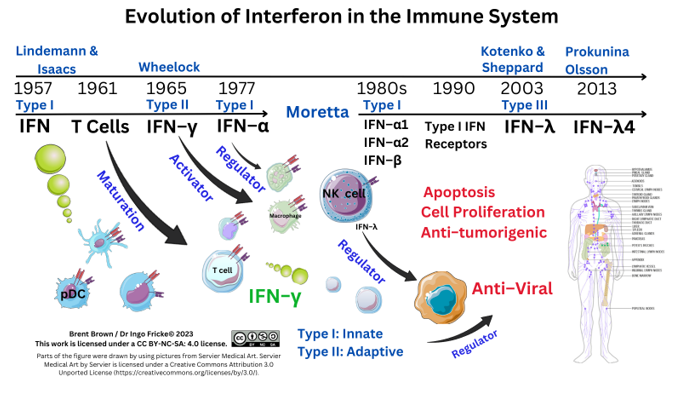
1. Introduction
Interferons (IFNs) are secreted glycoproteins with historically unique anti–viral activity as well as cellular oncological regulatory properties induced by the regulation, maturation, development or chemotaxis of immune cells, e.g., dendritic cells (DCs), in the early phase of infection. Different IFN types can stimulate the innate/adaptive compartments of the immune system to produce IFN influenced by other pleiotropic proteins released by immune cells, including cytokines (IL) and chemokines (CC, CXC), which act as specific cellular autocrine/paracrine signals in a hormonal manner [
1].
The nomenclature of interferons was historically derived as alpha (α, from leukocytes), beta (β, from fibroblasts), and gamma (γ, from mitogen–activated lymphocytes) stimulated to proliferate. After the initial discovery of the first IFN protein in 1957, three main types of IFN are known today: Type I (α/ β), Type II (γ), with the discovery of Type III (λ) in 2003, and each having distinct anti–proliferative and anti–viral activities through additional subtypes. At least three IFN types have distinct cellular functions and are expressed when differentially expressed genes (DEGs) are transcribed and translated by IFN regulatory factors (IRFs) and other proteins. This occurs in health and disease and is regulated by IFN–stimulated genes (ISGs), IFN–inducible proteins (IFI), together with IFI transmembrane proteins (IFITM), as well as cytoplasmic interferon regulatory factors (IRF).
Immunisation and therapeutic treatment have historically targeted IFN for therapeutic benefit in preclinical development, from phase 1/ 2 to phase 3 and beyond, according to the overall safety profile, and success rates by regulatory and monitoring agencies such as the United States Food and Drug Administration Agency (FDA), the European Centre for Disease Control (ECDC), and other organisations like the European Medicines Agency (EMA) (see
Supplementary Materials). However, the literature on type I/ II /III IFN is needed to compare overall mechanisms so far.
Either natural IFN or recombinant IFN compounds including human type I IFN and/or type II/III IFN concentrations within host cell populations can stimulate both innate/adaptive immune system branches honing an effective response during disease through cellular production from 2 cell phenotypes. One includes T cell synthesis, with the other natural killer (NK) cell synthesis of type II IFN also produced by other antigen–presenting cells (APCs), like monocytes and macrophages (M1ϕ/M2ϕ). The immune system also senses pathogenic antigens through pattern recognition receptors (PRRs), as well as cellular endosomal expressed Toll–like (TLR) receptors. Cancer pathologies can respond to type II IFN cell synthesis, whilst viral evolution may affect type I/II/III IFN homeostatic immune cell function. This aspect during viral epidemics/pandemics is considered, evidenced with Dengue Fever virus (DENV), Ebola virus (EBOV), and recently Monkeypox virus (MPXV) [
2,
3]. It is plausible that regulation of IFN is modulated and affects early therapeutic and/or clinical disease onset–delaying effects during viral evoked diseases like Influenza A virus (IAV), Measles virus (MeV), as well as Human Immunodeficiency virus (HIV); however, this can be affected by other bacterial infections such as lower respiratory tract bacterial infections caused by
Haemophilus influenzae,
Streptococcus pneumoniae and
Staphylococcus aureus, as well as oncological diseases, like hepatic melanoma [
4]. Other reviews ascertain regulatory IFN proteins affected by viral proteins (VP), synthesised by Coronaviridae (e.g., SARS–CoV–2) as well as Flaviviridae (e.g., DENV, Yellow Fever) [
5]. Individual VP mutations affect other cytosolic PRRs proteins (e.g., retinoic acid–inducible gene I, RIG–I/ mitochondrial anti–viral signalling protein (MAVS) pathways in at least two other virus families (
Filoviridae/
Nairovidiae) [
2,
3,
5]. Viral mutations occur in both DNA/RNA viruses, like the positive–sense single–stranded RNA virus (+ssRNA) Influenza A (Alphainfluenzavirus). This has 198 quantified potential antigen subtype combinations of the viral antigen expressed haemagglutinin/neuraminidase (HA/NA) proteins, affecting immune cell phenotypes. Therefore, viral antigens co–exist circulating in nature, with increases in antigen circulation necessitating clarification of IFN regulatory factors within host cells.
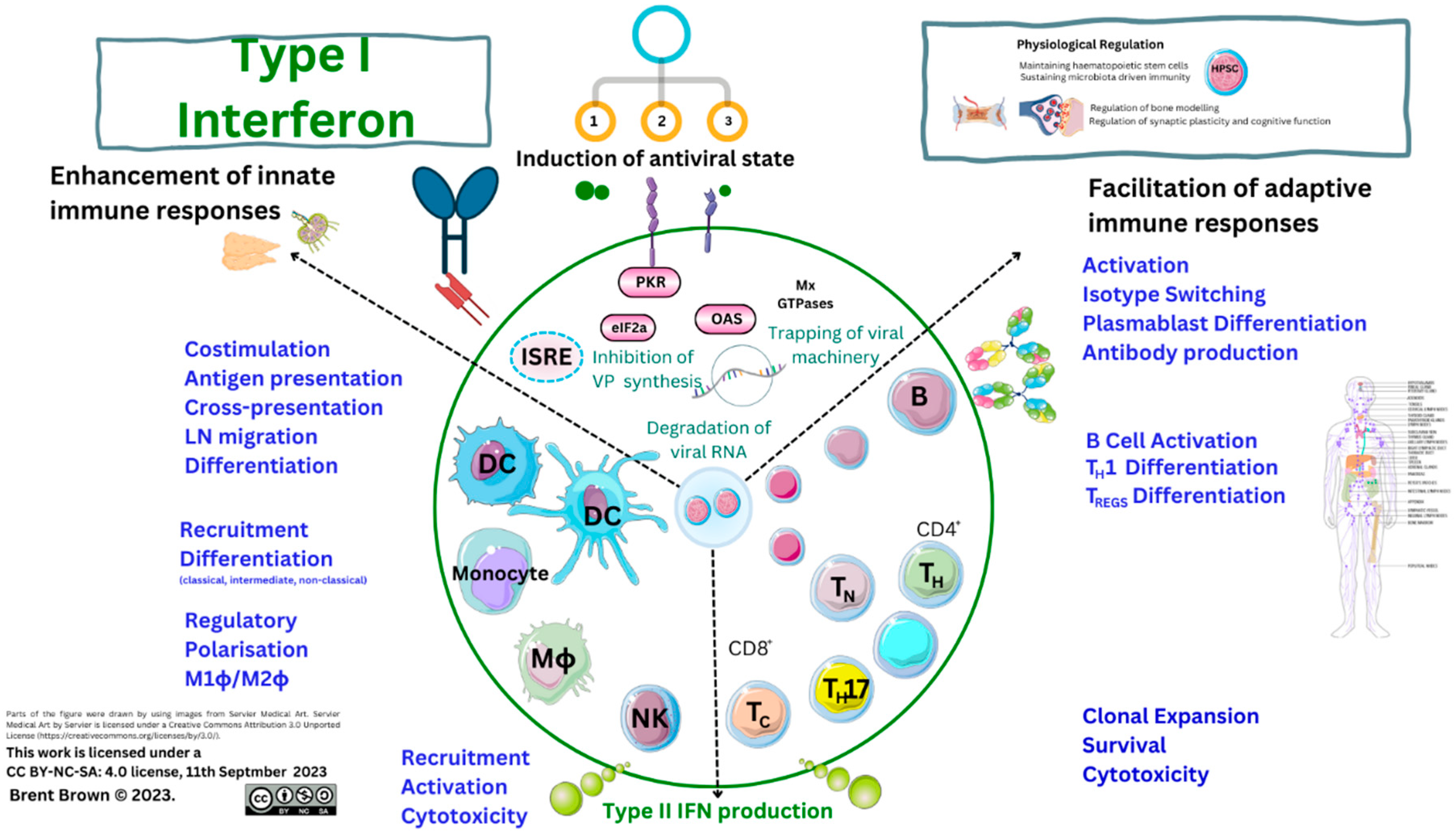
Three or more types of IFN have differential inhibitory or stimulatory mechanisms on the immune system causal in lysing infectious viruses effectively through stimulating effector cell activity through a myriad of proteins. This is effected through IFN receptors (IFNR) at the cell plasma membrane (PM) surface interacting with at least 18 types of IFN binding to 6 IFNR proteins expressed by dendritic cells (DCs), and others, having variable phenotypes. Interferon receptors are also expressed by B lymphocytes, monocytes, and M1ϕ/M2ϕ, as well as T lymphocytes. Receptor expression is also within the cellular PM on glial cells, neurons, and others. Interferon receptors (IFNR), therefore initiate downstream/upstream cellular effects, as well as T cell secretion of type II IFN–γ, upon host cellular viral infection. Plant products also generate IFN–stimulating proteins. Timing of cellular IFN synthesis and cellular secretion affects viral infection, propagation, and replication; but also, IFN acts differentially through other protein pathways to effect pathogen cellular lysis in organs, tissues and cell systems by regulating other cell cycle proteins, like p38. Immunodeficiency disorders or individual protein mutations may also cause errors in IFN/IFNR signalling throughout development.
