1. Introduction
Dyslipidemia is a disease characterised by a high level of lipids [
1] in the bloodstream. It has emerged as a potential risk factor in the insurgence of atherosclerosis, a chronic inflammatory disorder of the arterial walls [
1].
Atherosclerosis is commonly defined as the process of hardening of the arteries. It may cause many cardiovascular diseases, such as stroke and coronary artery disease [
2,
3]. The link between dyslipidemia and atherosclerosis has been studied in recent years [
4], and as a result, the pathophysiological mechanisms driving this lethal disease have been partially elucidated. Dyslipidemia is often associated with type 2 diabetes mellitus as a comorbidity [
5,
6].
Lipids, i.e. cholesterol and triglycerides, contribute to cellular processes in many ways; thus, the imbalance of their normal levels may cause dysfunctions. In particular, high levels of lipids can lead to the formation of lipid-rich plaques bound to the arteries. Consequently, such plaques can modify the elasticity of arterial walls, starting a set of events culminating in atherosclerosis [
7]. In particular, elevated levels of low-density lipoprotein cholesterol (LDL) [
8], contribute to atherogenesis by accumulating in the subendothelial space, triggering an inflammatory response and promoting the recruitment of immune cells. High-density lipoprotein cholesterol (HDL) is thought to exert protective effects against atherosclerosis [
9]. HDL promotes reverse cholesterol transport and removes cholesterol from peripheral tissues, including atherosclerotic plaques. However, dysfunctional HDL particles may lose their protective properties in dyslipidemia, exacerbating atherosclerotic plaque formation [
10].
While there exist a considerable number of studies elucidating the link between dyslipidemia and atherosclerosis, in particular in the context of patients affected by type 2 diabetes mellitus [
11,
12], the analysis of sex and age as factors is still an open field. This is particularly relevant for diabetic patients. A set of recent studies has provided many insights into the molecular causes of the different insurgence and progression in older males for males and females [
5,
6,
12]. The underlying pathophysiology of diabetes is exacerbated by the ageing process, by accelerating the progression of many comorbidities [
13,
14,
15,
16,
17,
18,
19,
20].
Man et al., [
21] proposed a clinical study to investigate sexual dimorphism in the incidence and complications of atherosclerosis. By combining multiple histological and imaging, the authors discuss the role of sex as a biological variable in atherosclerosis. The paper also evidences the importance of understanding the differences between men and women in the development and complications of atherosclerosis, as well as how risk factors, plaque size, and plaque characteristics may vary between sexes.
The review of Fairweather [
22] emphasizes that sex hormones significantly alter the immune response during atherosclerosis, resulting in different disease phenotypes in men and women. For instance, women tend to exhibit increased antibody and autoantibody responses in response to infection and damage, while men typically have elevated innate immune activation.
In parallel, a growing body of evidence suggests that age-related changes in the vasculature and various molecular pathways contribute to the initiation and progression of atherosclerosis. For instance, with advancing age, structural and functional changes occur in the blood vessels, collectively called vascular ageing. The accumulation of senescent cells, which exhibit a senescence-associated secretory phenotype (SASP), further contributes to the chronic inflammatory milieu in the arterial wall, fueling atherogenesis. Finally, epigenetic changes, such as DNA methylation, histone modifications, and non-coding RNA regulation, influence gene expression patterns during ageing and atherosclerosis.
Starting from demographic data which evidence a different incidence of atherosclerosis and previous studies [
5], we hypothesise that there exist some molecular differences due to age and sex of the basal expression of genes related to dyslipidemia-associated atherosclerosis [
23]. We consider genes for which there is evidence of correlation with dyslipidemia as reported in the T2DiACoD database [
24]. We analyse the expression of such genes and their variations with age and sex in blood, artery aorta and adipose tissue.
Our results show the existence of some genes whose expression changes with age and sex and are thus related to the risk of presenting dyslipidemia associated atherosclerosis. We also analyse these genes on a network level by gathering information from the STRING [
25] database for deriving the protein interaction networks with a multiscale approach [
26]. Using the proposed analysis, we suggest that ageing and sex may stratify the risk of dyslipidemia-associated atherosclerosis. This indicates the need for further research on the mechanisms of the age-associated increase [
27].
2. Materials and Methods
2.1. Data Sources
The proposed framework is based on the following data sources: (i) T2DiACoD database [
24]; (ii) GTexVisualiser for accessing GTEx database [
28,
29], (iii) voyAGEr [
30] for the analysis of sex differences , and (iv) STRING network database [
31]. T2DiACoD contains genes and non-coding transcripts related to complications associated with type 2 diabetes mellitus. We here consider only 115 genes related with atherosclerosis.
For each gene, we extract expression data associated with age class (20-29, 30-39, 40-49, 50-59, 60-69, 70-79 years) and sex metadata from the GTEx data portal [
29].
The current version of the GTEx database (v8 accessed on July 25th stores 17382 samples of 54 tissues of 948 donors, see at
https://gtexportal.org/home/tissueSummaryPage). GTExVisualizer is a web interface able to: (i) query the GTEx data portal; (ii) analyse genes using sex/age expression patterns, also integrated with network-based modules; and (iii) report results as gene networks. It also allows the user to obtain basic statistics that evidence differences in gene expression among sex/age groups.
2.2. Data Analysis
Median values of gene expression are calculated for age classes: 20-29, 30-39, 40-49, 50-59, 60-69, 70-79 years. We then selected those genes whose average expression values are monotonically increasing or decreasing in the age intervals. Differences among age intervals are measured by using a Kruskal-Wallis test. A less than 0.01 (after correction for multiple tests) was considered significant. The calculated has been corrected with the Bonferroni correction method.
We extracted the protein interaction networks by querying the STRING database [
29] induced by increasing or decreasing genes. The STRING protein interaction network database is used to build protein networks. We considered only physical interactions with a reliability value higher than
.
We calculated the functional enrichment [
32] on the STRING web server.
Differences in age and sex were evaluated by using the voyAGEr web portal [
30]. It uses GTEx data to build a linear model (ShARP-LM) that analyses tissue-specific differential gene expression. The gene expression is linearly modelled considering factors: age, sex and age-sex interaction effects. By using voyAGEr, we can identify the age periods when significant gene expression changes occur.
