |
| Y144del | ||
| Y145H | ||
| E156del | E156del | |
| F157del | F157del | |
| R158G | R158G | |
| A222V | ||
| L452R | L452R | |
| T478K | T478K | |
| N501Y | ||
| A570D | ||
| D614G | D614G | D614G |
| P681R | P681R | |
| P681H | ||
| T716I | ||
| L821L | ||
| D950N | D950N | |
| S982A | ||
| D1118H | ||
| L1141F |
References
- Nerpel, A.; Yang, L.; Sorger, J.; Käsbohrer, A.; Walzer, C.; Desvars-Larrive, A. SARS-ANI: a global open access dataset of reported SARS-CoV-2 events in animals. Scientific Data 2022, 9, 438. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.M.; Moreno, G.K.; Halfmann, P.J.; Hodcroft, E.B.; Baker, D.A.; Boehm, E.C.; Weiler, A.M.; Haj, A.K.; Hatta, M.; Chiba, S. Transmission of SARS-CoV-2 in domestic cats imposes a narrow bottleneck. PLoS Pathogens 2021, 17, e1009373. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.-i.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K. Transmission of SARS-CoV-2 in domestic cats. New England Journal of Medicine 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z. , et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Bosco-Lauth, A.M.; Hartwig, A.E.; Porter, S.M.; Gordy, P.W.; Nehring, M.; Byas, A.D.; VandeWoude, S.; Ragan, I.K.; Maison, R.M.; Bowen, R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci U S A 2020, 117, 26382–26388. [Google Scholar] [CrossRef]
- Sila, T.; Sunghan, J.; Laochareonsuk, W.; Surasombatpattana, S.; Kongkamol, C.; Ingviya, T.; Siripaitoon, P.; Kositpantawong, N.; Kanchanasuwan, S.; Hortiwakul, T. Suspected Cat-to-Human Transmission of SARS-CoV-2, Thailand, July–September 2021. Emerging Infectious Diseases 2022, 28, 1485. [Google Scholar] [CrossRef]
- Siegrist, A.A.; Richardson, K.L.; Ghai, R.R.; Pope, B.; Yeadon, J.; Culp, B.; Behravesh, C.B.; Liu, L.; Brown, J.A.; Boyer, L.V. Probable Transmission of SARS-CoV-2 from African Lion to Zoo Employees, Indiana, USA, 2021. Emerging Infectious Diseases 2023, 29, 1102. [Google Scholar] [CrossRef]
- OIE. Statement from the Advisory Group on SARS-CoV-2 Evolution in Animals. 2022. Available online: https://www.oie.int/app/uploads/2022/01/statement-agve-omicron.pdf (accessed on 11 January 2022).
- Kannekens-Jager, M.M.; de Rooij, M.M.; de Groot, Y.; Biesbroeck, E.; de Jong, M.K.; Pijnacker, T.; Smit, L.A.; Schuurman, N.; Broekhuizen-Stins, M.J.; Zhao, S. SARS-CoV-2 infection in dogs and cats is associated with contact to COVID-19-positive household members. Transboundary and Emerging Diseases 2022, 69, 4034–4040. [Google Scholar] [CrossRef]
- Goryoka, G.W.; Cossaboom, C.M.; Gharpure, R.; Dawson, P.; Tansey, C.; Rossow, J.; Mrotz, V.; Rooney, J.; Torchetti, M.; Loiacono, C.M. One Health investigation of SARS-CoV-2 infection and seropositivity among pets in households with confirmed human COVID-19 cases—Utah and Wisconsin, 2020. Viruses 2021, 13, 1813. [Google Scholar] [CrossRef]
- Barrs, V.R.; Peiris, M.; Tam, K.W.; Law, P.Y.; Brackman, C.J.; To, E.M.; Yu, V.Y.; Chu, D.K.; Perera, R.A.; Sit, T.H. SARS-CoV-2 in quarantined domestic cats from COVID-19 households or close contacts, Hong Kong, China. Emerging Infectious Diseases 2020, 26, 3071. [Google Scholar] [CrossRef]
- Bienzle, D.; Rousseau, J.; Marom, D.; MacNicol, J.; Jacobson, L.; Sparling, S.; Prystajecky, N.; Fraser, E.; Weese, J.S. Risk factors for SARS-CoV-2 infection and illness in cats and dogs. Emerging Infectious Diseases 2022, 28, 1154. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmeier, E.; Chan, T.; Meli, M.L.; Willi, B.; Wolfensberger, A.; Reitt, K.; Hüttl, J.; Jones, S.; Tyson, G.; Hosie, M.J. A Risk Factor Analysis of SARS-CoV-2 Infection in Animals in COVID-19-Affected Households. Viruses 2023, 15, 731. [Google Scholar] [CrossRef] [PubMed]
- SPI-M-O: Consensus Statement on COVID-19. 2021. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/993321/S1267_SPI-M-O_Consensus_Statement.pdf (accessed on 31 January 2022).