Therefore, regulation of type I/II/III IFN responses can have resultant detrimental and/or beneficial immune system effects during pathology. The subtypes of IFN directly affect and influence the two branches of the innate and adaptive immune response requiring clarity. Each IFN type fulfils unique immunological roles during 5 types of pathology including viral, fungal, bacterial, mycobacterial as well as oncogenic diseases. Immune system modulation and/or evasion may represent evolutionary development within animal hosts varying. Therefore, here is the analysis of genetic, molecular, and cellular analysis of type I/II/III IFN mechanisms of action to date, in sections 3-6, that will require further research.
2. Methods
Currently, indications are that more than 100,000 PubMed results show prior IFN research. National clinical trials (NCTs) investigated the utility of IFN as a potential therapeutic divided into other types that include type I IFN–α (380), type I IFN–β (116), type I IFN–omega (6) and type I (epsilon (1) (5 type II IFN–γ (173)), and type III IFN–λ (17) currently (see
Supplementary Materials).
3. Interferon Types
3.1. Overview to Interferon Cellular Types
Type I IFN proteins are synthesised/secreted by translation from cellular nuclear transcription factors (TFs) resulting in differential anti–viral activity against host pathogens that may vary. Each IFN protein is known as a small molecular weight (MW) molecule in humans; for example, type I IFN–α1/13, IFN–α2, IFN–α8 and IFN–α21 are composed of 187–189 amino–acids, while type III IFN–λ is within the MW range 179–200 amino acids. Chemokines in comparison are smaller MW proteins (e.g., CCL2, 99 amino–acids), with pleiotropic effects directing immune cell migration throughout tissues. Small MW proteins are induced through gene synthesis transcription and subtypes can be differentially modified earlier in response to pathogenic antigens both inside and outside the cell. Interferon subtypes can be synthesised by myeloid cells like plasmacytoid dendritic cells (pDCs) producing higher concentrations of type I IFN (IFN–α/IFN–β), effecting anti–viral responses in hosts; but also, within skin epithelial cell tissues through tumour necrosis factor (TNF) related apoptosis–inducing ligand (TRAIL), and at least 10 intra/extracellular PM and vesicular TLRs [
6]. On the other hand, type II IFN–γ is secreted by at least two effector cells (NK/T cells) together with two antigen presenting cells (DCs and Mϕ), each with different phenotypes characterised by cluster of differentiation (CD) marker. Type III IFN subtypes also influence host immune responses within epithelial layers. It is considered through regulating cellular cycle function that each IFN performs roles, with type I IFN–β potentially regulating Mϕ cell cycle (M1ϕ/M2ϕ), and metabolism; while type I IFN–α could be considered similar in the regulation of homeostatic function and observed commonly in health as well as inflammation and AI disorders.
Type I IFNs include IFN–β, IFN–δ, IFN–ε, IFN–κ, IFN–τ, IFN–ω, and IFN–ζ amongst others; whereas type III IFN is composed of IFN–λ (IFN–λ1, IFN–λ2, IFN–λ3, IFN–λ4), known originally as IL29, IL28A, and IL28B with IFN–λ4 discovered in 2014 [
7]. Two types of type III IFN (λ2, λ3), are considered to have 96% amino–acid homology [
8]. Other classifications of subtype exist and most vary between host animal species encoded by IFN genes. To clarify, human IFN consists of at least 18 subtypes, some others of which are type I IFN–α4, IFN–α7, and IFN–α14; whilst in pigs and bats diversity of IFN–ω is worthy of consideration, with less type I IFN–α described as discussed further [
9,
10,
11]. Amongst type I IFN–α subtypes, a recombinant IFN–α2b therapeutic version in humans is utilised as below [
12,
13]. However, research studies in 2015 indicate that IFN–α2 is non–glycosylated missing one aspartic acid (D) amino–acid at position 44 in humans without functional change [
14]. Furthermore, it is indicated two recombinant type I IFN–α2α / IFN–α2β preparations contain a neutral lysine (L) and alanine (K) substitution at position 23 when observations were that the type I IFN–α2 is conserved and less prone to mutations [
15].
Recently, it was shown that type I IFN may contain pro–inflammatory glycans unknown affecting predominant antibody (IgG) binding to immune cell FcγR PM receptors (CD16/CD32/CD64), all of which influence an effective innate system response [
15,
16]. As a result, this can influence 3 functional branches of the adaptive T cell response (helper (T
H), cytotoxic (T
C), and NK cells). Modulation of sialic acid residues present in other receptors like the specific intercellular adhesion molecule–3 grabbing non–integrin (CD209 or DC–SIGN) or fucose residues may also occur unknown to date. Therefore, the overall homeostatic properties of type I IFN can be considered further.
Before and after 2019, pharmacokinetic properties of recombinant type I IFN–α2 engineering indicated the synthetic IFN production vector could affect pharmacokinetic half–life when glycoengineering indicated
Pichia pastoris as an option, together with the purification method of recombinant IFN, whereas all subtypes of type I IFN–β are N–glycosylated [
17,
18,
19]. In comparison, other studies show the addition of a glycosyl group on IFN–λ4 may increase anti–inflammatory actions and anti–viral efficacy [
20]. It is notable that glycosylated IFNs vary in stability and display antimicrobial effects with research comparatively unknown [
21]. Glycosylated IFN also bind to carbohydrates/ and PM receptors with higher/lower binding affinities to receptors. Respective IFN receptors include type I IFN receptors (IFNAR1/2), type II IFN receptors (IFN–γR1/IFN–γR2), as well as type III IFN receptors (IFN–λR1/IL10R2), each composed of two subunit domains [
22,
23]. Below is a depiction of 2 type I IFNs (See
Figure 1).
