Submitted:
04 August 2023
Posted:
07 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
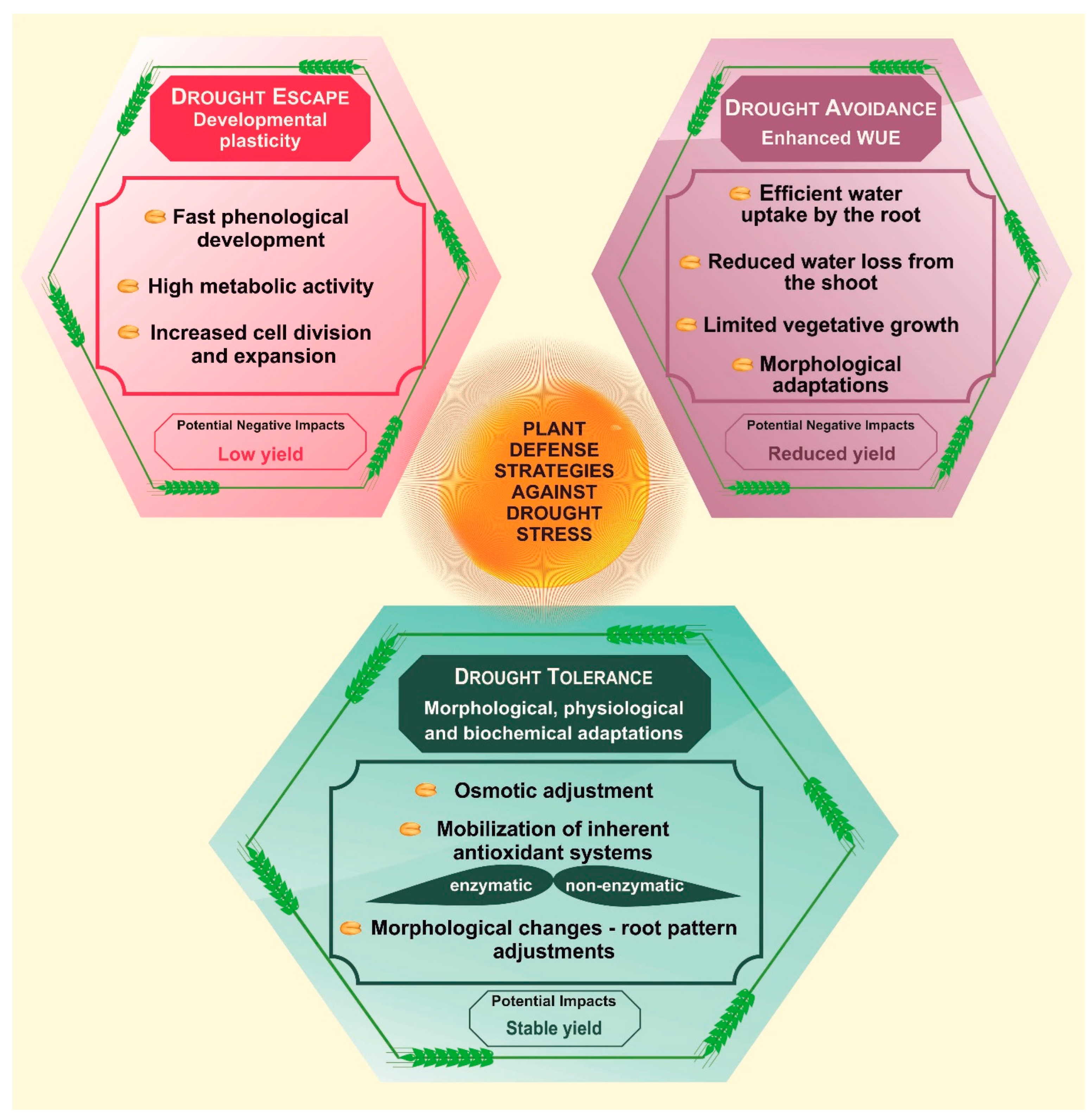
2. Plant Defense Strategies against Drought
2.1. Drought Escape Strategy
2.2. Drought Avoidance Strategy
2.3. Drought Tolerance Strategy
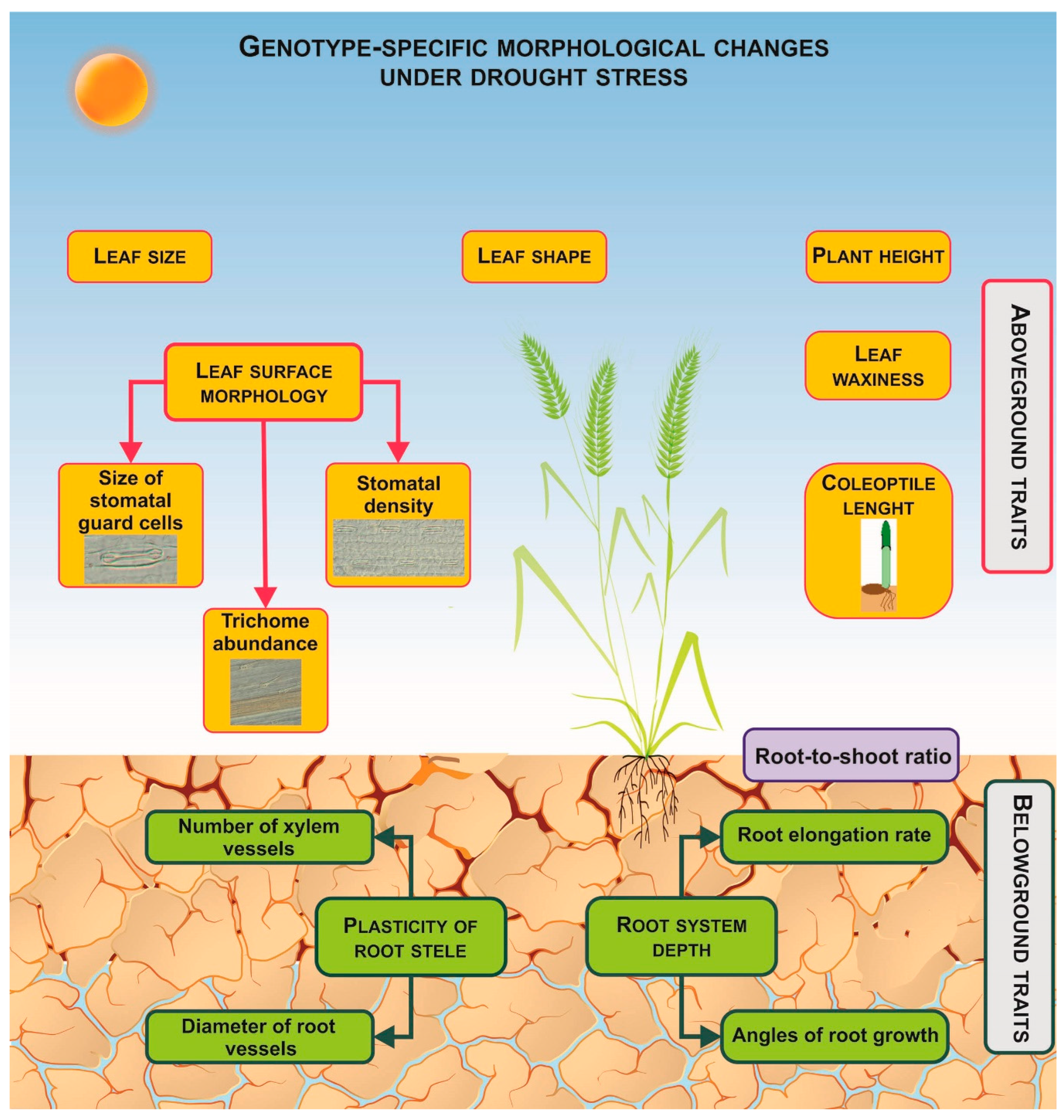
3. Genotype-Dependent Morphological Responses
3.1. Root System Traits
3.2. Aboveground Traits
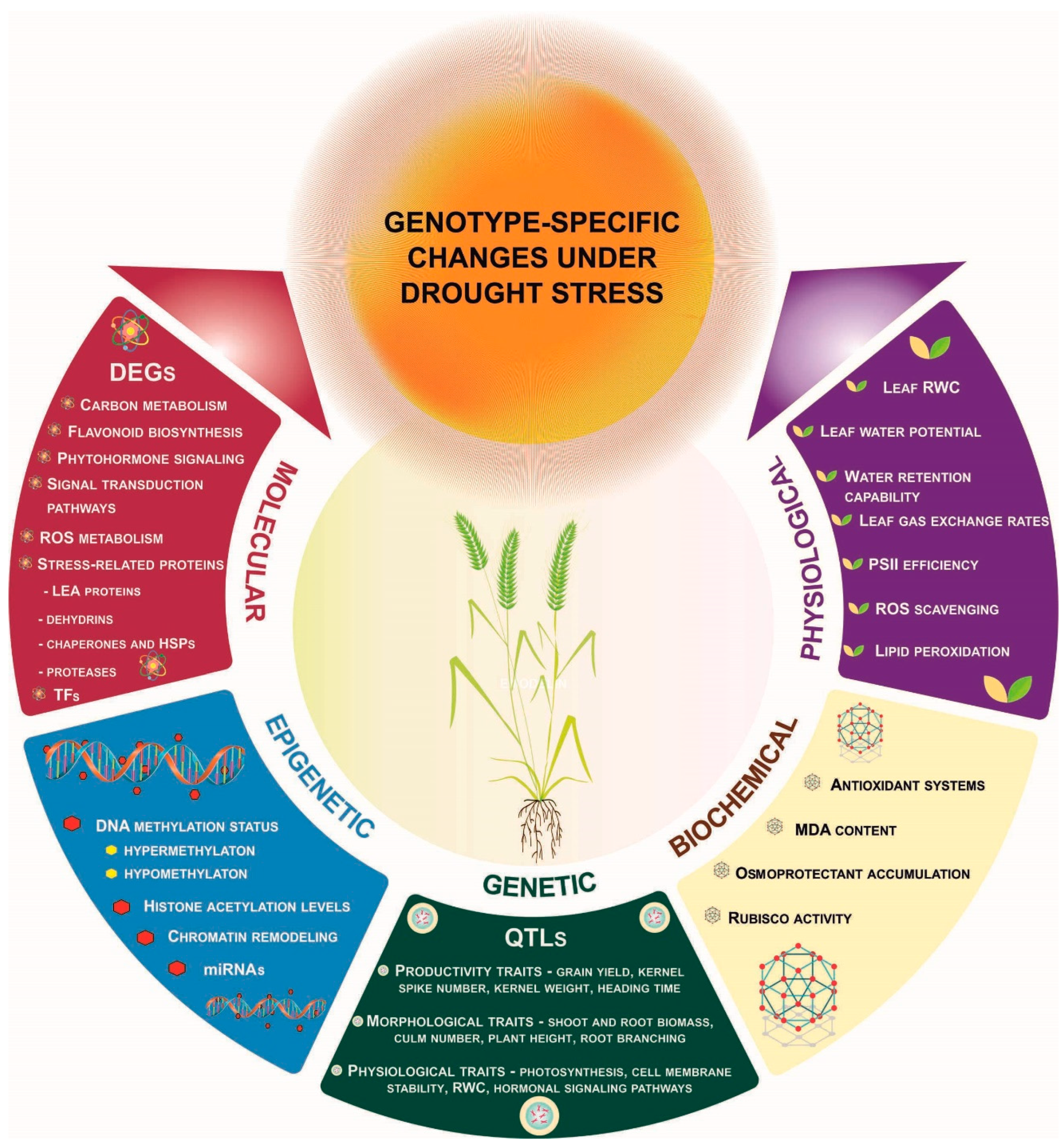
4. Genotype-Dependent Physiological and Biochemical Responses
5. Genotype-Dependent Molecular Responses
6. Genotype-Dependent Genetic Basis of Drought Tolerance
7. Genotype-Dependent Epigenetic Responses
8. Exploring the Drought Resistance of Bulgarian Wheat Genotypes
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 2009 103, 551–560. [Google Scholar] [CrossRef]
- Kapoor, D.; Bhardwaj, S.; Landi, M.; Sharma, A.; Ramakrishnan, M.; Sharma, A. The impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 2020, 10, 5692. [Google Scholar] [CrossRef]
- Khadka, K.; Raizada, M.N.; Navabi, A. Recent progress in germplasm evaluation and gene mapping to enable breeding of drought-tolerant wheat. Front. Plant Sci. 2020, 11, 1149. [Google Scholar] [CrossRef] [PubMed]
- Moravec, V.; Markonis, Y.; Rakovec, O.; Svoboda, M.; Trnka, M.; Kumar, R.; Hanel, M. Europe under multi-year droughts: How severe was the 2014-2018 drought period? Environ. Res. Lett. 2021, 16, 034062. [Google Scholar] [CrossRef]
- Kazandjiev, V.; Georgieva, V.; Malasheva, P.; Atanassov, D. Evapotranspiration and drought in different agricultural zones of Bulgaria. In Challenges in Agro-Climate and Ecosystem, Saifullah, M., Tardio, G.B., Mickovski, S., Eds.; IntechOpen, 2022. [CrossRef]
- Georgieva, V.; Kazandjiev, V.; Bozhanova, V.; Mihova, G.; Ivanova, D.; Todorovska, E.; Uhr, Z.; Ilchovska, M.; Sotirov, D.; Malasheva, P. Climatic changes—A challenge for the Bulgarian farmers. Agriculture 2022, 12, 2090. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef] [PubMed]
- De Leonardis, A.M.; Marone, D.; Mazzucotelli, E.; Neffar, F.; Rizza, F.; Di Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. Durum wheat genes up-regulated in the early phases of cold stress are modulated by drought in a developmental and genotype dependent manner. Plant Sci. 2007, 172, 1005–1016. [Google Scholar] [CrossRef]
- Pastori, G.M.; Foyer, C.H. Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol. 2002, 129, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Tardieu, F.; Tuberosa, R. Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 2010, 13, 206–212. [Google Scholar] [CrossRef]
- Mitura, K.; Cacak-Pietrzak, G.; Feledyn-Szewczyk, B.; Szablewski, T.; Studnicki, M. Yield and grain quality of common wheat (Triticum aestivum L.) depending on the different farming systems (organic vs. integrated vs. conventional). Plants 2023, 12, 1022. [Google Scholar] [CrossRef] [PubMed]
- Rockström, J.; Falkenmark, M. Agriculture: Increase water harvesting in Africa. Nature 2015, 519, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, Q.; Sun, P.; Song, C. Impact of droughts on winter wheat yield in different growth stages during 2001–2016 in Eastern China. Int. J. Disaster Risk Reduct. 2018, 3, 376–391. [Google Scholar] [CrossRef]
- Rich, S.M.; Wasson, A.P.; Richards, R.A.; Katore, T.; Prashar, R.; Chowdhary, R.; Saxena, D.C.; Mamrutha, H.M.; Zwart, A.; Misra, S.C.; et al. Wheats developed for high yield on stored soil moisture have deep vigorous root systems. Funct. Plant Biol. 2016, 43, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Portmann, F.T.; Siebert, S.; Döll, P. MIRCA2000-Global monthly irrigated and rainfed crop areas around the year 2000: A new high-resolution data set for agricultural and hydrological modeling. Glob. Biogeochem. Cycles 2010, 24, 1–24. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Christopher, J.T.; Hammer, G.L.; Devoil, P. Experimental and modelling studies of drought-adaptive root architectural traits in wheat (Triticum aestivum L.). Plant Biosyst. 2010, 144, 458–462. [Google Scholar] [CrossRef]