- Adler, J.M.; Weber, C.; Wernike, K.; Michelitsch, A.; Friedrich, K.; Trimpert, J.; Beer, M.; Kohn, B.; Osterrieder, K.; Müller, E. Prevalence of anti-severe acute respiratory syndrome coronavirus 2 antibodies in cats in Germany and other European countries in the early phase of the coronavirus disease-19 pandemic. Zoonoses and Public Health 2022, 69, 439–450. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira-Filho, E.F.; de Carvalho, O.V.; Carneiro, I.O.; Fernandes, F.; Vaz, S.N.; Pedroso, C.; Gonzalez-Auza, L.; Urbieta, V.C.; Kühne, A.; Mayoral, R. Frequent infection of cats with SARS-CoV-2 irrespective of pre-existing enzootic coronavirus immunity, Brazil 2020. Frontiers in Immunology 2022. [Google Scholar] [CrossRef] [PubMed]
- Bessière, P.; Vergne, T.; Battini, M.; Brun, J.; Averso, J.; Joly, E.; Guérin, J.-L.; Cadiergues, M.-C. SARS-CoV-2 Infection in Companion Animals: Prospective Serological Survey and Risk Factor Analysis in France. Viruses 2022, 14, 1178. [Google Scholar] [CrossRef]
- Smith, S.L.; Anderson, E.R.; Cansado-Utrilla, C.; Prince, T.; Farrell, S.; Brant, B.; Smyth, S.; Noble, P.M.; Pinchbeck, G.L.; Marshall, N. , et al. SARS-CoV-2 neutralising antibodies in dogs and cats in the United Kingdom. Curr Res Virol Sci 2021, 2, 100011. [Google Scholar] [CrossRef]
- Tyson, G.; Jones, S.; Logan, N.; McDonald, M.; Marshall, L.; Murcia, P.; Willett, B.; Weir, W.; Hosie, M. SARS-CoV-2 Seroprevalence and Cross-Variant Antibody Neutralization in Cats, United Kingdom. Emerging Infectious Diseases 2023, 29, 1223. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerging Microbes and Infections 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Barroso-Arévalo, S.; Sánchez-Morales, L.; Pérez-Sancho, M.; Domínguez, L.; Sánchez-Vizcaíno, J.M. First detection of SARS-CoV-2 B.1.617.2 (Delta) variant of concern in a symptomatic cat in Spain. Frontiers in Veterinary Science 2022, 9. [Google Scholar] [CrossRef]
- Neira, V.; Brito, B.; Agüero, B.; Berrios, F.; Valdés, V.; Gutierrez, A.; Ariyama, N.; Espinoza, P.; Retamal, P.; Holmes, E.C. A household case evidences shorter shedding of SARS-CoV-2 in naturally infected cats compared to their human owners. Emerging Microbes & Infections 2021, 10, 376–383. [Google Scholar] [CrossRef]
- Liew, A.Y.; Carpenter, A.; Moore, T.A.; Wallace, R.M.; Hamer, S.A.; Hamer, G.L.; Fischer, R.S.; Zecca, I.B.; Davila, E.; Auckland, L.D. Clinical and epidemiologic features of SARS-CoV-2 in dogs and cats compiled through national surveillance in the United States. Journal of the American Veterinary Medical Association 2023, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmeier, E.; Chan, T.; Agüí, C.V.; Willi, B.; Wolfensberger, A.; Beisel, C.; Topolsky, I.; Beerenwinkel, N.; Stadler, T.; Jones, S. Detection and Molecular Characterization of the SARS-CoV-2 Delta Variant and the Specific Immune Response in Companion Animals in Switzerland. Viruses 2023, 15, 245. [Google Scholar] [CrossRef] [PubMed]