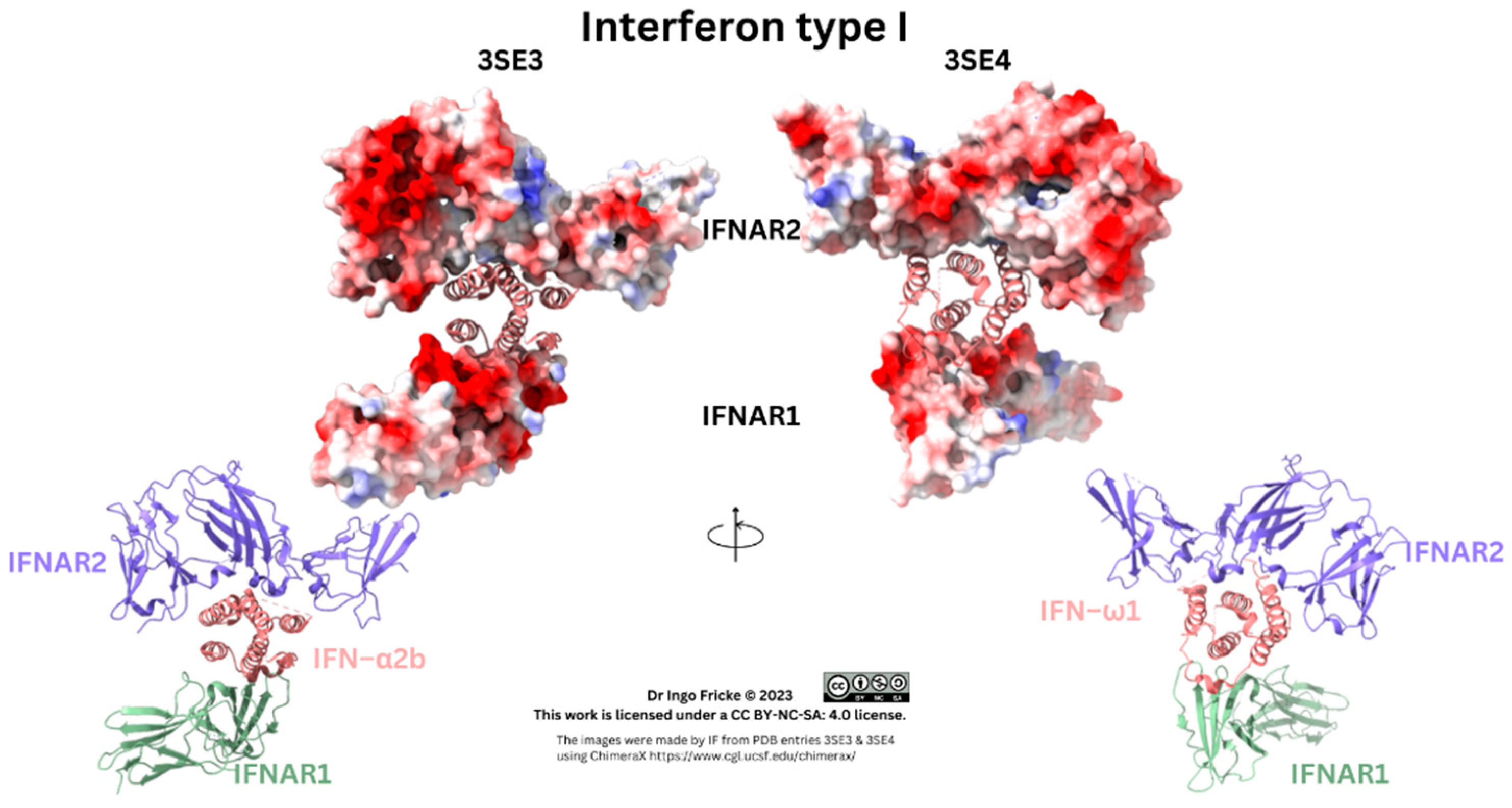
Figure 1.
Type I Interferon Receptor/Ligand Binding. Pictures were made using existing PDB files, namely 3SE3 and 3SE4, depicted as ribbon, and ribbon with surfaces of the receptor, with its electrostatic potential.
Figure 1.
Type I Interferon Receptor/Ligand Binding. Pictures were made using existing PDB files, namely 3SE3 and 3SE4, depicted as ribbon, and ribbon with surfaces of the receptor, with its electrostatic potential.
As above, the receptor/IFN complex was clarified in 2011, and comparisons of type I IFN–α assays allowed type I IFN–α receptor binding studies to show IFN binding to IFNAR1 occurred with higher (µm) affinity, whilst binding to IFNAR2 was lower affinity in a smaller (nm) range [
24]. However, IFN–β and IFN–λ are produced by various cells, with IFN–α generally synthesised by immune cells, but specifically pDCs during infection with receptors throughout the bodily system [
25]. Other reviews establish type I IFN downregulation, while research into type III IFNs is in the early stages; however, other authors suggest type III IFNs may have further biological mechanisms [
26,
27]. Much remains unknown with regard to type III IFN–λ signalling proteins. Specific data on IFN therapy comes through national clinical trials (NCTs) conducted throughout history before/after the first cloning of IFN receptors in 1990 and types of recombinant IFN–α2 (see Supplementary Data S1) [
28]. During the recent pandemic, type I IFNs were evidenced to have an effect in reducing SARS–CoV–2 viral genome load requiring further detail [
29,
30,
31,
32,
33].
3.2. Mechanisms of Action of the Three Types of Interferon in the Immune System
Cellular effects of IFN regulating immune system cells vary with affinity of 3 types of IFN and subtypes through 6 subunit receptor domains differentially expressed in organs, systems, tissues and cells. In brief, type I IFN–α research to date indicates unusual variance during host infections with evidential beneficial/detrimental effects. However, IFN regulates the differentiation and maturation of myeloid cell lineages as well as B/T cells, NK cells and others by being metabolised and secreted from cells. This training of immune responses occurs through inhibition as well as DC stimulation of cell maturation/differentiation by regulating costimulatory molecules like CD80/CD86 increasing major histocompatibility complex (MHC) antigen presentation, as well as stimulation of T cell phenotypes expressing adhesion molecules (e.g., CD62) [
34]. Also, DC tolerogenic and maturation phenotypes are known to occur through pDCs into three conventional types of DCs (cDC1, cDC2, cDC3) [
35]. These cells reversibly differentiate into myeloid/monocytic lineages during inflammatory processes during endothelial cell injury or cancer [
36].
It is suggested the dual role of type I IFN is of an inhibitory cytokine (IL–10) in monocytes whilst stimulating a T cell response [
37]. This is mediated through the suppressor of cytokine signalling–1 (SOCS–1) protein acting independently regulating the expression of IFNAR2 expression as well as IFN transduction through conserved phosphotyrosine residue on the tyrosine kinase (TYK) enzymes in effect regulating anti–viral/anti–proliferative effects and type III IFN [
38,
39,
40]. This was evidenced from 2002 in more than 40 countries where recombinant type I IFN–α2 was used as a therapeutic to treat various types of leukaemia (B/T cell lymphomas) [
41,
42]. In comparison, type II IFN–γ is largely produced by only cells of the immune system primarily induced by APCs which phagocytose pathogens produced from adaptive immune cells NK and T
C cells that utilise MHC class II proteins to effect cytolysis. Two primary T cell phenotypes produce type II IFN–γ with the majority expressing CD4 and/or CD8 molecule proteins [
43].
Historically, type II IFN is a measure of T cell activity of adaptive immunity. The activity of type III IFN expression is also measured by expression of subunit receptors in tissues/cells through cellular mRNA expression to indicate gene transcript location. However, it is currently considered that the RNA for another type III IFN,
IFNLR2 (
IL10R2), is present in lungs, intestines, and liver tissues as well as B cells, neutrophils, Mϕs and pDCs, but not in NK cells [
44]. Additionally, type III IFN is considered to have a higher affinity for one subunit (IFN–λR1), with less affinity for the other subunit (IL10R2) possibly explaining some of the differential activity of IL–10 which shares this receptor.
In the past, type III IFN was considered to be predominant on non–haematopoietic cells (e.g., intestinal epithelial cells). Type III IFN has lower affinity binding affinity to its respective receptors compared to type I IFN [
45]. Other reviews examine the relevance of single nucleotide point (SNP) mutations of type III IFN pathways during disease [
44]. The relevance of type III IFN is becoming clearer. Research
in vivo indicates that during type III IFN–λ2 (IL28A) deficiency, there is an effect on three crucial immune system branches. Namely, germinal B cell centre formation, where B cells develop that secrete antibodies (Abs) of 4 main types eventually (IgM, IgG, IgA, IgE). Therefore, as IFN can affect the innate branch of the immune system, this can affect the other adaptive branches where increased activity through two other T cell branches denoted by helper T cells (T
H/CD4
+) as well as cytotoxic T cells (T
C/CD8
+). Moreover, type III IFN–λ3 is similarly highlighted as relevant to B cell proliferation and antibody production [
44,
46].