- Wasson, A.P.; Richards, R.A.; Chatrath, R.; Misra, S.C.; Prasad, S.S.; Rebetzke, G.J.; Kirkegaard, J.A.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.S.; Rebetzke, G.J.; Reynolds, M. Integration of phenotyping and genetic platforms for a better understanding of wheat performance under drought. J. Exp. Bot. 2014, 65, 6167–6177. [Google Scholar] [CrossRef] [PubMed]
- Giordano, M.; Petropoulos, S.A.; Rouphael, Y. Response and defence mechanisms of vegetable crops against drought, heat and salinity stress. Agriculture, 2021, 11, 463. [Google Scholar] [CrossRef]
- Shavrukov, Y.; Kurishbayev, A.; Jatayev, S.; Shvidchenko, V.; Zotova, L.; Koekemoer, F.; De Groot, S.; Soole, K.; Langridge, P. Early flowering as a drought escape mechanism in plants: How can it aid wheat production? Front. Plant Sci. 2017, 8, 1950. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Ramegowda, V.; Kumar, A.; Pereira, A. Plant adaptation to drought stress. F1000Res. 2016, 5, F1000. [Google Scholar] [CrossRef]
- Franks, S.J. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol. 2011, 190, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Sherrard, M.E.; Maherali, H. The adaptive significance of drought escape in Avena barbata, an annual grass. Evolution 2006, 60, 2478–2489. [Google Scholar] [CrossRef] [PubMed]
- Kooyers, N.J. The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 2015, 234, 155–162. [Google Scholar] [CrossRef]
- Du, H.; Huang, F.; Wu, N.; Li, X.; Hu, H.; Xiong, L. Integrative regulation of drought escape through ABA dependent and independent pathways in rice. Mol. Plant. 2018, 11, 584–597. [Google Scholar] [CrossRef] [PubMed]
- Dohleman, F.G.; Long, S.P. More productive than maize in the midwest: How does miscanthus do it? Plant Physiol. 150, 2104–2115. [CrossRef] [PubMed]
- Tardieu, F.; Simonneau, T.; Muller, B. The physiological basis of drought tolerance in crop plants: A scenario-dependent probabilistic approach. Annu. Rev. Plant Biol. 2018, 69, 733–759. [Google Scholar] [CrossRef] [PubMed]
- Álvaro, F.; Isidro, J.; Villegas, D.; García del Moral, L.F.; Royo, C. Breeding effects on grain filling, biomass partitioning, and remobilization in Mediterranean durum wheat. Agron. J. 2008, 100, 361–370. [Google Scholar] [CrossRef]
- Hatfield, J.L.; Dold, C. Water-use efficiency: Advances and challenges in a changing climate. Front. Plant Sci. 2019, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Park, S.H.; Jenks, M.A. Changes in leaf cuticular waxes of sesame (Sesamum indicum L.) plants exposed to water deficit. J. Plant Physiol. 2007, 164, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Delfin, E.F.; Drobnitch, S.T.; Comas, L.H. Plant strategies for maximizing growth during water stress and subsequent recovery in Solanum melongena L. (eggplant). PLoS ONE 2021, 16, e0256342. [Google Scholar] [CrossRef]
- Mori, M.; Inagaki, M.N.; Inoue, T.; Nachit, M.M. Association of root water-uptake ability with drought adaptation in wheat. Cereal Res. Commun. 2011, 39, 551–559. [Google Scholar] [CrossRef]
- Caine, R.S.; Yin, X.; Sloan, J.; Harrison, E.L.; Mohammed, U.; Fulton, T.; Biswal, A.K.; Dionora, J.; Chater, C.C.; Coe, R.A.; et al. Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 2019, 221, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Y.; Turner, N.C.; Liu, Y.X.; Kadambot, H.M.S.; Xiong, Y.C. Effects of drought stress on morphological, physiological and biochemical characteristics of wheat species differing in ploidy level. Funct. Plant Biol. 2016, 44, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ma, B.; Palta, J.A.; Ding, T.; Cheng, Z.; Lv, G.; Xiong, Y. Wheat breeding highlights drought tolerance while ignores the advantages of drought avoidance: A meta-analysis. Eur. J. Agron. 2021, 122, 126196. [Google Scholar] [CrossRef]