3. Results
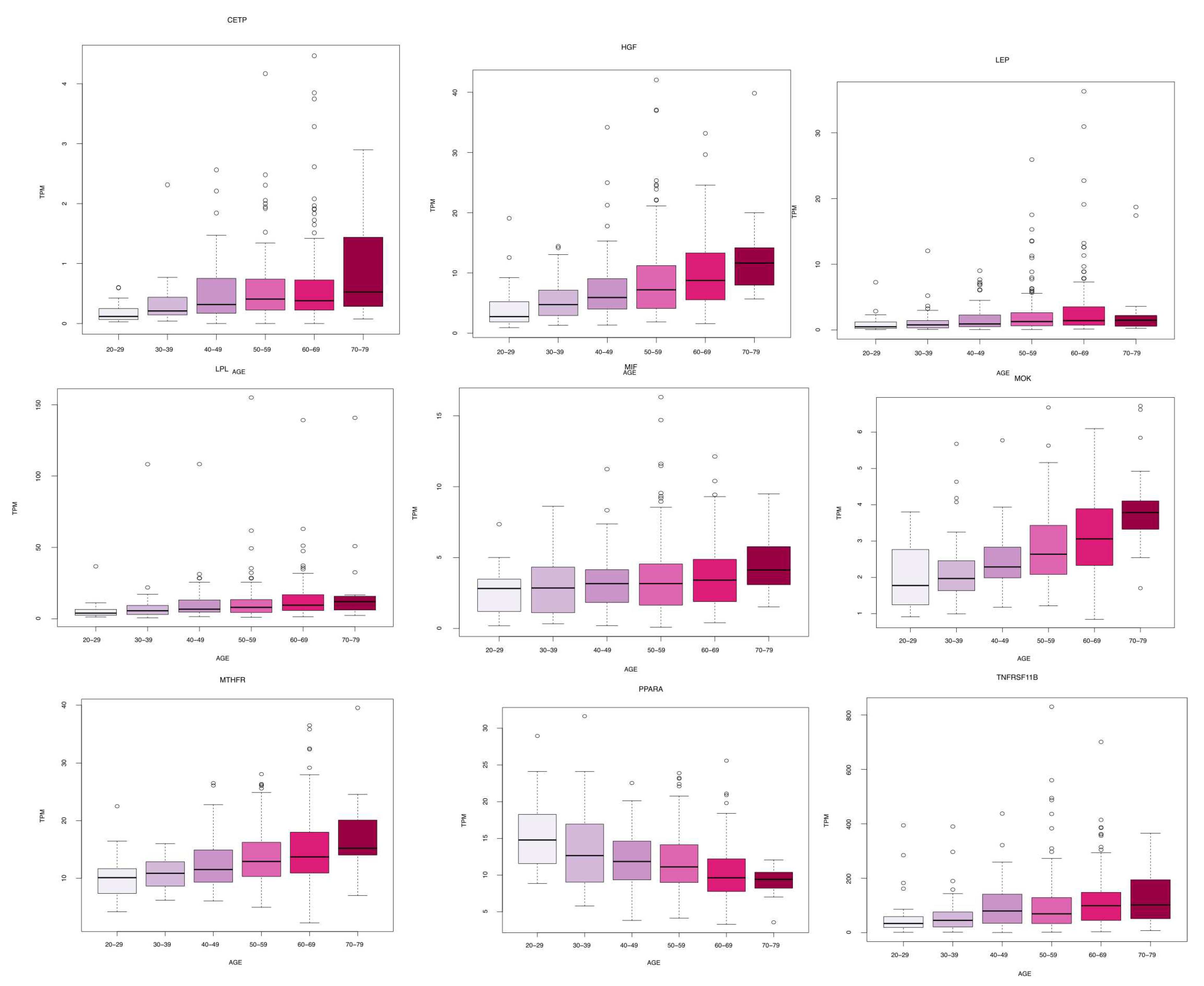
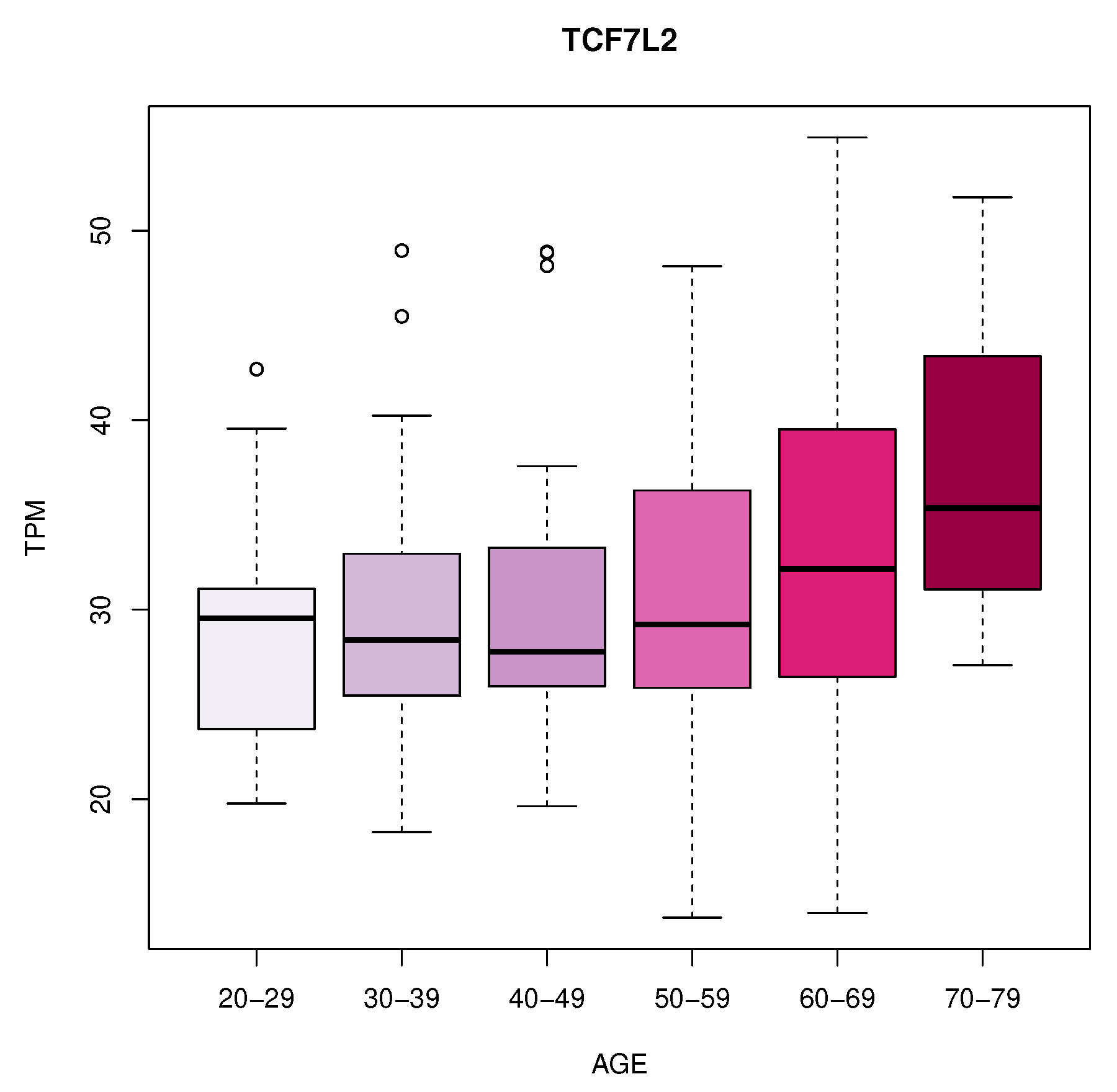
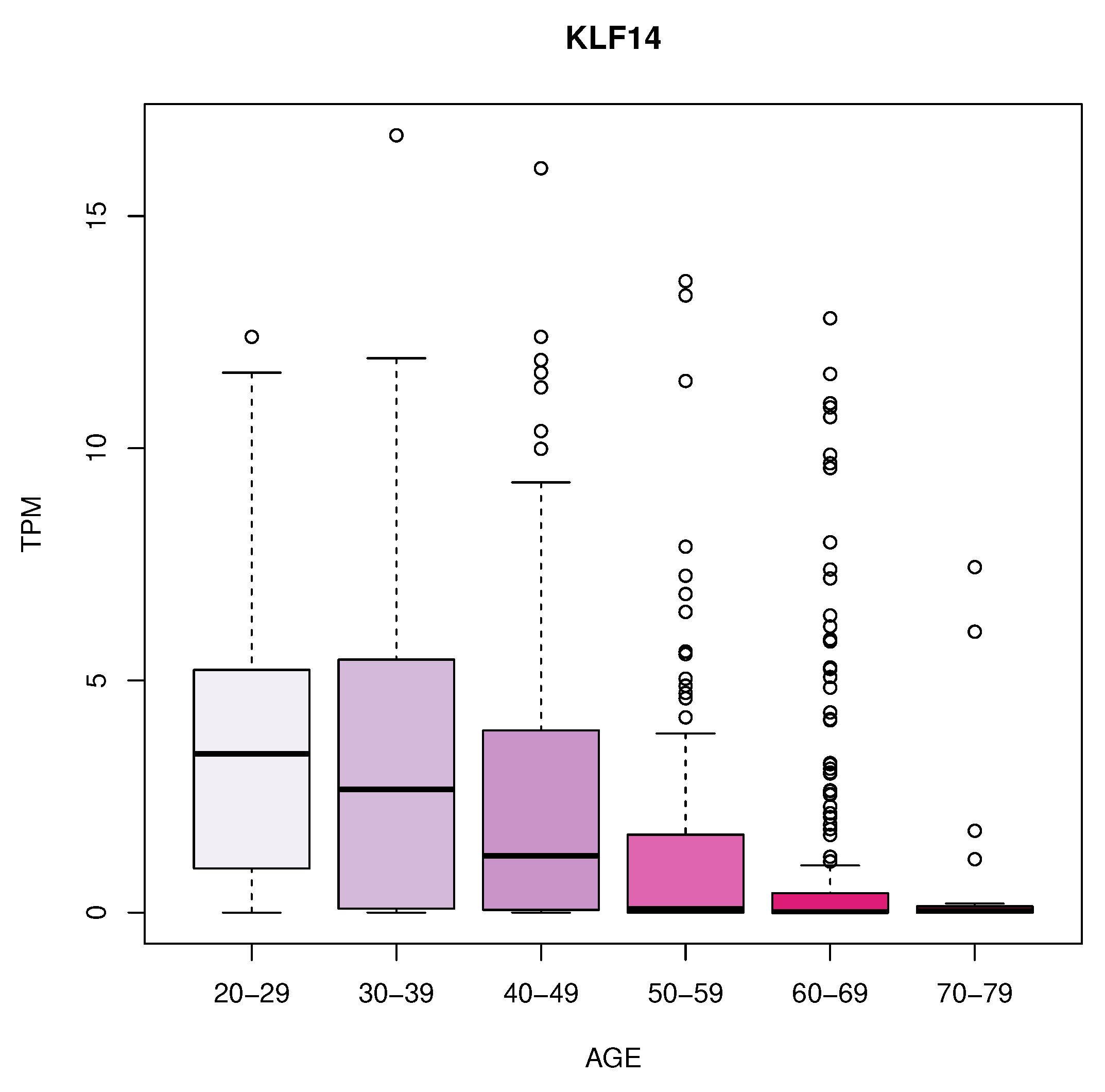
We identified eleven genes whose expression changes with age in blood, artery tibial and artery aorta tissues. In blood, we found a decrease in KLF14 expression. In Artery Tibial, we found a decrease in the expression of PPARA gene, while MTHFR, HGF, LEP, LPL, TNFRSF11B, MOK, CETP, and