Immune system modulation and/or evasion may represent evolutionary development within animal host immune systems and vary. Recently, two three types of cellular signalling are considered alongside IFN that are cytokines (interleukins, IL) and chemokines (CC/CXC). Individual cellular expression is stimulated by many pathogenic organisms, like Smallpox (VARV), Human Immunodeficiency virus (HIV), but also bacterial pathogens (Streptococci), and others like Respiratory Syncytial virus (RSV) that also cause viral–induced pathology., Viral mutations occur in DNA/RNA viruses like the positive–sense single–stranded RNA virus (+ssRNA) Influenza A (Alphainfluenzavirus), having 198 potential subtype combinations of haemagglutinin/neuraminidase (HA/NA) protein antigens that can differentially affect immune cell phenotypes. Different serotypes of Gram–negative (–ve) (e.g., Coccobacilli, Haemophilus influenzae, HI) are known to shed intracellular/extracellular protein membranes during infection denoted as 3 types (A, F or non–capsulated (ncHI) (see Supplementary materials). Other viruses like Influenza A avian virus (H5N1) can affect variable animal hosts.
An effective increase in pathogen antigen circulation may inhibit or stimulate/sensitise the immune system affecting the lysis of infectious viruses through regulatory host IFN synthesis or unknown metabolic factors. The three shared methods of immune system kinetics comprise of firstly pathogenic DNA/RNA 5’ capping, through the incorporation of methyl (CH3–) group to the 5’ genome with pattern recognition receptors (PRR) including TLRs affected. Secondly, cellular mitochondrial metabolic changes affect the synthesis rate of reactive oxygen species (ROS), whilst pathogens also utilise inter–cellular channelling nanotubes or porous membranes [
47]. The third objective considered could be unknown modulation of type I/III IFN or indeed the rate of IFN synthesis subtypes by immune cells or infected cells which is a historically well researched therapeutic that has initiated remission during oncological disorders.
3.3. Cellular Signalling Mechanisms of Interferons
Interferon cellular action occurs through transmembrane protein receptors, as above, utilising predominantly janus kinase (JAK), and STAT protein phosphorylation activation pathways. However, seven STAT proteins (STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, STAT6) are described in mammals as central to immune cell regulation with STAT1/STAT2 pertinent to IFN signalling [
48,
49]. The IFN–λR1/IL–10RB receptors for IFN–λ are notably shared with IL–22 implicated in disease [
50]. Less is known about type III IFN. Data (
www.proteinatlas.org) implies the IFN–λ receptor (IFN–λR1) is preferentially expressed by both pDCs and B cells, with IFNAR1 by both neutrophils and three phenotypes of monocytes (classical, intermediate and non–classical), whilst IFNAR2 evenly distributed on all immune cells.
During the 1990s, STAT proteins were found to bind to JAK proteins. Various laboratories were known when four scientists including James Darnell, George Stark, as well as Ian Kerr discovered the molecular basis. These were classified into four types (JAK1, JAK2, JAK3) when Muller discovered that the tyrosine kinase (TYK2) enzymes bridged the gap between JAK/STAT proteins to be essential in type I IFN signalling [
49]. Thereafter, two pathways are described including initially “canonical” or high–affinity binding of type I IFNs to the IFNAR2 to form a trimer with IFNAR1 [
49]. The second pathway described is “non canonical”, referring to three independent kinase enzyme pathways, that include activation of MAP kinase (MAPK), mammalian target of rapamycin (mTOR), but also phosphatidylinositol 3–kinases PI3K, a serine/threonine kinase [
49]. In the canonical model, activation and phosphorylation of JAK1/TYK2 occurs by phosphorylation to form a STAT1/STAT2 trimer with other interferon regulator factors (e.g., IRF1/3/7/9), producing interferon stimulating growth factors (e.g., ISGF3) translocating to nuclear interferon sensitive response elements (ISRE) effecting IFN synthesis [
51]. However, the original “non–canonical” pathway is considered to be where STAT1 or other proteins, like MAPK or PI3K, homodimerise. Other reviews summarise STAT proteins as containing a conserved DNA binding domain –SH2 recognising phosphotyrosine motifs of cytokine receptors [
49]. Below is shown the IFN signalling pathways (see
Figure 2).
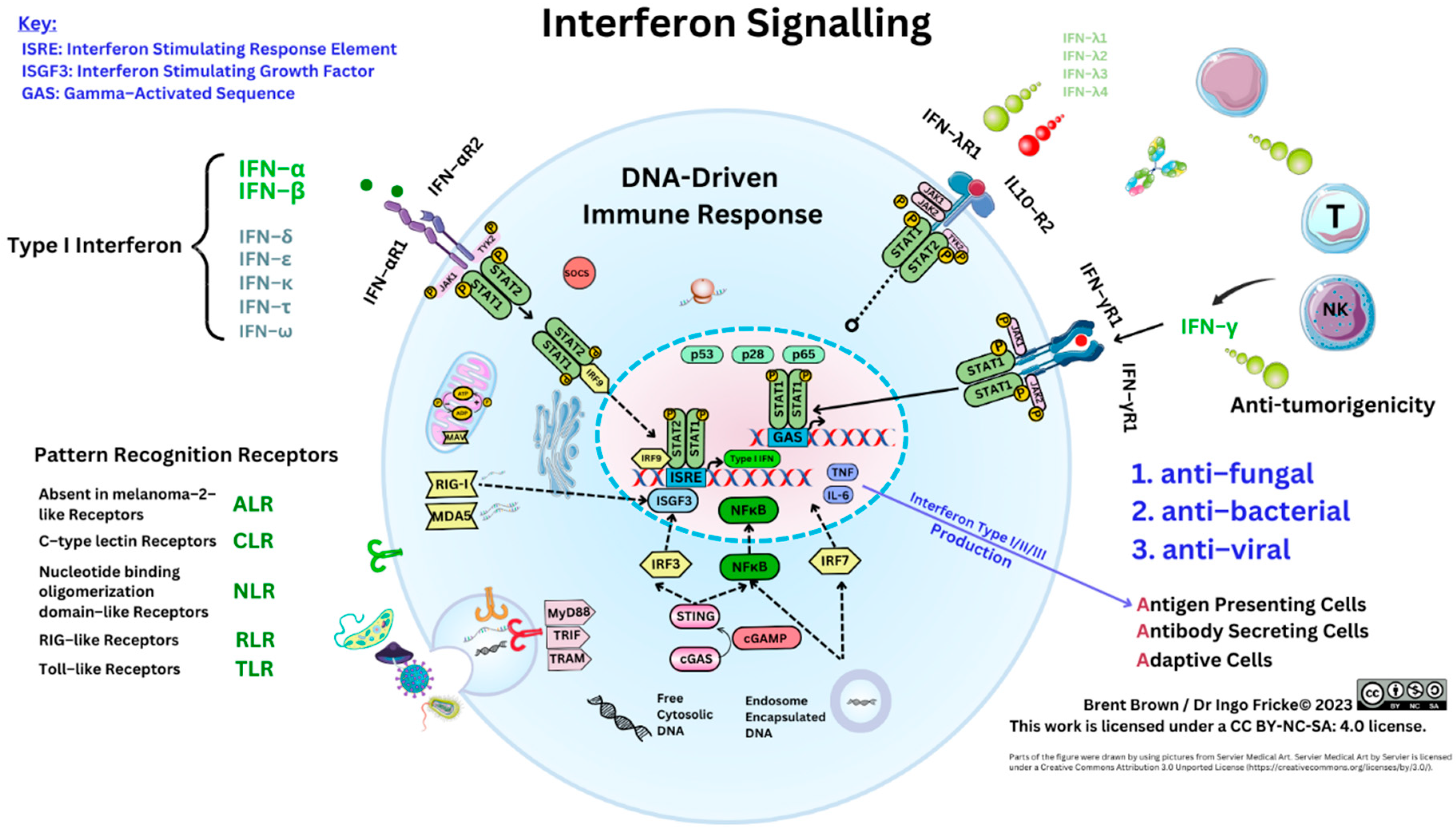
Figure 2.
Systemic Interferon Signalling.
Figure 2.
Systemic Interferon Signalling.