- Loss, S.P.; Siddique, K.H.M. Morphological and physiological traits associated with wheat yield increases in Mediterranean environments. Adv. Agron. 1994, 52, 229–276. [Google Scholar] [CrossRef]
- Kulkarni, M.; Soolanayakanahally, R.; Ogawa, S.; Uga, Y.; Selvaraj, M.G.; Kagale, S. Drought response in wheat: Key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 2017, 5, 106. [Google Scholar] [CrossRef] [PubMed]
- Puijalon, S.; Bouma, T.J.; Douady, C.J.; van Groenendael, J.; Anten, N.P.; Martel, E.; Bornette, G. Plant resistance to mechanical stress: Evidence of an avoidance-tolerance trade-off. New Phytol. 2011, 191, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.M. Osmoregulation as a selection criterion for drought tolerance in wheat. Aust. J. Agric. Res. 1983, 34, 607–614. [Google Scholar] [CrossRef]
- Izanloo, A.; Condon, A.G.; Langridge, P.; Tester, M.; Schnurbusch, T. Different mechanisms of adaptation to cyclic water stress in two South Australian bread wheat cultivars. J. Exp. Bot. 2008, 59, 3327–3346. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Hussain, M.; Siddique, K.H.M. Drought stress in wheat during flowering and grain-filling periods. CRC Crit. Rev. Plant Sci. 2014, 33, 331–349. [Google Scholar] [CrossRef]
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94. [Google Scholar] [CrossRef] [PubMed]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2021, 11, 552969. [Google Scholar] [CrossRef]
- Velinov, V.; Vaseva, I.; Zehirov, G.; Zhiponova, M.; Georgieva, M.; Vangheluwe, N.; Beeckman, T.; Vassileva, V. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 815. [Google Scholar] [CrossRef] [PubMed]
- Bengough, A.G.; McKenzie, B.M.; Hallett, P.D.; Valentine, T.A. Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 2011, 62, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, V.; Signarbieux, C.; Anders, I.; Feller, U. Genotypic variation in drought stress response and subsequent recovery of wheat (Triticum aestivum L.). J. Plant Res. 2011, 124, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Meister, R.; Rajani, M.S.; Ruzicka, D.; Schachtman, D.P. Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 2014, 19, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Manske, G.G.B.; Vlek, P.L.G. Root architecture - wheat as a model plant. In Plant Roots: The Hidden Half, 3rd ed.; Waisel, Y., Eshel, A., Kafkafi, U., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 249–259. [Google Scholar]
- Placido, D.F.; Sandhu, J.; Sato, S.J.; Nersesian, N.; Quach, T.; Clemente, T.E.; Staswick, P.E.; Walia, H. The LATERAL ROOT DENSITY gene regulates root growth during water stress in wheat. Plant Biotechnol. J. 2020, 18, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Golan, G.; Hendel, E.; Mendez Espitia, G.E.; Schwartz, N.; Peleg, Z. Activation of seminal root primordia during wheat domestication reveals underlying mechanisms of plant resilience. Plant Cell Environ. 2018, 41, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Whalley, P.A.; Ashton, R.W.; Evans, J.; Hawkesford, M.J.; Griffiths, S.; Huang, Z.D.; Zhou, H.; Mooney, S.J.; Whalley, W.R. A comparison between water uptake and root length density in winter wheat: Effects of root density and rhizosphere properties. Plant Soil 2020, 451, 345–356. [Google Scholar] [CrossRef]
- Palta, J.; Watt, M. Vigorous crop root systems: Form and function for improving the capture of water and nutrients. In Applied Crop Physiology: Boundaries between Genetic Improvement and Agronomy. Academic, San Diego, 2009; pp. 309–325.