MIF genes increased with age. Finally, we found an increased expression of the TCF7L2 gene in the aorta tissue.
Table 1 summarizes these results while
Figure 1,
Figure 2 and
Figure 3 depicts the gene expression as boxplots.
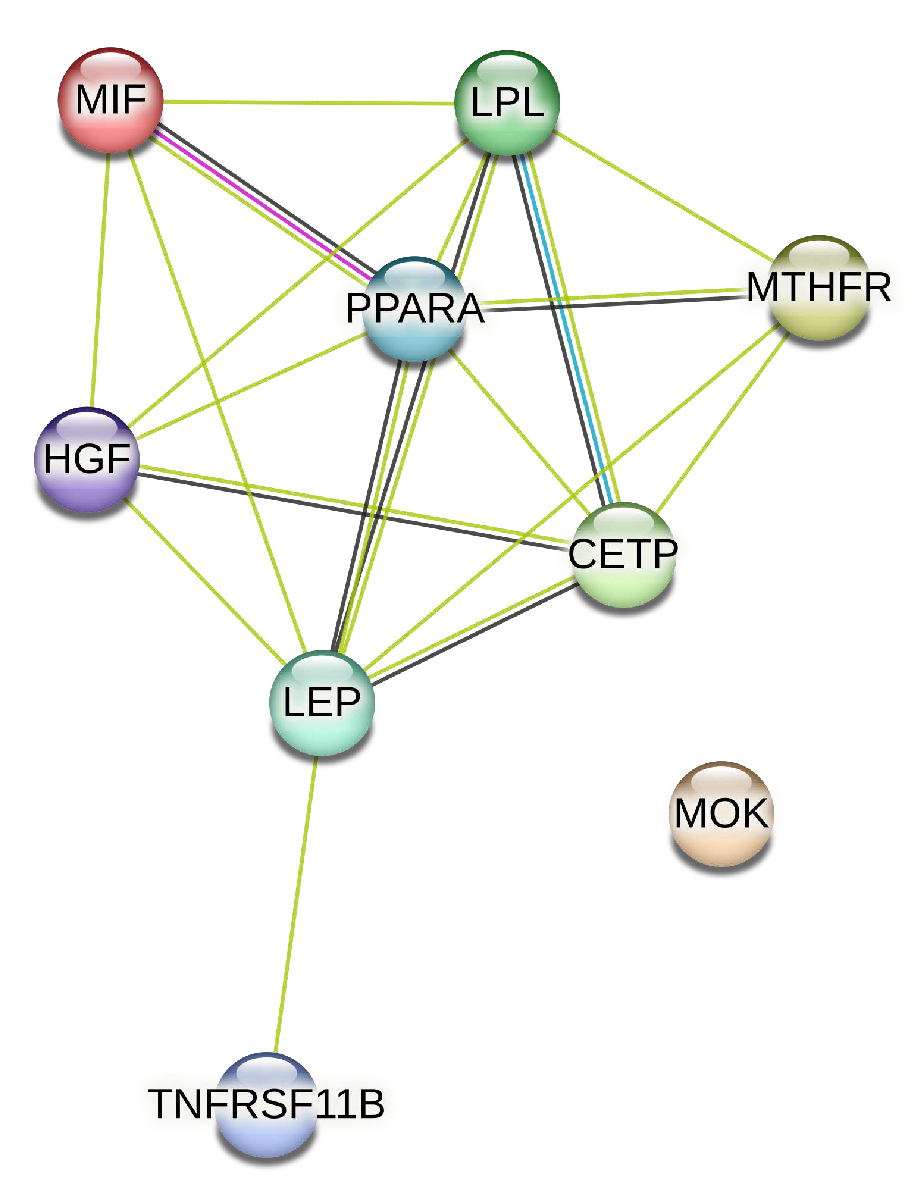
We extracted from the STRING database the protein interaction network associated with the genes of artery tibial, represented in
Figure 4.