- Garigliany, M.; Van Laere, A.S.; Clercx, C.; Giet, D.; Escriou, N.; Huon, C.; van der Werf, S.; Eloit, M.; Desmecht, D. SARS-CoV-2 Natural Transmission from Human to Cat, Belgium, March 2020. Emerging Infectious Diseases 2020, 26, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F. , et al. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet Rec 2021, 188, e247. [Google Scholar] [CrossRef]
- Klaus, J.; Meli, M.L.; Willi, B.; Nadeau, S.; Beisel, C.; Stadler, T.; Team, E.S.-C.-S.; Egberink, H.; Zhao, S.; Lutz, H. Detection and genome sequencing of SARS-CoV-2 in a domestic cat with respiratory signs in Switzerland. Viruses 2021, 13, 496. [Google Scholar] [CrossRef]
- Keller, M.; Hagag, I.T.; Balzer, J.; Beyer, K.; Kersebohm, J.C.; Sadeghi, B.; Wernike, K.; Höper, D.; Wylezich, C.; Beer, M. Detection of SARS-CoV-2 variant B.1.1.7 in a cat in Germany. Research in Veterinary Science 2021, 140, 229–232. [Google Scholar] [CrossRef]
- Zoccola, R.; Beltramo, C.; Magris, G.; Peletto, S.; Acutis, P.; Bozzetta, E.; Radovic, S.; Zappulla, F.; Porzio, A.M.; Gennero, M.S. First detection of an Italian human-to-cat outbreak of SARS-CoV-2 Alpha variant–lineage B.1.1.7. One Health 2021, 13, 100295. [Google Scholar] [CrossRef]
- Ferasin, L.; Fritz, M.; Ferasin, H.; Becquart, P.; Corbet, S.; Ar Gouilh, M.; Legros, V.; Leroy, E.M. Infection with SARS-CoV-2 variant B.1.1.7 detected in a group of dogs and cats with suspected myocarditis. Vet Rec 2021, 189, e944. [Google Scholar] [CrossRef]
- Carvallo, F.R.; Martins, M.; Joshi, L.R.; Caserta, L.C.; Mitchell, P.K.; Cecere, T.; Hancock, S.; Goodrich, E.L.; Murphy, J.; Diel, D.G. Severe SARS-CoV-2 Infection in a Cat with Hypertrophic Cardiomyopathy. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Chetboul, V.; Foulex, P.; Kartout, K.; Klein, A.M.; Sailleau, C.; Dumarest, M.; Delaplace, M.; Gouilh, M.A.; Mortier, J.; Le Poder, S. Myocarditis and Subclinical-Like Infection Associated With SARS-CoV-2 in Two Cats Living in the Same Household in France: A Case Report With Literature Review. Frontiers in Veterinary Science 2021. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D. From people to Panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. MBio 2020, 11, e02220. [Google Scholar] [CrossRef]
- Bartlett, S.L.; Diel, D.G.; Wang, L.; Zec, S.; Laverack, M.; Martins, M.; Caserta, L.C.; Killian, M.L.; Terio, K.; Olmstead, C. SARS-CoV-2 infection and longitudinal fecal screening in Malayan tigers (Panthera tigris jacksoni), Amur tigers (Panthera tigris altaica), and African lions (Panthera leo krugeri) at the Bronx Zoo, New York, USA. Journal of Zoo and Wildlife Medicine 2021, 51, 733–744. [Google Scholar] [CrossRef]