- Ober, E.S.; Alahmad, S.; Cockram, J.; Forestan, C.; Hickey, L.T.; Kant, J.; Maccaferri, M.; Marr, E.; Milner, M.; Pinto, F.; et al. Wheat root systems as a breeding target for climate resilience. Theor. Appl. Genet. 2021, 134, 1645–1662. [Google Scholar] [CrossRef] [PubMed]
- Palta, J.A.; Chen, X.; Milroy, S.P.; Rebetzke, G.J.; Dreccer, M.F.; Watt, M. Large root systems: Are they useful in adapting wheat to dry environments? Funct. Plant Biol. 2011, 38, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Manschadi, A.M.; Hammer, G.L.; Christopher, J.T.; Devoil, P. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 2008, 303, 115–129. [Google Scholar] [CrossRef]
- Nakamoto, T.; Oyanagi, A. The direction of growth of seminal roots of Triticum aestivum L. and experimental modification thereof. Ann. Bot. 1994, 73, 363–367. [Google Scholar] [CrossRef]
- Slack, S.; York, L.M.; Roghazai, Y.; Lynch, J.; Bennett, M.; Foulkes, J. Wheat shovelomics II: Revealing relationships between root crown traits and crop growth. BioRxiv 2018, 0917. [Google Scholar] [CrossRef]
- Alahmad, S.; El Hassouni, K.; Bassi, F.M.; Dinglasan, E.; Youssef, C.; Quarry, G.; Aksoy, A.; Mazzucotelli, E.; Juhász, A.; Able, J.A.; et al. A major root architecture QTL responding to water limitation in durum wheat. Front. Plant Sci. 2019, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Du, Y.; Wang, J.; Wu, A.; Qiao, S.; Xu, B.; Zhang, S.; Siddique, K.H.; Chen, Y. Moderate drought stress affected root growth and grain yield in old, modern and newly released cultivars of winter wheat. Front. Plant Sci. 2017, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Grzesiak, M.T.; Hordyńska, N.; Maksymowicz, A.; Grzesiak, S.; Szechyńska-Hebda, M. Variation among spring wheat (Triticum aestivum L.) genotypes in response to the drought stress. II-Root system structure. Plants 2019, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- McDonald, G. The effects of root angle on root growth and yield of wheat in the Australian cereal belt. In Food Security from Sustainable Agriculture: Proceedings of 15th Agronomy Conference.; p. 2010.
- Ouyang, W.; Yin, X.; Yang, J.; Struik, P.C. Comparisons with wheat reveal root anatomical and histochemical constraints of rice under water-deficit stress. Plant Soil 2020, 2020 452, 547–568. [Google Scholar] [CrossRef]
- Kadam, N.N.; Yin, X.; Bindraban, P.S.; Struik, P.C.; Jagadish, K.S. Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol. 2015, 167, 1389–1401. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.A.; Passioura, J.B. A breeding program to reduce the diameter of the major xylem vessel in the seminal roots of wheat and its effect on grain yield in rain-fed environments. Aust. J. Agric. Res. 1989, 40, 943–950. [Google Scholar] [CrossRef]
- Ehdaie, B.; Layne, A.P.; Waines, J.G. Root system plasticity to drought influences grain yield in bread wheat. Euphytica 2012, 186, 219–232. [Google Scholar] [CrossRef]
- Correa, J.; Postma, J.A.; Watt, M.; Wojciechowski, T. Soil compaction and the architectural plasticity of root systems. J. Exp. Bot. 2019, 70, 6019–6034. [Google Scholar] [CrossRef] [PubMed]
- Nezhadahmadi, A.; Prodhan, Z.H.; Faruq, G. Drought tolerance in wheat. Sci. World J. 2013, 2013, 610721. [Google Scholar] [CrossRef] [PubMed]
- Farhad, M.; Hakim, M.A.; Alam, M.A.; Barma, N.C.D. Screening wheat genotypes for coleoptile length: A trait for drought tolerance. Am. J. Agric. Forest 2014, 2, 237–245. [Google Scholar] [CrossRef]
- Petrov, P.; Petrova, A.; Dimitrov, I.; Tashev, T.; Olsovska, K.; Brestic, M.; Misheva, S. Relationships between leaf morpho-anatomy, water status and cell membrane stability in leaves of wheat seedlings subjected to severe soil drought. J. Agron. Crop Sci. 2018, 204, 219–227. [Google Scholar] [CrossRef]
- Sewore, B.M.; Abe, A.; Nigussie, M. Evaluation of bread wheat (Triticum aestivum L.) genotypes for drought tolerance using morpho-physiological traits under drought-stressed and well-watered conditions. PLoS ONE 2023, 18, p–e0283347. [Google Scholar] [CrossRef] [PubMed]
- Kilic, H.; Yagbasanlar, T. The effect of drought stress on grain yield, yield components and some quality traits of durum wheat (Triticum turgidum) cultivars. Not. Bot. Horti. Agrobot. 2010, 38, 164–170. [Google Scholar] [CrossRef]
- Liwani, U.; Magwaza, L.S.; Odindo, A.O.; Sithole, N.J. Growth, morphological and yield responses of irrigated wheat (Triticum aestivum L.) genotypes to water stress. Acta Agric. Scand. B Soil Plant Sci. 2019, 69, 369–376. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, L.; Shen, Q.; Yang, J.; Han, X.; Tian, F.; Wu, J. Effects of water stress on photosynthesis, yield, and water use efficiency in winter wheat. Water 2020, 12, 2127. [Google Scholar] [CrossRef]
- Willick, I.R.; Lahlali, R.; Vijayan, P.; Muir, D.; Karunakaran, C.; Tanino, K.K. Wheat flag leaf epicuticular wax morphology and composition in response to moderate drought stress are revealed by SEM, FTIR-ATR and synchrotron X-ray spectroscopy. Physiol. Plant. 2018, 162, 316–332. [Google Scholar] [CrossRef] [PubMed]
- Richards, R.A.; Rawson, H.M.; Johnson, D.A. Glaucousness in wheat: Its development and effect on water-use efficiency, gas exchange and photosynthetic tissue temperatures. Aust. J. Plant Physiol. 1986, 13, 465–473. [Google Scholar] [CrossRef]
- David, O.A.; Osonubi, O.; Olaiya, C.O.; Agbolade, J.O.; Ajiboye, A.A.; Komolafe, R.J.; Chukwuma, D.M.; Akomolafe, G.F. Anatomical response of wheat cultivars to drought stress. Ife J. Sci. 2017, 19, 323–331. [Google Scholar] [CrossRef]
- Jäger, K.; Fábián, A.; Eitel, G.; Szabó, L.; Deák, C.; Barnabás, B.; Papp, I. A morpho-physiological approach differentiates bread wheat cultivars of contrasting tolerance under cyclic water stress. J. Plant Physiol. 2014, 171, 1256–1266. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Bruce, S.E.; Kirkegaard, J.A. Longer coleoptiles improve emergence through crop residues to increase seedling number and biomass in wheat (Triticum aestivum L.). Plant Soil 2005, 272, 87–100. [Google Scholar] [CrossRef]
- Rebetzke, G.J.; Richards, R.A.; Fettell, N.A.; Long, M.; Condon, A.G.; Forrester, R.I.; Botwright, T.L. Genotypic increases in coleoptile length improves stand establishment, vigour and grain yield of deep-sown wheat. Field Crops Res. 2007, 100, 10–23. [Google Scholar] [CrossRef]