The activity of type I IFN occurs via activation of the nitric oxide synthase (NOS), and inactivation of an enzyme protein kinase R (PKR) that can be regulated by cellular viral DNA/RNA. Further activation of the enzyme oligoadenylate synthetases (OAS) along with peptide presentation by class I/II (MHC–I/II) occurs. Albeit, VP protein fragments are metabolised with short amino–acid peptide chains presented known as epitopes to immune cell receptors (e.g., T cell phenotypes CD4/CD8) [
52]. There are four members of OAS enzymes, of which three (OAS1/2/3) produce 2′–5′–linked oligoadenylates and a similar OAS ligand (OASL) binding to RNAase L regulating the degradation of viral or cellular RNA. Activation of adenosine deaminase 1 (ADAR1), a dsRNA binding protein, is known to catalyse the process of deamination of adenosine usually involved in viral RNA replication, but also maturation and development of leukocytes to effect apoptosis of infected [
53,
54]. Protein kinase R downregulates translation of viral RNA encoding pathogenic protein domains; whereas OAS activation can degrade and lyse RNA with ADAR1 enabling RNA editing Viral NSPs may activate the phosphatidylinositol 3–kinase (PI3K) pathway inhibiting type I IFN synthesis, as well as activating cellular stress–response proteins (e.g., heat–shock proteins) involved in cell proliferation regulation, survival, and differentiation as well as immune cell regulation. Therefore, temporal initial inhibition of regulatory apoptotic pathways can occur while a pathogen replicates, prior to induction of innate immune system host cells [
55].
In brief, STAT1 proteins are regulators of cell cyclin–dependent kinase inhibitors (CDKI), P21 and P27, but also caspases (1/3/11) that sense and are activated during cellular apoptosis [
49]. However, STAT1 is also involved in antigen presentation as well as B cell development through regulating Fas (CD95), and Bcl–2 affecting granulocyte development [
49]. In an immunological context, STAT1 is described to be activated by three cytokines (IL–2, IL–6, TNF), but also interferons. In contrast, STAT2 does not homo–polymerise but can be activated by type I IFN. Thirdly, STAT3 is described to be activated by the IL–6 and IL–10 family of cytokines regulated by CD95 acting as a molecular switch controlling immune cell differentiation, growth and apoptosis as observed in certain cancer types. Transcription of STAT3 is seen to occur constitutively in certain cancer types, like head and neck cancer, as well as haematological tumours amongst others [
49,
56,
57]. However, STAT3 inhibition has been described to affect cytokine receptor, IL–4Rα, expression by naïve CD4 T cells expressing the migration adhesion molecule, CD62L, required to transverse endothelial cell membrane layers [
57]. During gene knockout experiments of CD95, it is evidenced that overexpression of STAT1 inhibits STAT3 transcription of the
IL17a promoter gene transcript necessary facilitating synthesis of IL–17 from T
H17 cells largely unknown to date [
58]. Conversely,
in vivo, the role of STAT3 is intertwined with STAT5 where overexpression of STAT5 is suggested with the cytokine GM–CSF to activate the differentiation of both neutrophils while inhibiting myeloid lineages (monocyte/Mϕs) [
56]. Overall, viral antagonism is affected by extraneous factors, but also cellular PM as well as vesicular TLRs that could plausibly also have mutations leading to a sensitised and/or delayed immune system response dependent on the homeostatic function of IFN proteins. For example, in this review (n=5/1288) individuals were indicated to have autosomal recessive (AR) disorder. These may result from deficiencies in the genes involved in IFN regulation (
OAS1/OAS2/RNASEL), with type II IFN
in vitro able to upregulate expression of OAS1/2/3 in the myeloid cell lineage required to synthesise IFN through nuclear transcription of IFN and viral antigen presentation [
59,
60]. This one project further clarifies, in a subset of multi–inflammatory syndrome associated disorders in children (MIS–C), without COVID–19 pneumonia, but with antibodies to SARS–CoV–2 that mononuclear phagocyte function could rely on IFN signalling during pathological disorders [
59].
The last key protein to be considered, ISG15, derived through the translation of the IFN–stimulated gene 15
(ISG15), is an intracellular/extracellular protein described as ”ubiquitin–like” [
61]. The protein, ISG15 has only been found in vertebrates and is induced by a range of cellular–associated injury or infection (bacterial/viral) factors and can initiate cytokine release (e.g., IL–1β, retinoic acid), during hypoxia or DNA damage induced by each of the type I/II/III IFNs [
61]. The function of ISG15 in relation to immune cells was described some years ago to direct 3 cellular factors. Firstly, it can increase stimulation of monocyte cytotoxicity, secondly stimulate type II IFN synthesis, thirdly, induce NK cell maturation and finally DC maturation. This variability in function remains largely unknown as to the mechanism of ISG15 exocytosis; although it is also considered that ISG15 is localised in neutrophil vesicle exosomes in TLR3–activated endothelial cells during apoptosis [
61]. Other authors ascertain that ISG15 can bind to the leukocyte function antigen (LFA–1,) and can induce IL–10 known to affect both NK and T cell differentiation, but also is induced by type I IFNs. The gene
ISG15 has two IFN–stimulated response elements (ISREs) in its promoter area that bind to IRF3/9 ISRE. Of these, IRF9 interacts with STAT1/2 to form the ISGF3 complex that induces ISG nuclear transcription, although IRF3 also complexes, however, the other IRFs (e.g., IRF4) can also induce
ISG15 translation [
61,
62].
3.4. Type I Interferons in Infections
Therefore, to begin it is necessary to examine type I IFN subtypes synthesised by human cells. Interferon regulation affects all bodily tissue system immune responses. Research prior to 2009 examined chronic hepatitis C (HCV) infection to find all type I IFN subtypes inhibited viral replication, but 3 subtypes harboured more activity (IFN–α17, IFN–α7, IFN–α8) [
63]. Whilst 2 years later, during human metapneumovirus infection (hMPV), similarly, four subtypes of type I IFN (IFN–α5, IFN–α6, α8, and α10) appeared to exhibit high anti–viral potency [
64]. In comparison, after 2012, investigations into Mumps viral (MuV) infection evidenced that all 12 subtypes of human type I IFN–α could be synthesised. It was then postulated that viral mutations affect IFN affinity for IFNAR1. Increased synthesis of type I IFN (α5, α8, α17, α21), in comparison to less induction of other type I IFNs (α2, α4, α6, α7, α16) was observed [
65]. Genetic MuV mutations were indicated with IFN–α10 and IFN–α14 appearing to be host synthesised in response to different MuV strains [
65].
In 2020, type I IFN synthesis variability was also observed
in vitro with Influenza infection of human respiratory epithelial cells, compared to
in vivo, to find induction of type I IFN (α1, α6, α14, α16), whilst other type I IFN subtypes (α5, α8, α21) were pertinent to lesser virulent strains [
66]. Non–structural proteins (NSP) are produced (e.g., Zika virus (ZIKV)) and packaged in vesicles within the endosomal/exosomal cellular pathway after translation in host cells that may affect either host type I//IIIII IFN gene transcription [
67]. This molecular event seems to occur with some of the SARS–CoV–2 proteins encoded where viral particle replication rate together with IFN synthesis rate is therefore a regulatory checkpoint.
Whereas, in comparison other viruses like Monkeypox virus (MPXV), HIV–1/HIV–2, as well as Henipaviridae (NiV), can affect host cell nuclear activity antagonising synthesis and exocytosis of type I and possibly type III IFNs unknown [
3,
68]. As exemplified, by the translation of viral proteins, IFN–encoding mRNA may be cleaved or IFN gene transcription altered [
26,
68]. During
Filoviridae (EBOV/Marburg virus) infection, comparisons were made between the function of VP24/VP35, which appeared to affect the rate of IFN synthesis in specific cell types more than others [
3,
69,
70]. Whereas the EBOV VP35 protein did not suppress IFN production in pDCs, sensitised type I IFN–mediated immune responses could attenuate EBOV virulence [
71,
72]. Investigators induced a loss–of–function (LOF) mutation in the EBOV gene encoding VP35 to observe EBOV antigens with decreased virulence [
73]. However, during
Flaviviridae infection (ZIKV), type I IFN–ε expression within both mucosal and glandular epithelial cells is suggested to be protective [
74]. Research involving type I IFN–β seems to involve mycobacterial research on leprosy implying that this dsDNA mycobacteria species does differentially activate cyclic GMP–AMP synthase (cGAS), but can antagonise the OASL ligand required for IFN signalling [
75]. Moreover, within retroviral (HIV–1) infection, suppression of type I IFN synthesis could occur and cellular transmission by producing a viral infectivity factor (Vif). This occurs by triggering the cytoplasmic stimulator of interferon gene (STING), by interacting with cellular tyrosine (Tyr/Y) phosphatase enzymes known as Src homology region 2 domain–containing phosphatase–1 (SHP–1) within STAT pathways regulating various IRFs [
49,
51,
76]. This leads to STING dephosphorylation at the Y162 amino–acid position [
77]. Another study highlighted the ability of HIV–1 to evade TLR8 detection via translation and signalling of the snapin VP to neighbouring cells [
78]. Therefore, it is necessary to clarify how IFN regulation may be compared.