- Mishra, A.; Kumar, N.; Bhatia, S.; Aasdev, A.; Kanniappan, S.; Sekhar, A.T.; Gopinadhan, A.; Silambarasan, R.; Sreekumar, C.; Dubey, C.K. Sars-CoV-2 Delta variant among asiatic lions, India. Emerging Infectious Diseases 2021, 27, 2723. [Google Scholar] [CrossRef]
- Giraldo-Ramirez, S.; Rendon-Marin, S.; Jaimes, J.A.; Martinez-Gutierrez, M.; Ruiz-Saenz, J. SARS-CoV-2 clinical outcome in domestic and wild cats: A systematic review. Animals 2021, 11, 2056. [Google Scholar] [CrossRef] [PubMed]
- Klaus, J.; Zini, E.; Hartmann, K.; Egberink, H.; Kipar, A.; Bergmann, M.; Palizzotto, C.; Zhao, S.; Rossi, F.; Franco, V. SARS-CoV-2 infection in dogs and cats from southern Germany and northern Italy during the first wave of the COVID-19 pandemic. Viruses 2021, 13, 1453. [Google Scholar] [CrossRef] [PubMed]
- Murcia, P.; Streiker, D.; Philipe, A.D.S.; Robertson, D.; Jarrett, R.; Willett, B.; Hosie, M.; Biek, R.; Allan, K.; Weir, W. Send cat and dog samples to test for SARS-CoV-2. The Veterinary Record 2020, 186, 571. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.; Salimans, M.; Jansen, C.; Wertheim-van Dillen, P.; Van der Noordaa, J. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Davis, C.; Logan, N.; Tyson, G.; Orton, R.; Harvey, W.T.; Perkins, J.S.; Mollett, G.; Blacow, R.M.; Consortium, C.-G.U.; Peacock, T.P. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathogens 2021, 17, e1010022. [Google Scholar] [CrossRef]
- Willett, B.J.; Grove, J.; MacLean, O.A.; Wilkie, C.; De Lorenzo, G.; Furnon, W.; Cantoni, D.; Scott, S.; Logan, N.; Ashraf, S. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nature Microbiology 2022, 7, 1161–1179. [Google Scholar] [CrossRef]
- APHA. SARS-CoV-2 in Animals – Case Definition, Testing and International Reporting Obligations. 1 May 2021. Available online: http://apha.defra.gov.uk/documents/ov/Briefing-Note-0921.pdf (accessed on 1 May 2021).
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B. , et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evolution 2021, 7. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W. GISAID’s role in pandemic response. China CDC Weekly 2021, 3, 1049. [Google Scholar] [CrossRef] [PubMed]
- International Renal Interest Society. IRIS Staging of CKD (modified in 2023). 2023. Available online: http://www.iris-kidney.com/guidelines/staging.html (accessed on 14 June 2023).
- Office for National Statistics (ONS). Coronavirus (COVID19) Infection Survey technical article: Cumulative incidence of the number of people who have been infected with COVID-19 by variant and age, England: 09 February 2023. 09 February 2023. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/articles/coronaviruscovid19infectionsurveytechnicalarticlecumulativeincidenceofthenumberofpeoplewhohavebeeninfectedwithcovid19byvariantandageengland/9february2023 (accessed on 1 March 2023).