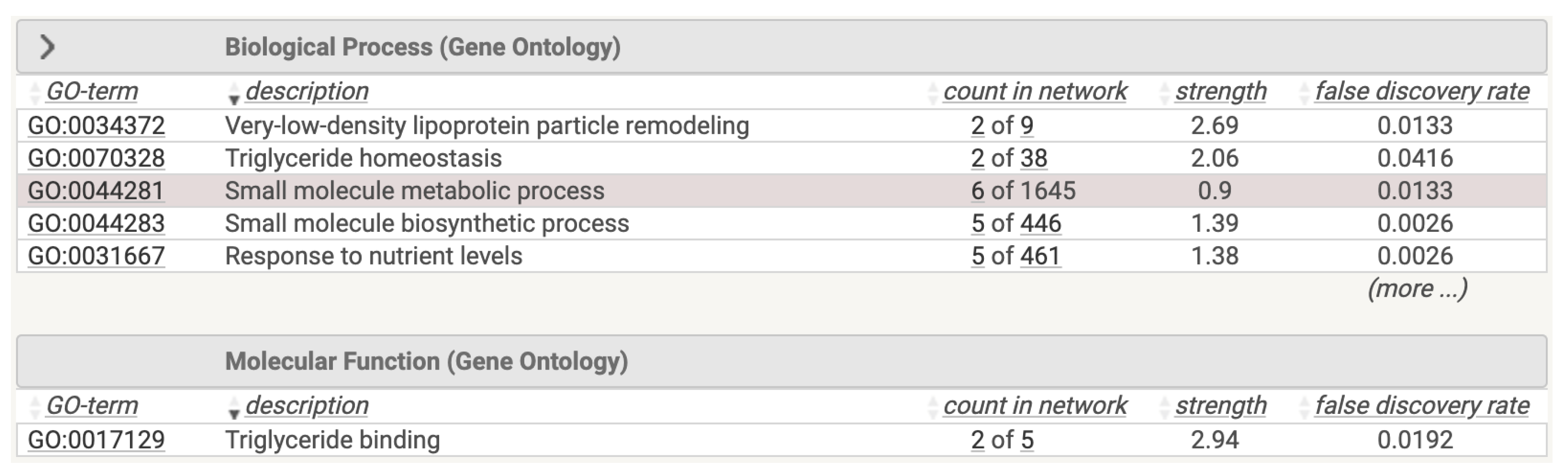
We performed a functional analysis of this network on the STRING database, reporting the results depicted in
Figure 5.
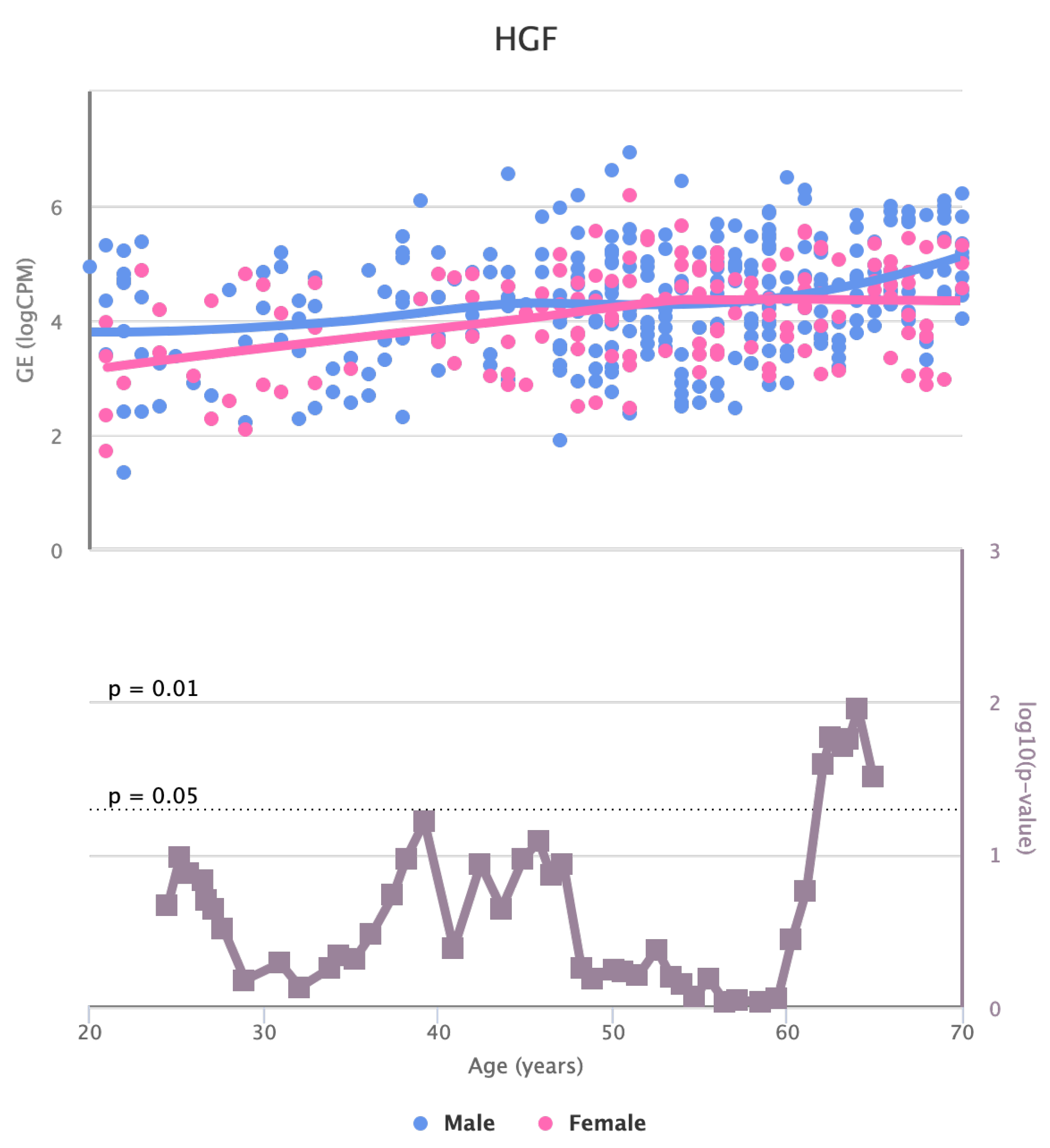
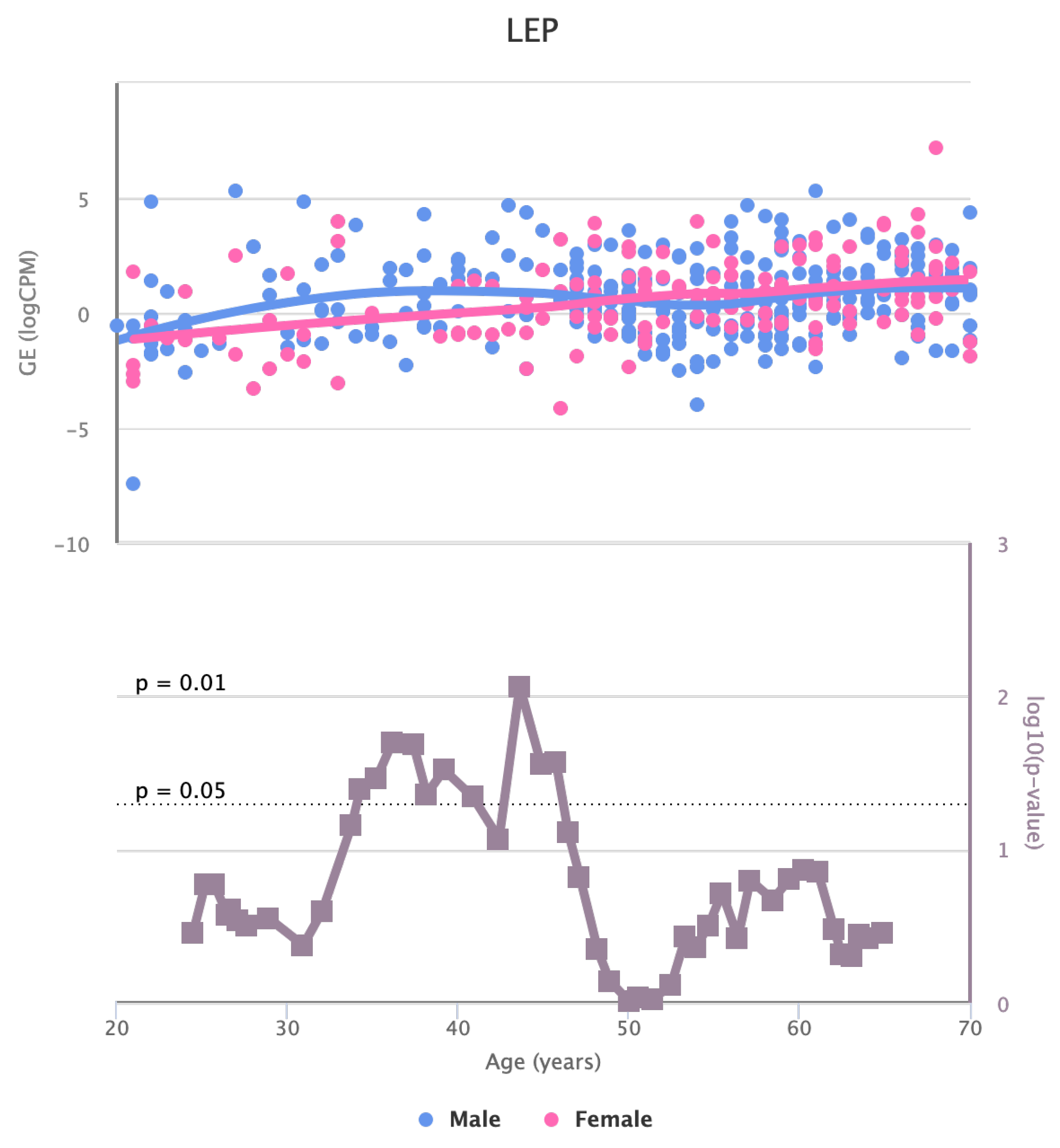
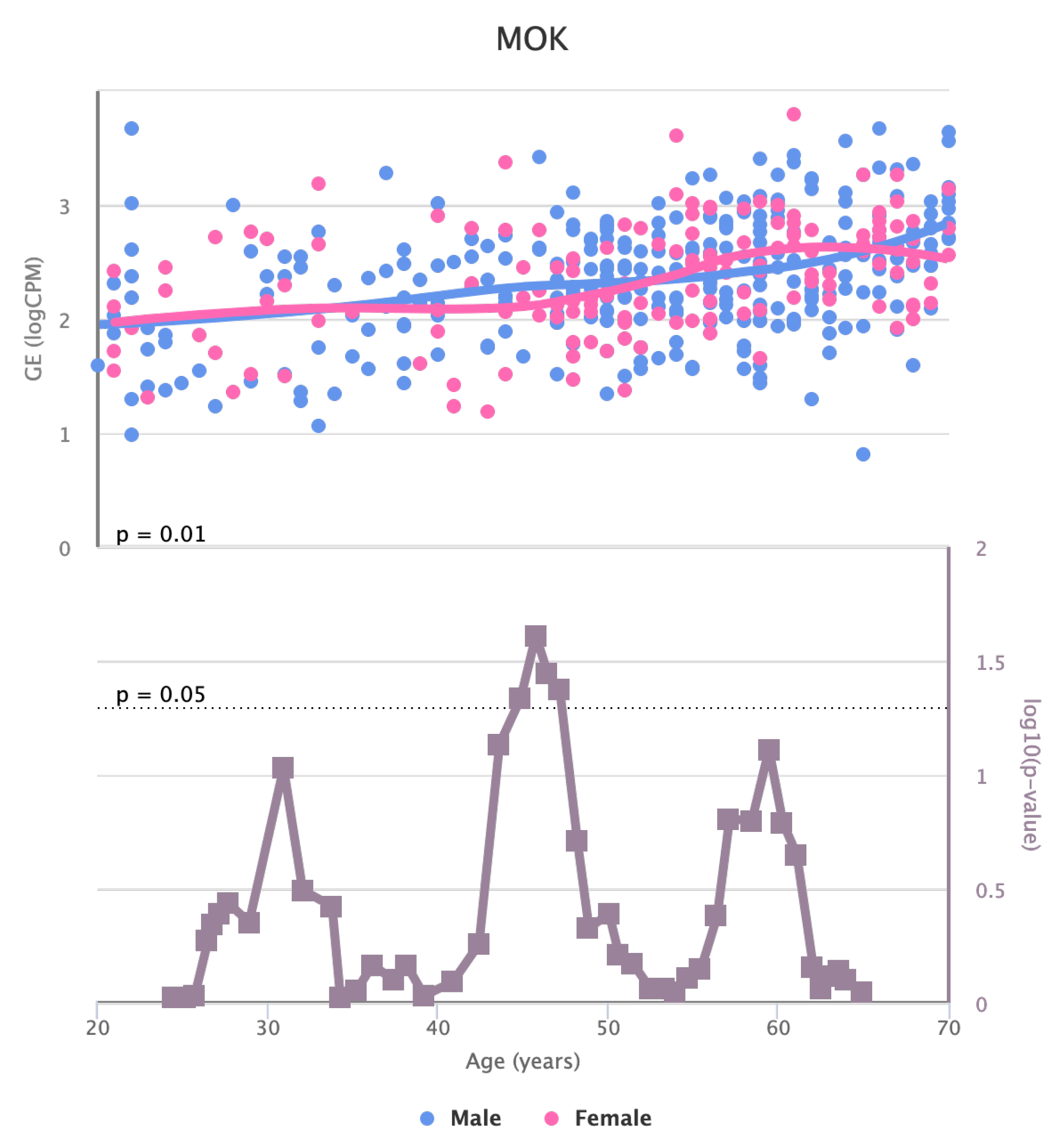
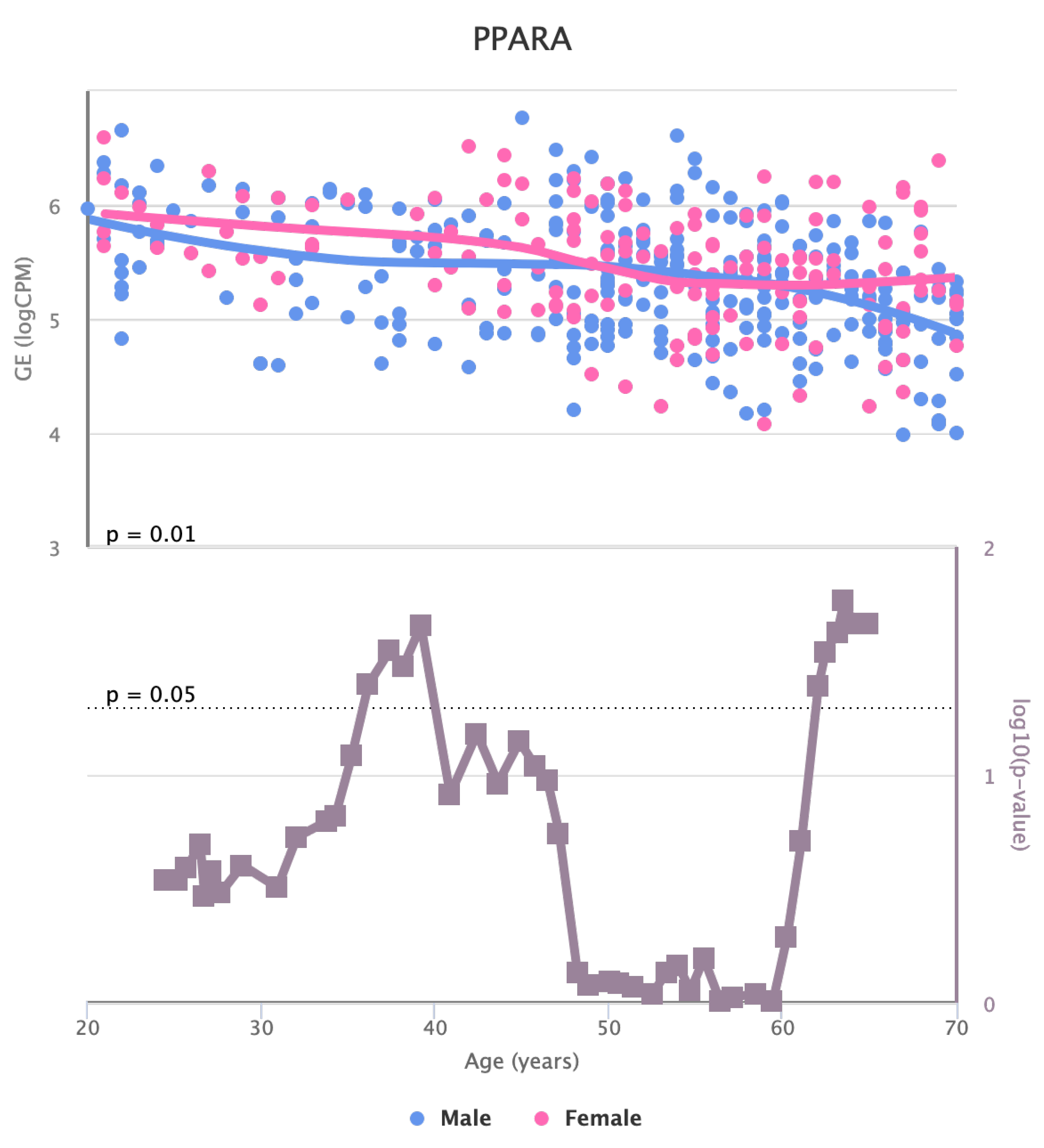
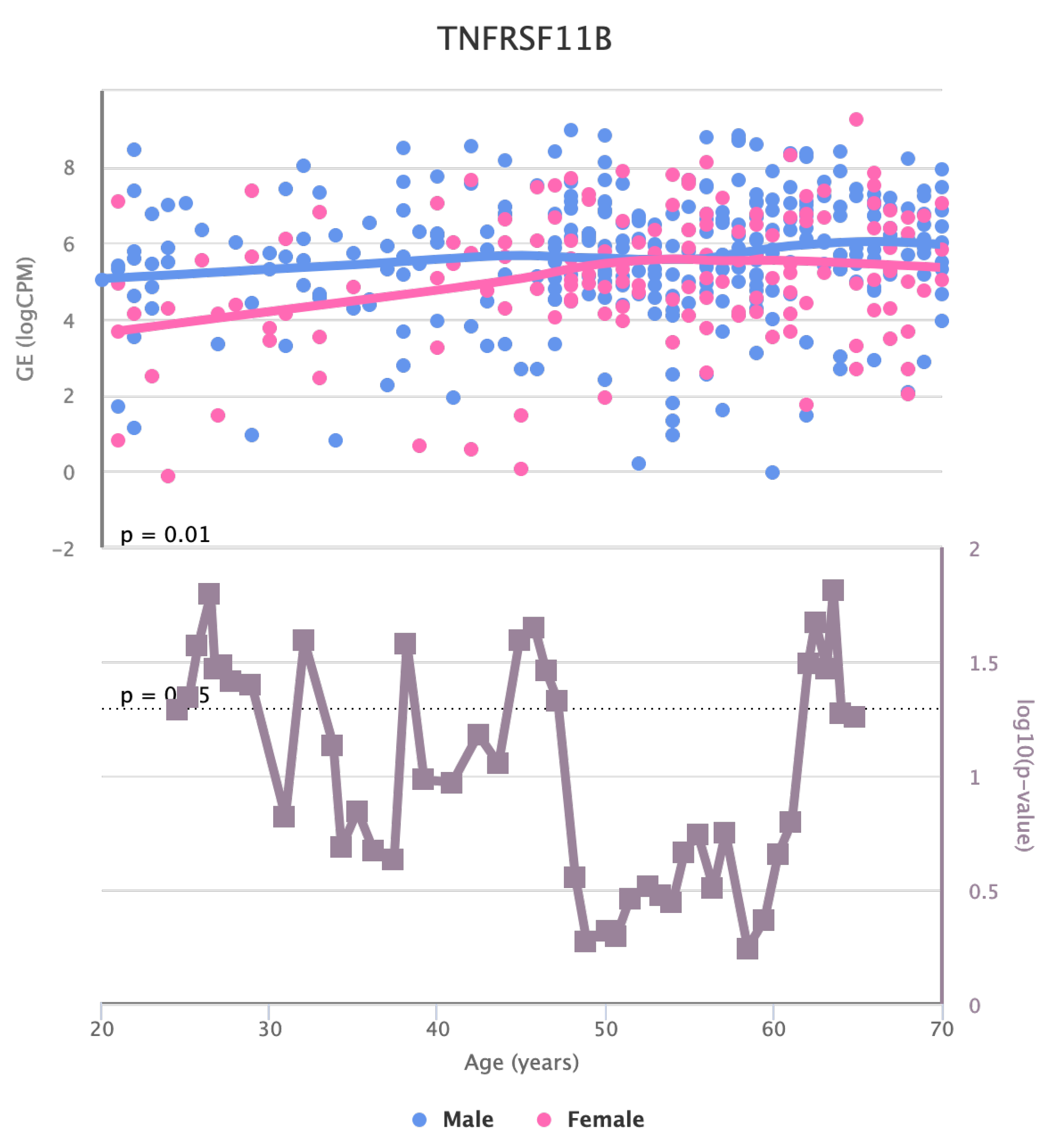
Finally, we also identified for HGF, LEP, MOK, MTHFR, PPARA, and TNFRSF11B genes a significant change in the basal expression with sex as reported in
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10 and
Figure 11.
4. Discussion
The bioinformatic analysis we carried out in this work evidenced the existence of a set of genes in artery tibial tissue presenting changes in basal expression with age [
33]. The modification of gene expression and other parameters may constitute a risk factor for the insurgence and development of some diseases. We here analysed genes related to dyslipidemia-based atherosclerosis.
In artery tibial, we found changes in KLF14 gene, which encodes a transcription factor that regulates several cellular processes. The decrease in its expression in blood with age suggests that it might play a role in age-related changes in the blood and may have implications in ageing processes or age-related diseases [