More recent developments seem to indicate type I IFN subtypes (α6, α8, α14) were pertinent to regulation of HIV infection
in vitro/
in vivo [
12] . Although, type I IFN (α5) could potently inhibit Influenza (H3N2) in epithelial layers [
79]. However, in 2023, the role of type I IFN affecting STAT2 proteins was defined that could affect the adaptive branch of the immune system through effector memory (T
EM) T cells alongside classical monocytes through defective IFN signalling as well as potentially IFNAR2 [
79,
80,
81,
82]. Conversely, the other type I IFN–β utilised in therapeutics, through differentially expressed genes (DEG) changes examined TNF–α
in vitro stimulation of monocytes and T cells, during 2013, to find metabolic programming could occur. Clarification came that type I IFN–β could modify 2 immune cell checkpoint proteins, CD38/CD83, through upregulation on monocytes at 2 days, but not T cells confirming that type I IFN–β can modify the STAT3 signalling pathway [
83].
3.4. Type II Interferon and Immunological Disorders
Interferon cellular signalling and synthesis can be influenced by many factors that are genetic mutations, and cellular transcription/translation of TFs, affecting the resultant immune cell secretion as well as naturally produced auto–antibodies (aAbs). Changes can exhibit pathological consequences in individuals during either an ineffective immune response (e.g., immunodeficiency) or an overactive immune response. For instance, Mendelian susceptibility to mycobacterial disease (MSMD) was first reported in 1996 to be an inherited human IFN–γR1 and IFN–γR2 mutational deficiency; therefore, causal with resultant effect on type II IFN receptor signalling with reduced synthesis observed and less effective immune responses. Type II IFN is a crucial NK cell and T cell cytokine required to be produced naturally. Mutations in genes affecting type II IFN–γ signalling through IFN–γR1/IFN–γR2 were reported in two cases in 2020 [
84]. Mutations in this trimer interface of type IFN–γ/IFNγ–R1/IFN–γR2 can be deficient abrogating downstream nuclear signals. It was shown that this synthesis by both NK/T cells in some cases may be independent of circulating viral antigens [
84]. Further overviews appeared around 2000 of two individuals elucidating that type II IFN production through MSMD research can be affected by a number of other point mutations in many genes (IFNGR1, IFNGR2, IRF8, IL–12RB, IL–12RB1, STAT1), each within the IFN signalling pathway [
84]. Type II IFN–γ production therefore does influence outcome during mycobacterial infection or repeated other infection.
However, during 2012, STAT1 LOF was observed within the IFN pathway indicative that MSMD could occur as 4 inherited phenotypes [
85]. Research shows that increased host susceptibility occurs to viral, bacterial, and mycobacteria infections with the resultant immune responses affected [
85]. Furthermore, two cases in 2012 evidenced that granulocytes could display reduced production of interferon signature gene protein (ISG15) resulting in less type II IFN–γ lymphocyte responses with recurrent mycobacterial illness [
86]. It was suggested and hypothesised then that ISG15 alongside free ISG15 protein could have cytokine–like properties and be synthesised by B cells as well as monocytes in the sera of healthy individuals. More recently, in 2021, a categorization was proposed for the other type I interferonopathies where other inherited diseases can cause auto–inflammation due to dysregulation in this crucial IFN pathway [
87]. With regards to type II IFN, genetic variations can affect proteins encoding human leukocyte antigens (HLA) producing other proteins processing antigens, like MHC I/II surface receptors, which vary between populations. As recently as 2021, anti–IFN–γ autoantibodies (AIGA) were suggested to be affected by HLA antigens in some diagnosed cases (n=600) that could explain differential immune responses to infections like mycobacteria. Specifically, it was suggested that the following alleles (DRB1*16:02–DQB1*05:02 and HLA–DRB1*15:02–DQB1*05:01) encoding MHC type II peptide presenting molecules could be variable [
88,
89]. Therefore, each of the factors will be discussed further.
3.5. Errors in Interferon STAT Pathway Signalling
Initially, four errors in STAT1 signalling were defined to be genetic factors affecting protein production and immune system function. These were defined as follows: “AR complete” STAT1 deficiency, along with “autosomal dominant (AD)”, but also “partial”, along with “gain of function (GOF)” and observed in pathological reports (n=6) in children [
85].
During 2006, errors in TYK signalling emerged when one patient was observed to have recurrent viral and mycobacterial infections along with increased levels of IgE susceptible to bacterial staphylococcal infections. In 2015, other cases emerged (n=7) indicating the IFNAR1 could be downregulated, but that 2 key cytokine receptors (IL–10R2 and IL–12Rβ1) were affected, with reduced expression of the IFNLR subunit affecting both IL–12 and IL–23 receptors during mycobacterial infection [
90,
91]. Conversely, isolated reports from 2015 into chronic mucocutaneous candidiasis (CMC), evidenced mutations in STAT1 to be independent of STAT3 affecting T
H17 cell differentiation producing IL–17 [
92]. During 2020, only the second report of an individual case reported from a family with a heterozygous deficiency for the type I IFN receptor (
IFNAR2) during a similar clinical pathology to haemophagocytic lymphohistiocytosis (HLH) occurred. This indicates that type I IFN–α does affect NK degranulation and function as well as controlling inhibition of type II IFN. Interestingly, the same donor cells were used
in vitro to confirm that STAT1 phosphorylation is required for IFN signalling. This project noted that IFN signalling did not occur in monocytes by flow cytometry with type I IFN gene transcripts abrogated nearly completely (
RSAD2,
IFI44,
ISG15,
SIGLEC1,
CXCL10,
IFI27), with type II IFN regulated genes observed [
93]. Therefore, it could be considered given the scarcity of reports prior that further research would be needed. Summary reviews in 2020 detail the complexity of errors in IFN signalling occurring affecting type I/II IFN signalling through gamma activated sequences (GAS) causal in delivering an effective immune response to infections and cell cycle regulation in cancer through adaptive T cell phenotypes [
94]. The role of type II IFN cannot equally be understated as 3 types of IFN regulate and signal through STAT proteins.
More recently, reports indicate that TLR3 deficiencies in people may occur as an AR disorder found during Influenza infection [
95]. Moreover, in 2020, it was seen that the function of STAT1 in monocytes could display a dual role of abrogating or reducing type II IFN–γ and type I IFN–α function during infections. These can result in serious complications immunologically, by LOF of monocytes, with recurrent infections independent of type III IFN–λ [
96]. More recently, within the Shigella bacterial species outer surface protein C (OspC) member, OspC2 inhibition of type III IFN–λ1 synthesis was observed during infection [
97]. In 2021, one report discovered through Enterovirus infection that deficiencies in cytoplasmic TLR3 together with a RIG–I like receptor, named Melanoma differentiation–associated protein 5 (MDA5), may explain that activation of TLR3 is required for endosomal sensing type I/III IFN and MDA5 is required for cytoplasmic pattern recognition independently [
98]. These were interesting observations.
Deficiencies in many STAT proteins affect all aspects of an effective immunological response. Through single–cell RNA sequencing (scRNA) experiments, the STAT2 deficiency recently (n=23) was quantified further elucidating the relationship between IFNAR2 and STAT1/STAT2 IFN signalling. There were indications STAT2 deficiency results in loss of sensitivity to type I IFN. Gene transcripts at a single cell level showed that STAT2 deficiency affects effector memory (T
EM) cells with reduced gene transcripts (
MX1,
IRF9,
USP18 and
ISG15), observed concurrently with three others (
STAT1/
IRF1/
ICAM1), that could affect classical monocyte response IFN signalling and adhesion during viral inflammatory disorders like severe Influenza, SARS–CoV–2, Enterovirus, but also Herpes Simplex virus (HSV–1) [
80,
99]. On the other hand, STAT3 deficiencies were also initially described as hyper–immunoglobulinaemia E in 1966. The protein, STAT3, was suggested as a factor during sporadic cases in individuals (n=98) of another rare AD disorder (Job’s syndrome) characterised by dermatitis and increased serum IgE. In this instance, IL–6 stimulation resulted in less CCL2 synthesis by leukocytes was suggested [
100]. Therefore, below is shown the role of STAT3 in the immune system (see
Figure 3).