- Martins, M.; do Nascimento, G.M.; Nooruzzaman, M.; Yuan, F.; Chen, C.; Caserta, L.C.; Miller, A.D.; Whittaker, G.R.; Fang, Y.; Diel, D.G. The Omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and Delta variants in a feline model of SARS-CoV-2 infection. Journal of Virology 2022, 96, e00961-22. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Morales, L.; Sánchez-Vizcaíno, J.M.; Pérez-Sancho, M.; Domínguez, L.; Barroso-Arévalo, S. The Omicron (B.1.1.529) SARS-CoV-2 variant of concern also affects companion animals. Frontiers in Veterinary Science 2022, 9, 940710. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nature Microbiology 2022, 7, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Munnink, B.B.O.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; Van Der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Michel, K. Management of anorexia in the cat. Journal of Feline Medicine and Surgery 2001, 3, 3–8. [Google Scholar] [CrossRef]
- Burges Watson, D.L.; Campbell, M.; Hopkins, C.; Smith, B.; Kelly, C.; Deary, V. Altered smell and taste: Anosmia, parosmia and the impact of long Covid-19. PLOS ONE 2021, 16, e0256998. [Google Scholar] [CrossRef]
- Hecht, G.; Sarbo, N.; Svoboda, W.; Mead, H.L.; Ruberto, I.; Altin, J.A.; Engelthaler, D.M.; Venkat, H.; Yaglom, H.D. “Sniffing” out SARS-CoV-2 in Arizona working dogs: an exploratory serosurvey. Frontiers in Veterinary Science 2023, 10. [Google Scholar] [CrossRef]
- Segalés, J.; Puig, M.; Rodon, J.; Avila-Nieto, C.; Carrillo, J.; Cantero, G.; Terrón, M.T.; Cruz, S.; Parera, M.; Noguera-Julián, M. Detection of SARS-CoV-2 in a cat owned by a COVID-19−affected patient in Spain. Proceedings of the National Academy of Sciences 2020, 117, 24790–24793. [Google Scholar] [CrossRef]
- Shirazi, S.; Mami, S.; Mohtadi, N.; Ghaysouri, A.; Tavan, H.; Nazari, A.; Kokhazadeh, T.; Mollazadeh, R. Sudden cardiac death in COVID-19 patients, a report of three cases. Future Cardiology 2020, 17, 113–118. [Google Scholar] [CrossRef]
- Long, J.D.; Strohbehn, I.; Sawtell, R.; Bhattacharyya, R.; Sise, M.E. COVID-19 Survival and its impact on chronic kidney disease. Translational Research 2022, 241, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biology 2019, 20, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Turakhia, Y.; Thornlow, B.; Hinrichs, A.S.; De Maio, N.; Gozashti, L.; Lanfear, R.; Haussler, D.; Corbett-Detig, R. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nature Genetics 2021, 53, 809–816. [Google Scholar] [CrossRef]
| Drug | Trade name | Route of administration | Dose | Dosing interval | Days of hospitalisation |
|---|---|---|---|---|---|
| Maropitant | Vetemex | Intravenous/subcutaneous | 1 mg/kg | 24h | 1 to 5 |
| Metoclopramide | Vomend | Subcutaneous | 0.5 mg/kg | 12h | 5 to 7 and post-discharge |
| Omeprazole | Intravenous | 1 mg/kg | 12h | 1 and 2 | |
| Mirtazapine | Oral | 1.9 - 3.8 mg | 48h | Throughout | |
| Vitamin B | Subcutaneous | 0.02 mg/kg | Once | 2 | |
| Buprenorphine | Intravenous/subcutaneous/oral | 0.01-0.02 mg/kg | 6-8 h | 2, 3, 5, 6 and post-discharge | |
| Meloxicam | Loxicam | Subcutaneous | 0.1 mg/kg | Once | 5 |
| Amoxycillin-clavulanate | Synulox | Oral | 100 mg | 12h | Post-discharge (for 5 days) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