- Schillinger, W.F.; Donaldson, E.; Allan, R.E.; Jones, S.S. Winter wheat seedling emergence from deep sowing depths. Agron. J. 1998, 90, 582–586. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed]
- Ghaffar, A.; Hussain, N.; Ajaj, R.; Shahin, S.M.; Bano, H.; Javed, M.; Khalid, A.; Yasmin, M.; Shah, K.H.; Zaheer, M.; et al. Photosynthetic activity and metabolic profiling of bread wheat cultivars contrasting in drought tolerance. Front. Plant Sci. 2023, 14, 1123080. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.; Jiang, D.; Snider, J.L.; Dai, T. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4615. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, V.; Demirevska, K.; Simova-Stoilova, L.; Petrova, T.; Tsenov, N.; Feller, U. Long-term field drought affects leaf protein pattern and chloroplast ultrastructure of winter wheat in a cultivar-specific manner. J. Agron. Crop Sci. 2012, 198, 104–117. [Google Scholar] [CrossRef]
- Qayyum, A.; Al Ayoubi, S.; Sher, A.; Bibi, Y.; Ahmad, S.; Shen, Z.; Jenks, M.A. Improvement in drought tolerance in bread wheat is related to an improvement in osmolyte production, antioxidant enzyme activities, and gaseous exchange. Saudi J. Biol. Sci. 2021, 28, 5238–5249. [Google Scholar] [CrossRef] [PubMed]
- Khamssi, N.N.; Najaphy, A. Agro-morphological and phenological attributes under irrigated and rain-fed conditions in bread wheat genotypes. Afr. J. Agric. Res. 2012, 7, 51–57. [Google Scholar] [CrossRef]
- Clarke, J.M.; McCaig, T.N. Evaluation of techniques for screening for drought resistance in wheat. Crop Sci. 1982, 22, 503–506. [Google Scholar] [CrossRef]
- Roostaei, M.; Mohammadi, S.A.; Amri, A.; Majidi, E.; Nachit, M.; Haghparast, R. Chlorophyll fluorescence parameters and drought tolerance in a mapping population of winter bread wheat in the highlands of Iran. Russ. J. Plant Physiol. 2011, 58, 351–358. [Google Scholar] [CrossRef]
- Larouk, C.; Gabon, F.; Kehel, Z.; Djekoun, A.; Nachit, M.; Amri, A. Chlorophyll fluorescence and drought tolerance in a mapping population of durum wheat. Contemporary Agric. 2021, 70, 123–134. [Google Scholar] [CrossRef]
- Grigorova, B.; Vassileva, V.; Klimchuk, D.; Vaseva, I.; Demirevska, K.; Feller, U. Drought, high temperature, and their combination affect ultrastructure of chloroplasts and mitochondria in wheat (Triticum aestivum L.) leaves. J. Plant Interact. 2012, 7, 204–213. [Google Scholar] [CrossRef]
- Basal, H.; Smith, C.W.; Thaxton, P.S.; Hemphill, J.K. Seedling drought tolerance in upland cotton. Crop Sci. 2005, 45, 766–771. [Google Scholar] [CrossRef]
- Kane, C.N.; Jordan, G.J.; Jansen, S.; McAdam, S.A. A permeable cuticle, not open stomata, is the primary source of water loss from expanding leaves. Front. Plant Sci. 2020, 11, 774. [Google Scholar] [CrossRef]
- Thapa, S.; Reddy, S.K.; Fuentealba, M.P.; Xue, Q.; Rudd, J.C.; Jessup, K.E.; Devkota, R.N.; Liu, S. Physiological responses to water stress and yield of winter wheat cultivars differing in drought tolerance. J. Agron. Crop Sci. 2018, 204, 347–358. [Google Scholar] [CrossRef]
- Demirevska, K.; Simova-Stoilova, L.; Vassileva, V.; Feller, U. Rubisco and some chaperone protein responses to water stress and rewatering at early seedling growth of drought sensitive and tolerant wheat varieties. Plant Growth Regul. 2008, 56, 97–106. [Google Scholar] [CrossRef]
- Thakur, P.; Prasad, L.C.; Prasad, R.; Chandra, K. Estimation of genetic variability, heat susceptibility index and tolerance efficiency of wheat (Triticum aestivum L.) for timely and late sown environments. Electron. J. Plant Breed. 2020, 11, 769–775. [Google Scholar] [CrossRef]
- Khalilzadeh, R.; Seyed Sharifi, R.; Jalilian, J. Antioxidant status and physiological responses of wheat (Triticum aestivum L.) to cycocel application and bio fertilizers under water limitation condition. J. Plant Interact. 2016, 11, 130–137. [Google Scholar] [CrossRef]
- Marček, T.; Hamow, K.A.; Végh, B.; Janda, T.; Darko, E. Metabolic response to drought in six winter wheat genotypes. PLoS ONE 2019, 14, e0212411. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, A.; Kumar, A.; Kaur, N. ROS and oxidative burst: Roots in plant development. Plant Divers. 2020, 42, 33–43. [Google Scholar] [CrossRef]
- Maevskaya, S.N.; Nikolaeva, M.K. Response of antioxidant and osmoprotective systems of wheat seedlings to drought and rehydration. Russ. J. Plant Physiol. 2013, 60, 343–350. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.; Demirevska, K.; Petrova, T.; Tsenov, N.; Feller, U. Antioxidative protection and proteolytic activity in tolerant and sensitive wheat (Triticum aestivum L.) varieties subjected to long-term field drought. Plant Growth Regul. 2009, 58, 107–117. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Tabarzad, A.; Ayoubi, B.; Riasat, M.; Saed-Moucheshi, A.; Pessarakli, M. Perusing biochemical antioxidant enzymes as selection criteria under drought stress in wheat varieties. J. Plant Nutr. 2017, 40, 2413–2420. [Google Scholar] [CrossRef]
- Zhang, W.; Gu, J.; Wang, Z.; Wei, C.; Yang, J.; Zhang, J. Comparison of structural and functional properties of wheat starch under different soil drought conditions. Sci. Rep. 2017, 7, 12312. [Google Scholar] [CrossRef]
- Fan, X.W.; Li, F.M.; Song, L.; Xiong, Y.C.; An, L.Z.; Jia, Y.; Fang, X.W. Defense strategy of old and modern spring wheat varieties during soil drying. Physiol. Plant. 2009, 136, 310–323. [Google Scholar] [CrossRef]
- Erdei, L.; Tari, I.; Csiszar, J.; Pecsvaradi, A.; Horvath, F.; Szabo, M.; Ordog, M.; Cseuz, L.; Zhiponova, M.; Szilak, L.; et al. Osmotic stress responses of wheat species and cultivars differing in drought tolerance: Some interesting genes (advices for gene hunting). Acta Biol. Szeged 2002, 46, 63–65. [Google Scholar]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Camaille, M.; Fabre, N.; Clément, C.; Ait Barka, E. Advances in wheat physiology in response to drought and the role of plant growth promoting rhizobacteria to trigger drought tolerance. Microorganisms 2021, 9, 687. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Al-Busaidi, W.M.; Al-Sadi, A.M.; Farooq, M. Bread wheat genotypes accumulating free proline and phenolics can better tolerate drought stress through sustained rate of photosynthesis. J. Soil Sci. Plant Nutr. 2022, 22, 165–176. [Google Scholar] [CrossRef]
- Saeedipour, S. Relationship of grain yield, ABA and proline accumulation in tolerant and sensitive wheat cultivars as affected by water stress. Proc. Natl. Acad. Sci. U.S.A. 2013, 83, 311–315. [Google Scholar] [CrossRef]