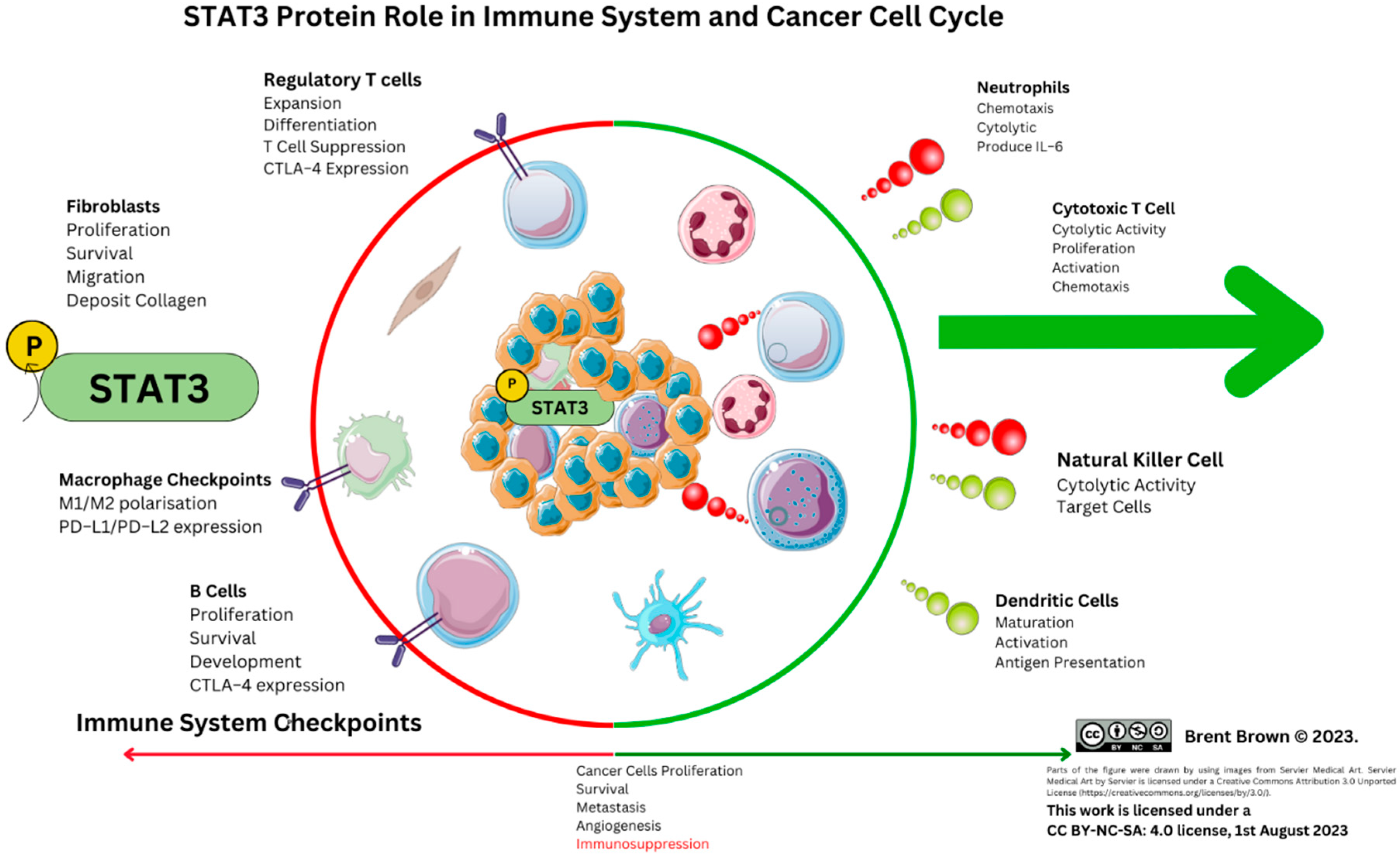
Figure 3.
STAT3 Protein role in immune system and cancer. Depicted are positive (green half of the circle) and negative (red half of the circle) effects exerted in the shown immune cells.
Figure 3.
STAT3 Protein role in immune system and cancer. Depicted are positive (green half of the circle) and negative (red half of the circle) effects exerted in the shown immune cells.
Therefore, STAT1 signalling was examined in individuals with GOF or overactive STAT1 signalling to examine the role in diagnosed CMC individuals (n=8) [
92]. Fungal and T
H17 cell immune response during recurrent infection remains unknown with clarification on DCs examined further. In 2023 DC analysis, through comparison of monocyte–derived DCs (moDCs) to tolerogenic DCs (tDCs), it is surmised that tDCs may express less immune cell checkpoint regulatory proteins (CD80, CD83, CD40); whilst moDC phenotypes expressed other inhibitory receptors like programmed death–ligand, PD–L1 [
101]. However, the dual activity action of PD–L1 with the T cell immunoglobulin (Ig) and mucin domain–containing protein (TIM3) was characterised as a receptor expressed on both T cell types (CD4
+/CD8
+) producing type II IFN with both considered checkpoint proteins remaining a target of cancer therapeutic development [
102,
103].
3.6. Other Types of Interferon Regulation Pathways
The other 3 crucial STAT proteins (STAT4/5/6) remain comparatively recently investigated. As recently as 2021, other authors concur that STAT4 has not been extensively examined and remains comparatively unknown. The considered role of STAT4 is that it is expressed constitutively by haematopoietic cells (HSPCs), including both NK/T cells and is consistent in health and disease [
104]. In 2020, STAT4 was clarified to be encoded by a further two gene transcripts (
α/β), with the STAT4α subunit able to induce cellular production of more type II IFN; whereas the STAT4β subunit could better respond to IL–12 stimulation [
105]. Other reviews ascertain that STAT4 is a pertinent protein as well as a relevant potential modulator of tumour suppression during hepatocellular carcinoma (HCC), but also correlates with serum hepatitis B antigen levels (HBsAg). An additional role in AI diseases (Sjögren’s syndrome (SS), Systemic Lupus Erythematosus (SLE), Psoriasis, Type 1 Diabetes 1 (T1D), Rheumatoid Arthritis (RA)), as well as both asthma and atherosclerosis, is suggested but discussed elsewhere [
105,
106,
107]. Of the last two STAT proteins (STAT5), conversely is essential for NK cell development and has two types of protein domains [
49]. This was shown comparatively recently utilising mouse cytomegalovirus (MCMV) infection
in vitro of human cells. Expression of STAT5 was observed as upregulated on memory NK cells rather than naïve NK cells [
108]. Furthermore, this was induced by IL–12 dependent on two cytokines (IL–2/IL–15) to produce granzyme A, with the possibility that the apoptotic PI3K pathway could be affected [
108,
109]. However, 2 subunit proteins of STAT5 possess varying functions. Overexpression of STAT5A
in vitro with type I IFN–β stimulation of CD4
+ T cells was seen to suppress PD–1 induction, in effect regulating other coinhibitory receptors [
110]. In comparison, STAT5B deficiency has been observed to produce reduced counts of T
REG cells with STAT5A unchanged [
111]. Furthermore, STAT5B deficiencies could manifest during lymphopenia together with reduced γδ T cells, as well as NK cells [
111]. Deficiency of STAT5B in individuals has been associated with other AI diseases, like idiopathic arthritis, thyroiditis, and thrombocytic purpura, with the role of T
REG cells unknown [
108,
109]. Lastly, the dimer STAT6 can be activated by phosphorylation and is considered to transduce signals from Mϕ maturation factors, (IL–4/IL–13), alongside B cell–driven maturation, and Ig subtype maturation in germinal centres [
49]. STAT6 can be activated independently by viruses, but also recruits APCs and T cells playing a part in innate immunity during allergic conditions, and immunity to helminthic parasites during TH2 cell–driven responses [
112]. The relevance of other genes translated into extracellular cytokine–like proteins induced by type I IFN, like ISG15, is of consideration. During deficiency, it could be seen that the encoded protein appears to play a role in regulating type II IFN mycobacterial immune responses and is expressed in acute arthritic conditions [
86]. The overall role of STAT proteins is shown below (see
Figure 4).