34,
35]. PPARA encodes a nuclear receptor involved in lipid metabolism and inflammation. The decrease in its expression in artery tibial with age may indicate alterations in lipid metabolism and inflammation pathways in this tissue during ageing [
36,
37,
38].
The changes in the expression of MTHFR, HGF, LEP, LPL, TNFRSF11B, MOK, CETP, and MIF in artery tibial with age suggests that they might be involved in age-related processes in this tissue [
38,
39].
TCF7L2 encodes a transcription factor in the Wnt signalling pathway and plays a crucial role in various cellular processes [
40]. The increased expression of TCF7L2 in the artery aorta with age indicates its potential involvement in ageing-related changes in this tissue and might have implications for cardiovascular health [
41].
Overall, this study provides valuable insights into the age-related changes in gene expression in different tissues. It is important to note that gene expression is just one aspect of the complex process of ageing, and further research is needed to understand the underlying mechanisms and potential implications of these findings for age-related diseases and health conditions [
42,
43]. Additionally, considering the dynamic nature of gene regulation, factors such as lifestyle, environmental influences, and genetic variations can also contribute to the observed changes in gene expression with age.
Further investigation is needed to understand the specific roles of these genes and their contributions to aging-related changes.
5. Conclusions
We identified eleven genes whose expression changes with age in blood, artery tibial, and artery aorta tissues. This may shed light on potential molecular mechanisms associated with aging and dyslipidemia-based atherosclerosis. The decrease in KLF14 expression in blood and PPARA expression in artery tibial, along with the increased expression of MTHFR, HGF, LEP, LPL, TNFRSF11B, MOK, CETP, and MIF in artery tibial, and TCF7L2 in the artery aorta, may indicate their involvement in age-related processes. The protein interaction network associated with these genes in artery tibial further provides valuable insights into their potential functional relationships.