- Pecetti, L.; Annicchiarico, P.; Gorham, J. Field heterogeneity of the stress affects genotypic response to salinity in durum wheat. Cereal Res. Commun. 1995, 23, 173–177. [Google Scholar]
- Wang, G.P.; Zhang, X.Y.; Li, F.; Luo, Y.; Wang, W. Overaccumulation of glycine betaine enhances tolerance to drought and heat stress in wheat leaves in the protection of photosynthesis. Photosynthetica 2010, 48, 117–126. [Google Scholar] [CrossRef]
- Bowne, J.B. , Erwin, T.A.; Juttner, J.; Schnurbusch, T.; Langridge, P.; Bacic, A.; Roessner, U. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant 2012, 5, 418–429. [Google Scholar] [CrossRef]
- Wang, X.; Mao, Z.; Zhang, J.; Hemat, M.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. Osmolyte accumulation plays important roles in the drought priming induced tolerance to post-anthesis drought stress in winter wheat (Triticum aestivum L.). Environ. Exp. Bot. 2019, 166, 103804. [Google Scholar] [CrossRef]
- Khoshro, H.H.; Taleei, A.; Bihamta, M.R.; Shahbazi, M.; Abbasi, A. Expression analysis of the genes involved in osmotic adjustment in bread wheat (Triticum aestivum L.) cultivars under terminal drought stress conditions. J. Crop Sci. Biotechnol. 2013, 16, 173–181. [Google Scholar] [CrossRef]
- Bogdan, J.; Zagdańska, B. Changes in the pool of soluble sugars induced by dehydration at the heterotrophic phase of growth of wheat seedlings. Plant Physiol. Biochem. 2006, 44, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Feng, K.; Peng, S.; Wang, J.; Zhang, Y.; Bian, J.; Nie, X. Comparative analysis of the transcriptional response of tolerant and sensitive wheat genotypes to drought stress in field conditions. Agronomy 2018, 8, 247. [Google Scholar] [CrossRef]
- Shamloo-Dashtpagerdi, R.; Shahriari, A.G.; Tahmasebi, A.; Vetukuri, R.R. Potential role of the regulatory miR1119-MYC2 module in wheat (Triticum aestivum L.) drought tolerance. Front. Plant Sci. 2023, 14, 1161245. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xie, Y.; Fan, S.; Wang, Z.; Wang, F.; Zhang, B.; Li, H.; Song, J.; Kong, L. Comparative analysis of root transcriptome profiles between drought-tolerant and susceptible wheat genotypes in response to water stress. Plant Sci. 2018, 272, 276–293. [Google Scholar] [CrossRef]
- Ergen, N.Z.; Thimmapuram, J.; Bohnert, H.J.; Budak, H. Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct. Integr. Genom. 2009, 9, 377–396. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 2017, 8, 592. [Google Scholar] [CrossRef]
- Dalal, M.; Sahu, S.; Tiwari, S.; Rao, A.R.; Gaikwad, K. Transcriptome analysis reveals interplay between hormones, ROS metabolism and cell wall biosynthesis for drought-induced root growth in wheat. Plant Physiol. Biochem. 2018, 130, 482–492. [Google Scholar] [CrossRef]
- Mia, M.S.; Liu, H.; Wang, X.; Zhang, C.; Yan, G. Root transcriptome profiling of contrasting wheat genotypes provides an insight to their adaptive strategies to water deficit. Sci. Rep. 2020, 10, 4854. [Google Scholar] [CrossRef]
- Nouraei, S.; Mia, M.S.; Liu, H.; Turner, N.C.; Yan, G. Transcriptome analyses of near isogenic lines reveal putative drought tolerance controlling genes in wheat. Front. Plant Sci. 2022, 13, 857829. [Google Scholar] [CrossRef]
- Liu, W.J.; Yuan, S.; Zhang, N.H.; Lei, T.; Duan, H.G.; Liang, H.G.; Lin, H.H. Effect of water stress on photosystem 2 in two wheat cultivars. Biol. Plant. 2006, 50, 597–602. [Google Scholar] [CrossRef]
- Yu, T.F.; Xu, Z.S.; Guo, J.K.; Wang, Y.X.; Abernathy, B.; Fu, J.D.; Chen, X.; Zhou, Y.B.; Chen, M.; Ye, X.G.; et al. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 2017, 7, 44050. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.G.; Banowetz, G.M.; Peterson, C.J.; Kronstad, W.E. Dehydrin expression and drought tolerance in seven wheat cultivars. Crop Sci. 2003, 43, 577–582. [Google Scholar] [CrossRef]
- Vaseva, I.I.; Grigorova, B.S.; Simova-Stoilova, L.P.; Demirevska, K.N.; Feller, U. Abscisic acid and late embryogenesis abundant protein profile changes in winter wheat under progressive drought stress. Plant Biol. 2010, 12, 698–707. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V.; Kumar, A.; Gupta, P.K. Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 2016, 129, 2019–2042. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, D.; Li, Q.; Mao, X.; Li, A.; Wang, J.; Chang, X.; Jing, R. Overexpression of wheat gene TaMOR improves root system architecture and grain yield in Oryza sativa. J. Exp. Bot. 2016, 67, 4155–4167. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, R.; Cheng, Y.; Lei, P.; Song, W.; Zheng, W.; Nie, X. Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 721253. [Google Scholar] [CrossRef]
- Des Marais, D.L.; Hernandez, K.M.; Juenger, T.E. Genotype-by-environment interaction and plasticity: Exploring genomic responses of plants to the abiotic environment. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 5–29. [Google Scholar] [CrossRef]
- Ishikawa, A. A strategy for identifying quantitative trait genes using gene expression analysis and causal analysis. Genes 2017, 8, 347. [Google Scholar] [CrossRef]
- Gupta, P.K.; Balyan, H.S.; Gahlaut, V. QTL analysis for drought tolerance in wheat: Present status and future possibilities. Agronomy 2017, 7, 5. [Google Scholar] [CrossRef]
- Bapela, T.; Shimelis, H.; Tsilo, T.J.; Mathew, I. Genetic improvement of wheat for drought tolerance: Progress, challenges and opportunities. Plants 2022, 11, 1331. [Google Scholar] [CrossRef]
- Malosetti, M.; Voltas, J.; Romagosa, I.; Ullrich, S.E.; Van Eeuwijk, F.A. Mixed models including environmental covariables for studying QTL by environment interaction. Euphytica 2004, 137, 139–145. [Google Scholar] [CrossRef]
- Mathews, K.L.; Malosetti, M.; Chapman, S.; McIntyre, L.; Reynolds, M.; Shorter, R.; Van Eeuwijk, F. Multi-environment QTL mixed models for drought stress adaptation in wheat. Theor. Appl. Genet. 2008, 117, 1077–1091. [Google Scholar] [CrossRef] [PubMed]
- Quarrie, S.A.; Gulli, M.; Calestani, C.; Steed, A.; Marmiroli, N. Location of a gene regulating drought-induced abscisic acid production on the long arm of chromosome 5A of wheat. Theor. Appl. Genet. 1994, 89, 794–800. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Mathews, K.L.; Rattey, A.; Chapman, S.C.; Drenth, J.; Ghaderi, M.; Reynolds, M.; Shorter, R. Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor. Appl. Genet. 2010, 120, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Czyczyło-Mysza, I.; Marcińska, I.; Skrzypek, E.; Cyganek, K.; Juzoń, K.; Karbarz, M. QTL mapping for germination of seeds obtained from previous wheat generation under drought. Open Life Sci. 2014, 9, 374–382. [Google Scholar] [CrossRef]