It is essential to recognize that gene expression changes are part of a complex interplay of various factors contributing to the ageing process. Thus, these findings lay the groundwork for future research to explore the broader implications of these gene expression alterations and how they may influence age-related diseases and overall health.
The main limitation of this work is the use of only in silico data; even we retain that our findings may stimulate further wet lab and clinical experiments.
Author Contributions
Conceptualization, P.H.G. and P.V.; methodology, P.H.G. ad E.P.; software, F.C.; validation, P.H.G. and P.V..; resources, P.V.; data curation, P.H.G. and F.C:; writing—original draft preparation, P.H.H.; writing—review and editing, P.H.G and P.V.; visualization, X.X.; supervision, P.V..; project administration, P.V.; funding acquisition, P.V.. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable
Informed Consent Statement
Not applicable
Data Availability Statement
Acknowledgments
Eng - This work was funded by the Next Generation EU - Italian NRRP, Mission 4, Component 2, Investment 1.5, call for the creation and strengthening of ’Innovation Ecosystems’, building ’Territorial R&D Leaders’ (Directorial Decree n. 2021/3277) - project Tech4You - Technologies for climate change adaptation and quality of life improvement, n. ECS0000009. This work reflects only the authors’ views and opinions, neither the Ministry for University and Research nor the European Commission can be considered responsible for them.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Farmer, J.A. Diabetic dyslipidemia and atherosclerosis: evidence from clinical trials. Current diabetes reports 2008, 8, 71–77. [Google Scholar] [CrossRef]
- Koba, S.; Hirano, T. Dyslipidemia and atherosclerosis. Nihon rinsho. Japanese journal of clinical medicine 2011, 69, 138–143. [Google Scholar]
- Stein, R.; Ferrari, F.; Scolari, F. Genetics, dyslipidemia, and cardiovascular disease: new insights. Current cardiology reports 2019, 21, 1–12. [Google Scholar] [CrossRef]
- Tietge, U.J. Hyperlipidemia and cardiovascular disease: inflammation, dyslipidemia, and atherosclerosis. Current opinion in lipidology 2014, 25, 94–95. [Google Scholar] [CrossRef] [PubMed]
- Succurro, E.; Marini, M.A.; Fiorentino, T.V.; Perticone, M.; Sciacqua, A.; Andreozzi, F.; Sesti, G. Sex-specific differences in prevalence of nonalcoholic fatty liver disease in subjects with prediabetes and type 2 diabetes. Diabetes Research and Clinical Practice 2022, 190, 110027. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Cortese, F.; Mannino, G.C.; Pedace, E.; Succurro, E.; Andreozzi, F.; Veltri, P. Differential network analysis between sex of the genes related to comorbidities of type 2 mellitus diabetes. Applied Network Science 2023, 8, 1–16. [Google Scholar] [CrossRef]
- Mizuno, Y.; Jacob, R.F.; Mason, R.P. Inflammation and the development of atherosclerosis—effects of lipid-lowering therapy. Journal of atherosclerosis and thrombosis 2011, 18, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.V.; Ala-Korpela, M. What is ‘LDL cholesterol’? Nature Reviews Cardiology 2019, 16, 197–198. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Brewer Jr, H.B.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nature reviews cardiology 2016, 13, 48–60. [Google Scholar] [CrossRef]
- Navab, M.; Reddy, S.T.; Van Lenten, B.J.; Anantharamaiah, G.; Fogelman, A.M. The role of dysfunctional HDL in atherosclerosis. Journal of lipid research 2009, 50, S145–S149. [Google Scholar] [CrossRef] [PubMed]
- Libby, P. The biology of atherosclerosis comes full circle: lessons for conquering cardiovascular disease. Nature Reviews Cardiology 2021, 18, 683–684. [Google Scholar] [CrossRef]
- Guzzi, P.H.; Cortese, F.; Mannino, G.C.; Pedace, E.; Succurro, E.; Andreozzi, F.; Veltri, P. Analysis of age-dependent gene-expression in human tissues for studying diabetes comorbidities. Scientific Reports 2023, 13, 10372. [Google Scholar] [CrossRef]
- LeRoith, D.; Biessels, G.J.; Braithwaite, S.S.; Casanueva, F.F.; Draznin, B.; Halter, J.B.; Hirsch, I.B.; McDonnell, M.E.; Molitch, M.E.; Murad, M.H.; others. Treatment of diabetes in older adults: an Endocrine Society clinical practice guideline. The Journal of Clinical Endocrinology & Metabolism 2019, 104, 1520–1574. [Google Scholar]
- Mercatelli, D.; Pedace, E.; Veltri, P.; Giorgi, F.M.; Guzzi, P.H. Exploiting the molecular basis of age and gender differences in outcomes of SARS-CoV-2 infections. Computational and Structural Biotechnology Journal 2021, 19, 4092–4100. [Google Scholar] [CrossRef]
- Bahour, N.; Cortez, B.; Pan, H.; Shah, H.; Doria, A.; Aguayo-Mazzucato, C. Diabetes mellitus correlates with increased biological age as indicated by clinical biomarkers. GeroScience 2022, 44, 415–427. [Google Scholar] [CrossRef]
- Munshi, M.N.; Meneilly, G.S.; Rodríguez-Mañas, L.; Close, K.L.; Conlin, P.R.; Cukierman-Yaffe, T.; Forbes, A.; Ganda, O.P.; Kahn, C.R.; Huang, E.; others. Diabetes in ageing: pathways for developing the evidence base for clinical guidance. The Lancet Diabetes & Endocrinology 2020, 8, 855–867. [Google Scholar]
- Dennis, J.M.; Mateen, B.A.; Sonabend, R.; Thomas, N.J.; Patel, K.A.; Hattersley, A.T.; Denaxas, S.; McGovern, A.P.; Vollmer, S.J. Type 2 diabetes and COVID-19–Related mortality in the critical care setting: a national cohort study in England, March–July 2020. Diabetes care 2021, 44, 50–57. [Google Scholar] [CrossRef]
- Care, F. Standards of medical care in diabetes-2019. Diabetes Care 2019, 42, S124–S138. [Google Scholar]
- Atlas, D.; others. International diabetes federation. IDF Diabetes Atlas, 7th edn. Brussels, Belgium: International Diabetes Federation 2015, 33. [Google Scholar]
- Antal, B.; McMahon, L.P.; Sultan, S.F.; Lithen, A.; Wexler, D.J.; Dickerson, B.; Ratai, E.M.; Mujica-Parodi, L.R. Type 2 diabetes mellitus accelerates brain aging and cognitive decline: Complementary findings from UK Biobank and meta-analyses. Elife 2022, 11, e73138. [Google Scholar] [CrossRef]
- Man, J.J.; Beckman, J.A.; Jaffe, I.Z. Sex as a biological variable in atherosclerosis. Circulation research 2020, 126, 1297–1319. [Google Scholar] [CrossRef]
- Fairweather, D. Sex differences in inflammation during atherosclerosis. Clinical Medicine Insights: Cardiology 2014, 8, CMC–S17068. [Google Scholar] [CrossRef]
- Roetker, N.S.; Pankow, J.S.; Bressler, J.; Morrison, A.C.; Boerwinkle, E. Prospective study of epigenetic age acceleration and incidence of cardiovascular disease outcomes in the ARIC study (Atherosclerosis Risk in Communities). Circulation: Genomic and Precision Medicine 2018, 11, e001937. [Google Scholar] [CrossRef]
- Rani, J.; Mittal, I.; Pramanik, A.; Singh, N.; Dube, N.; Sharma, S.; Puniya, B.L.; Raghunandanan, M.V.; Mobeen, A.; Ramachandran, S. T2DiACoD: a gene atlas of type 2 diabetes mellitus associated complex disorders. Scientific Reports 2017, 7, 1–21. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic acids research 2016, gkw937. [Google Scholar] [CrossRef]
- Gu, S.; Jiang, M.; Guzzi, P.H.; Milenković, T. Modeling multi-scale data via a network of networks. Bioinformatics 2022, 38, 2544–2553. [Google Scholar] [CrossRef]
- Health, T.L.D. Equitable precision medicine for type 2 diabetes, 2022.