- Maccaferri, M.; Mantovani, P.; Tuberosa, R.; DeAmbrogio, E.; Giuliani, S.; Demontis, A.; Massi, A.; Sanguineti, M.C. A major QTL for durable leaf rust resistance widely exploited in durum wheat breeding programs maps on the distal region of chromosome arm 7BL. Theor. Appl. Genet. 2008, 117, 1225–1240. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nature Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef]
- Yang, Y.; Chai, Y.; Zhang, X.; Lu, S.; Zhao, Z.; Wei, D.; Chen, L.; Hu, Y.G. Multi-locus GWAS of quality traits in bread wheat: Mining more candidate genes and possible regulatory network. Front. Plant Sci. 2020, 11, 1091. [Google Scholar] [CrossRef]
- Tsonev, S.; Christov, N.K.; Mihova, G.; Dimitrova, A.; Todorovska, E.G. Genetic diversity and population structure of bread wheat varieties grown in Bulgaria based on microsatellite and phenotypic analyses. Biotechnol. Biotechnol. Equip. 2021, 35, 1520–1533. [Google Scholar] [CrossRef]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef]
- Peleg, Z.V.I.; Fahima, T.; Krugman, T.; Abbo, S.; Yakir, D.A.N.; Korol, A.B.; Saranga, Y. Genomic dissection of drought resistance in durum wheat×wild emmer wheat recombinant inbreed line population. Plant, Cell & Environ. 2009, 32, 758–779. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Heidari, B.; Pakniyat, H.; McIntyre, C.L. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome 2017, 60, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Zandipour, M.; Majidi Hervan, E.; Azadi, A.; Khosroshahli, M.; Etminan, A. A QTL hot spot region on chromosome 1B for nine important traits under terminal drought stress conditions in wheat. Cereal Res. Commun. 2020, 48, 17–24. [Google Scholar] [CrossRef]
- Deng, S.; Wu, X.; Wu, Y.; Zhou, R.; Wang, H.; Jia, J.; Liu, S. Characterization and precise mapping of a QTL increasing spike number with pleiotropic effects in wheat. Theor. Appl. Genet. 2011, 122, 281–289. [Google Scholar] [CrossRef]
- Jin, J.; Liu, D.; Qi, Y.; Ma, J.; Zhen, W. Major QTL for seven yield-related traits in common wheat (Triticum aestivum L.). Front. Genet. 2020, 11, 1012. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Xu, Z.; Fan, X.; Zhou, Q.; Yu, Q.; Liu, X.; Liao, S.; Feng, B.; Wang, T. Identification of a major and stable QTL on chromosome 5A confers spike length in wheat (Triticum aestivum L.). Mol. Breed. 2021, 41, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Itam, M.O.; Mega, R.; Gorafi, Y.S.; Yamasaki, Y.; Tahir, I.S.; Akashi, K.; Tsujimoto, H. Genomic analysis for heat and combined heat-drought resilience in bread wheat under field conditions. Theor. Appl. Genet. 2022, 135, 337–350. [Google Scholar] [CrossRef]
- Said, A.A.; Moursi, Y.S.; Sallam, A. Association mapping and candidate genes for physiological non-destructive traits: Chlorophyll content, canopy temperature, and specific leaf area under normal and saline conditions in wheat. Front. Genet. 2022, 13, 980319. [Google Scholar] [CrossRef]
- Malik, S.; Malik, T.A. Genetic mapping of potential QTLs associated with drought tolerance in wheat. JAPS: J. Anim. Plant Sci. 2015, 25, 1032–1040. [Google Scholar]
- Pshenichnikova, T.A.; Osipova, S.V.; Smirnova, O.G.; Leonova, I.N.; Permyakova, M.D.; Permyakov, A.V.; Rudikovskaya, E.G.; Konstantinov, D.K.; Verkhoturov, V.V.; Lohwasser, U.; et al. Regions of chromosome 2A of bread wheat (Triticum aestivum L.) associated with variation in physiological and agronomical traits under contrasting water regimes. Plants 2021, 10, 1023. [Google Scholar] [CrossRef]
- Kobayashi, F.; Takumi, S.; Handa, H. Identification of quantitative trait loci for ABA responsiveness at the seedling stage associated with ABA-regulated gene expression in common wheat. Theor. Appl. Genet. 2010, 121, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Iehisa, J.C.; Matsuura, T.; Mori, I.C.; Takumi, S. Identification of quantitative trait locus for abscisic acid responsiveness on chromosome 5A and association with dehydration tolerance in common wheat seedlings. J. Plant Physiol. 2014, 171, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Mohamed, E.A.; Hussein, M.Y.; Sallam, A. Genomic regions associated with leaf wilting traits under drought stress in spring wheat at the seedling stage revealed by GWAS. Environ. Exp. Bot. 2021, 184, 104393. [Google Scholar] [CrossRef]
- Kocheva, K.; Nenova, V.; Karceva, T.; Petrov, P.; Georgiev, G.I.; Börner, A.; Landjeva, S. Changes in water status, membrane stability and antioxidant capacity of wheat seedlings carrying different Rht-B1 dwarfing alleles under drought stress. J. Agron. Crop Sci. 2014, 200, 83–91. [Google Scholar] [CrossRef]
- Iannucci, A.; Marone, D.; Russo, M.A.; De Vita, P.; Miullo, V.; Ferragonio, P.; Blanco, A.; Gadaleta, A.; Mastrangelo, A.M. Mapping QTL for root and shoot morphological traits in a durum wheat × T. dicoccum segregating population at seedling stage. Int. J. Genomics 2017, 2017, 6876393. [Google Scholar] [CrossRef]
- Schierenbeck, M.; Alqudah, A.M.; Thabet, S.G.; Lohwasser, U.; Simón, M.R.; Börner, A. Association mapping unravels the genetics controlling seedling drought stress tolerance in winter wheat. Front. Plant Sci. 2023, 14, 1061845. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Fei, Y.; Xue, Y.; Du, P.; Yang, S.; Deng, X. Expression analysis and promoter methylation under osmotic and salinity stress of TaGAPC1 in wheat (Triticum aestivum L). Protoplasma 2017, 254, 987–996. [Google Scholar] [CrossRef]
- Kaur, A.; Grewal, A.; Sharma, P. Comparative analysis of DNA methylation changes in two contrasting wheat genotypes under water deficit. Biol. Plant. 2018, 62, 471–478. [Google Scholar] [CrossRef]
- Duan, H.; Li, J.; Zhu, Y.; Jia, W.; Wang, H.; Jiang, L.; Zhou, Y. Responsive changes of DNA methylation in wheat (Triticum aestivum) under water deficit. Sci. Rep. 2020, 10, 7938. [Google Scholar] [CrossRef]
- Lim, A. Drought-induced epigenetic modulation and transcriptional variation of winter wheat. BS Thesis, Oklahoma State University, USA, 2020. Available online: Hdl.handle.net/11244/329964.
- Li, J.; Jia, W.; Wang, H.; Zhu, Y.; Duan, Z.; Jiang, L.; Zhou, Y.; Duan, H. Morpho-physiological adaptation and DNA methylation of wheat seedlings under osmotic stress. Crop Pasture Sci. 2020, 71, 349–355. [Google Scholar] [CrossRef]
- Gao, S.; Li, L.; Han, X.; Liu, T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.; Zhang, F.; Chen, J.; et al. Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genom. 2021, 22, 1–17. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Pei, X.; Chen, H.; Li, X.; Wang, J.; Wang, C. Comparative genome-wide analysis and expression profiling of histone acetyltransferases and histone deacetylases involved in the response to drought in wheat. J. Plant Growth Regul. 2022, 41, 1065–1078. [Google Scholar] [CrossRef]
- Li, S.; He, X.; Gao, Y.; Zhou, C.; Chiang, V.L.; Li, W. Histone acetylation changes in plant response to drought stress. Genes 2021, 12, 1409. [Google Scholar] [CrossRef] [PubMed]
- Demetriou, K.; Kapazoglou, A.; Tondelli, A.; Francia, E.; Stanca, M.A.; Bladenopoulos, K.; Tsaftaris, A.S. Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol Plant. 2009, 136, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Gautam, T.; Pal, S.; Chaturvedi, D.; Rakhi, Jan, I. ; Balyan, H.S.; Gupta, P.K. Identification and characterization of SET domain family genes in bread wheat (Triticum aestivum L.). Sci. Rep. 2020, 10, 14624. [Google Scholar] [CrossRef]
- Kong, L.; Liu, Y.; Wang, X.; Chang, C. Insight into the role of epigenetic processes in abiotic and biotic stress response in wheat and barley. Int. J. Mol. Sci. 2020, 21, 1480. [Google Scholar] [CrossRef]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Saez, A.; Rodrigues, A.; Santiago, J.; Rubio, S.; Rodriguez, P.L. HAB1–SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 2008, 2008 20, 2972–2988. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Y.; Zheng, X.; Liu, C.; Dong, S.; Li, R.; Zhang, G.; Wei, Y.; Qu, H.; Li, Y.; et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 2019, 76, 646–659. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Bhardwaj, A.R.; Agarwal, M.; Katiyar-Agarwal, S. Discovery of small RNAs in wheat: A survey. Indian J. Plant Physiol. 2017, 22, 411–421. [Google Scholar] [CrossRef]
- Shanker, A.K.; Maheswari, M. Small RNA and drought tolerance in crop plants. Indian J. Plant Physiol. 2017, 22, 422–433. [Google Scholar] [CrossRef]
- Bakhshi, B.; Fard, E.M.; Gharechahi, J.; Safarzadeh, M.; Nikpay, N.; Fotovat, R.; Azimi, M.R.; Salekdeh, G.H. The contrasting microRNA content of a drought tolerant and a drought susceptible wheat cultivar. J. Plant Physiol. 2017, 216, 35–43. [Google Scholar] [CrossRef]
- Gómez-Martín, C.; Zhou, H.; Medina, J.M.; Aparicio-Puerta, E.; Shi, B.; Hackenberg, M. Genome-wide analysis of microRNA expression profile in roots and leaves of three wheat cultivars under water and drought conditions. Biomolecules 2023, 13, 440. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xin, Z.; Wang, Z.; Yang, Q.; Guo, S.; Guo, X.; Cao, L.; Lin, T. Identification and comparative analysis of differentially expressed miRNAs in leaves of two wheat (Triticum aestivum L.) genotypes during dehydration stress. BMC Plant Biol. 2015, 15, 1–15. [Google Scholar] [CrossRef]
- Akpinar, B.A.; Kantar, M.; Budak, H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct. Integr. Genom. 2015, 15, 587–598. [Google Scholar] [CrossRef]
- Liu, H. , Able, A.J. and Able, J.A. Integrated analysis of small RNA, transcriptome, and degradome sequencing reveals the water-deficit and heat stress response network in durum wheat. Int. J. Mol. Sci. 2020, 21, 6017. [Google Scholar] [CrossRef]
- Liu, H.; Able, A.J.; Able, J.A. Small RNAs and their targets are associated with the transgenerational effects of water-deficit stress in durum wheat. Sci. Rep. 2021, 11, 3613. [Google Scholar] [CrossRef]
- Li, N.; Liu, T.; Guo, F.; Yang, J.; Shi, Y.; Wang, S.; Sun, D. Identification of long non-coding RNA-microRNA-mRNA regulatory modules and their potential roles in drought stress response in wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 1011064. [Google Scholar] [CrossRef]
- Todorovska, E.; Abu Mhadi, N.; Christov, N.; Bozhanova, V.; Atanassov, A. Cereal genetics and genomics in Bulgaria - challenges and perspectives. C. R. Acad. Bulg. Sci. 2018, 71, 143–160. [Google Scholar]
- Mihova, G.; Baychev, V.; Alexandrov, T.; Petrova, T.; Stanoeva, Y.; Ivanova, V. Breeding of cereal crops at Dobrudzha Agricultural Institute-General Toshevo, Bulgaria. J. Agricult. Food Environmen. Sci., JAFES.
- Dimitrov, E.; Uhr, Z.; Chipilski, R. Study of yield and stability by common winter wheat varieties by changing climatic conditions in Sadovo region. Bulg. J. Agric. Sci. 2022, 28. [Google Scholar]
- Simova-Stoilova, L.; Vaseva, I.; Grigorova, B.; Demirevska, K.; Feller, U. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiol. Biochem. 2010, 48, 200–206. [Google Scholar] [CrossRef]
- Vassileva, V.; Simova-Stoilova, L.; Demirevska, K.; Feller, U. Variety-specific response of wheat (Triticum aestivum L.) leaf mitochondria to drought stress. J. Plant Res. 2009, 122, 445–454. [Google Scholar] [CrossRef]
- Vaseva, I.; Akiscan, Y.; Simova-Stoilova, L.; Kostadinova, A.; Nenkova, R.; Anders, I.; Feller, U.; Demirevska, K. Antioxidant response to drought in red and white clover. Acta Physiol. Plant. 2012, 34, 1689–1699. [Google Scholar] [CrossRef]
- Vassileva, V.; Vaseva, I.; Dimitrova, A. Expression profiling of DNA methyltransferase genes in wheat genotypes with contrasting drought tolerance. Bulg. J. Agric. Sci. 2019, 25, 845–851. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