- Guzzi, P.H.; Lomoio, U.; Veltri, P. GTExVisualizer: a web platform for supporting ageing studies. Bioinformatics 2023, 39, btad303. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; others. The genotype-tissue expression (GTEx) project. Nature genetics 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Schneider, A.L.; Saraiva-Agostinho, N.; Barbosa-Morais, N.L. voyAGEr: free web interface for the analysis of age-related gene expression alterations in human tissues. bioRxiv 2022, 2022–12. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; others. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic acids research 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Cho, Y.R.; Mina, M.; Lu, Y.; Kwon, N.; Guzzi, P.H. M-finder: Uncovering functionally associated proteins from interactome data integrated with go annotations. Proteome science 2013, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ai, R.; Jin, X.; Tang, B.; Yang, G.; Niu, Z.; Fang, E.F. Ageing and Alzheimer’s Disease: Application of Artificial Intelligence in Mechanistic Studies, Diagnosis, and Drug Development. In Artificial Intelligence in Medicine; Springer, 2021; pp. 1–16. [Google Scholar]
- Spólnicka, M.; Pośpiech, E.; Adamczyk, J.G.; Freire-Aradas, A.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Lareu, M.V.; Phillips, C.; et al. Modified aging of elite athletes revealed by analysis of epigenetic age markers. Aging (Albany NY) 2018, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Li, L.; Zheng, X.L.; Yin, W.D.; Tang, C.K. The role of Krüppel-like factor 14 in the pathogenesis of atherosclerosis. Atherosclerosis 2017, 263, 352–360. [Google Scholar] [CrossRef]
- Biswas, D.; Ghosh, M.; Kumar, S.; Chakrabarti, P. PPARa-ATGL pathway improves muscle mitochondrial metabolism: implication in aging. The FASEB Journal 2016, 30, 3822–3834. [Google Scholar] [CrossRef]
- Mangoni, M.; Petrizzelli, F.; Liorni, N.; Bianco, S.D.; Biagini, T.; Napoli, A.; Adinolfi, M.; Guzzi, P.H.; Novelli, A.; Caputo, V.; others. Investigating mitochondrial gene expression patterns in Drosophila melanogaster using network analysis to understand aging mechanisms. Applied Sciences 2023, 13, 7342. [Google Scholar] [CrossRef]
- Darci-Maher, N.; Alvarez, M.; Arasu, U.T.; Selvarajan, I.; Lee, S.H.T.; Pan, D.Z.; Miao, Z.; Das, S.S.; Kaminska, D.; Örd, T.; others. Cross-tissue omics analysis discovers ten adipose genes encoding secreted proteins in obesity-related non-alcoholic fatty liver disease. EBioMedicine 2023, 92. [Google Scholar] [CrossRef]
- Bell, E.J.; Decker, P.A.; Tsai, M.Y.; Pankow, J.S.; Hanson, N.Q.; Wassel, C.L.; Larson, N.B.; Cohoon, K.P.; Budoff, M.J.; Polak, J.F.; others. Hepatocyte growth factor is associated with progression of atherosclerosis: The Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis 2018, 272, 162–167. [Google Scholar] [CrossRef]
- Li, J.; Zhou, L.; Ouyang, X.; He, P. Transcription factor-7-like-2 (TCF7L2) in atherosclerosis: a potential biomarker and therapeutic target. Frontiers in Cardiovascular Medicine 2021, 8, 701279. [Google Scholar] [CrossRef]
- He, L.H.; Gao, J.H.; Yu, X.H.; Wen, F.J.; Luo, J.J.; Qin, Y.S.; Chen, M.X.; Zhang, D.W.; Wang, Z.B.; Tang, C.K. Artesunate inhibits atherosclerosis by upregulating vascular smooth muscle cells-derived LPL expression via the KLF2/NRF2/TCF7L2 pathway. European Journal of Pharmacology 2020, 884, 173408. [Google Scholar] [CrossRef]
- Costopoulos, C.; Liew, T.V.; Bennett, M. Ageing and atherosclerosis: Mechanisms and therapeutic options. Biochemical pharmacology 2008, 75, 1251–1261. [Google Scholar] [CrossRef]
- Tyrrell, D.J.; Goldstein, D.R. Ageing and atherosclerosis: vascular intrinsic and extrinsic factors and potential role of IL-6. Nature Reviews Cardiology 2021, 18, 58–68. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Boxplots of gene expression in artery tibial tissue. All the genes present a significant increase or decrease of basal levels with age class (20-29, 30-39, 40-49, 50-59, 60-69, 70-79) in different tissues. Differences have been tested by using a Kruskal-Wallis test. Figure evidences a decrease in the expression of PPARA, while MTHFR, HGF, LEP, LPL, TNFRSF11B, MOK, CETP, and MIF increased with age.
Figure 1.
Boxplots of gene expression in artery tibial tissue. All the genes present a significant increase or decrease of basal levels with age class (20-29, 30-39, 40-49, 50-59, 60-69, 70-79) in different tissues. Differences have been tested by using a Kruskal-Wallis test. Figure evidences a decrease in the expression of PPARA, while MTHFR, HGF, LEP, LPL, TNFRSF11B, MOK, CETP, and MIF increased with age.
Figure 2.
Increased expression of TCF7L2 in the aorta tissue.
Figure 2.
Increased expression of TCF7L2 in the aorta tissue.
Figure 3.
Expression with age of KLF14 gene in blood.
Figure 3.
Expression with age of KLF14 gene in blood.
Figure 4.
Protein Interaction Network Associated with genes presenting changes with age in artery tibial tissue.
Figure 4.
Protein Interaction Network Associated with genes presenting changes with age in artery tibial tissue.
Figure 5.
Functional Enrichment of the Genes
Figure 5.
Functional Enrichment of the Genes
Figure 6.
Changes in the Expression of HGF gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 6.
Changes in the Expression of HGF gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 7.
Changes in the Expression of LEP gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 7.
Changes in the Expression of LEP gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 8.
Changes in the Expression of MOK gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 8.
Changes in the Expression of MOK gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 9.
Changes in the Expression of MTHFR gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 9.
Changes in the Expression of MTHFR gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 10.
Changes in the Expression of PPARA gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 10.
Changes in the Expression of PPARA gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 11.
Changes in the Expression of PPARA gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Figure 11.
Changes in the Expression of PPARA gene with age due to sex effects. The bottom part of the figure reports the associated p-value.
Table 1.
Genes presenting a significant increase or decrease of basal levels with age class (20-29, 30-39, 40-49, 50-59, 60-69, 70-79) in different tissues. Differences have been tested by using a Kruskal-Wallis test.
Table 1.
Genes presenting a significant increase or decrease of basal levels with age class (20-29, 30-39, 40-49, 50-59, 60-69, 70-79) in different tissues. Differences have been tested by using a Kruskal-Wallis test.
| Tissue |
Increasing |
Decreasing |
| Blood |
|
KLF14 |
| Artery Tibial |
MTHFR |
PPARA |
| |
HGF |
|
| |
LEP |
|
| |
LPL |
|
| |
TNFRSF11B |
|
| |
MOK |
|
| |
CETP |
|
| |
MIF |
|
| Aorta |
TCF7L2 |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).