Figure 4.
Protein and Cytokine STAT Protein Interactions Summary known today.
Figure 4.
Protein and Cytokine STAT Protein Interactions Summary known today.
4. The Role of Type I/II/III Interferon Signalling Since Discovery
4.1. Timing of Interferon Synthesis and Immune Cell Pathways
The chronology of IFN signalling post–cellular infection can be affected in three stages. Initial early IFN synthesis from DCs, second a delayed response, and thirdly an absent IFN response through various cellular and nuclear factors. The first can occur with temporal viral load regulation, enhanced regulation of pro–inflammatory responses in the acute/chronic phase, as in many bacterial as well as viral diseases (e.g., EBOV/COVID–19) [
1]. The second is followed by a dysregulated DC maturation process, T cell maturation, or other cells presenting antigens during acute/chronic inflammation (DC, monocyte/Mϕ), that can be affected by IFN signalling through respective receptors and STAT proteins (e.g., STAT1/STAT3/STAT5) signalling [
49]. The third may occur through either inborn genetic errors unknown, or the production of aAbs introduced above, as well as pathogen infections (e.g., H. Pylori). The homeostatic early synthesis of type I IFN underpins many of the current research therapeutics to date affecting regulation of the immune system.
4.2. Autoantibodies, Interferon and Errors
Systemic production of aAbs, including against type I IFN, has long been known to occur in different pathologies. For example, during AI polyendocrinopathy syndrome type I (APS–I), an AR syndrome occurs by immune cells affecting endocrine function resulting in candidiasis adrenal insufficiency. In this case, point mutations in the AI regulator gene (AIRE) affect the tolerogenic profile development of T cells [
113]. It was noted, in population studies in 2017 that aAb titres against type I IFN–ω were particularly high varying across populations with IFN–α2 [
113]. Subsequently, a notable study in 2017(n=8972) examined other aAbs against type I IFNs including IFNα2 to find aAbs occur naturally in 86% of people combined with four other cytokines measured (IL–1α, IL–6, IL–10, GM–CSF) to note natural occurrence in younger adults [
114]. Conversely, other studies examining this during non–COVID–19 acute respiratory failure indicate (n=284) that 1.1% were positive for antibodies against IFN–α2 and similarly with type III interferon recently [
115,
116].
Furthermore, single nucleotide point (SNP) mutations occur in other IFN signalling proteins, like STAT2, or other AR individuals, with pathological consequences, including HLH, but also bronchiolitis, and recurrent RSV amongst other pathologies [
99]. Causal factors in aAb production resulting in disease as being a genetic trait were observed in investigating type II IFN research. Exemplified in 2019, a population study (n=74) in Southeast Asia appears to indicate that aAbs against type II IFN–γ vary between populations and may be present as a risk factor in nontuberculous mycobacteria (NTM), alongside other opportunistic infections like Salmonella, Histoplasma and Cryptococcus [
117].
During the recent pandemic, research indicates variability and unknowns with regards to autoantibodies. It is indicated that approximately 10%–25% of those with chronic COVID–19 pneumonia possess aAbs to one or two type I IFNs (IFN–α2/IFN–ω) age over 25 or one of the 12 other subtypes of type I IFN–α respectively, but not type I IFN–β [
118,
119]. In other viral infections like West Nile virus (WNV), aAbs to type I IFN (IFN–α/IFN–ω) were detected in a cohort (n=441), indicated to occur in males over 65 years at a prevalence range of 0.3%– 1.0% and in a third of individuals hospitalised [
120]. Although SARS–CoV–2 is a well–characterised virus, earlier in the pandemic, three types of type I IFN do possess anti–viral regulatory properties (IFN–α8, IFN–β, IFN–ω), with type I IFN–ω having the most potent inhibitory activity against earlier B.1.351 lineages circulating up to 2021 [
79]. For reasons, explained below maybe this was an oversite in research, as another type I IFN–ε alongside type III IFN–ω proteins was observed at higher concentrations in infant nasopharyngeal samples (n=192) [
121]. This was concurrently observed in population studies showing variance in IFN subtypes inhibition ability declining between 4 strains of SARS–CoV–2 showing less type I IFN anti–viral activity [
79]. Combined genome–wide association studies (GWAS) did indicate the type I IFN (IFNAR) pathway could be associated potentially with severity (n=466) by coronaviruses in gene set enrichment analysis (GSEA) [
122]. Authors correctly pointed out there are few if any global population studies that do examine human leukocyte antigen (HLA) polymorphisms which are key in tissue typing. The HLA encodes two functional protein complexes required to present antigens (MHC class I/II) that are polymorphic proteins aside from IFN proteins as above.
4.3. Interferon–Inducible Transmembrane Proteins during Viral Infection
However, other IFN proteins induced by IFN are relevant and could be directly affected during pathology. For example, the IFIT family of proteins has 5 members regulating viral replication. Some authors consider other IFI proteins (IFIT1, IFIT3 and IFIT5) to regulate SARS–CoV–2 replication. Various IFIT proteins intracellularly sequester viral ss/dsRNA and also unmethylated RNA present during host cellular pathogen infection [
123]. During the 2009 Influenza (H1N1) pandemic, as above, IFITM1/2, became clearer as potentially limiting the rate of VP synthesis through type I IFN synthesis in other viruses like WNV) as well as DENV infections [
124]. A third protein, IFITM3, estimated to compose 50–80% of total IFITMs, present on T cell PMs, was seen to differentially regulate IFN synthesis through ubiquitination and methylation inhibited concurrently with point mutations in different tyrosine residues (e.g., Y20) [
125,
126]. Phosphorylation could be inhibited preventing endocytosis and ubiquitination occurring through E3–ubiquitin ligase, similarly to STAT proteins [
127]. Around this time, the CD225 domain of IFITM proteins was discovered to be required for inhibition of both Influenza/DENV replication [
128]. Indeed, IFITM3 was recently indicated to be increased in severely affected Influenza patients additionally pointing towards this as a potential factor in restricting viral replication in tissues [
129]. Therefore, IFITM3 could be considered important to Influenza infection immunisation responses and is confirmed to be present during SARS–CoV–2 infections to be a regulatory checkpoint
in vivo observed in gene knock–out non-human primates (NHPs) in the pulmonary tract [
130]. Finally, this class of IFIT/IFITM3 proteins are further implicated in modulating amyloid plaques during Alzheimer’s disease [
131,
132].
4.4. Interferon and Immunotherapy Regulation in Cancer
Interferons play roles in many pathologies. Both type I/II IFNs have long been considered to be immune cell regulatory affecting both cell cycle and cancer cell proliferation, with partial protective roles during tumorigenesis through expression of PD–L1, whilst tissue cells produce a suppressive cytokine IL–10, but also a metabolite indoleamine–pyrrole 2,3–dioxygenase (IDO1/2). The regulatory effects of IFN rely on complex interactions with metabolic and cytokine factors affecting immune cell function (see
Figure 5).
Figure 5.
The Role of Type I Interferon in the Immune System.
Figure 5.
The Role of Type I Interferon in the Immune System.
Some of these include tryptophan and kynurenine regulating cellular metabolism differentially in immune cells like Mϕ (M1ϕ/M2ϕ), as well as activating killer cytotoxic T cells expressing CD8
+. Effector immune cell function relies on the production of perforin, granzymes and other cytolytic enzymes from both T
C/NK cells in the tumour microenvironment (TME) where other cell cycle proteins are factors [
43]. Other reviews ascertain categories of therapeutics undergoing clinical trials targeting type I IFN pathways as TLR agonists, STING agonists, chemotherapeutics, oncolytic viruses and cancer–targeted drugs [
133]. In contrast, overexpression of IRF7 proteins also affects cancer and acute myeloid leukaemia (AML) progression [
134]. Similarly, inhibition of other vascular endothelial cell adhesion molecules (VCAM1/VLA–4) may reduce intracerebral invasion during AML
in vivo [
134]. Before 2020, it was known that type II IFN–γ affected an immune cell regulation checkpoint, PD–L1, as well as IDO1/2 and may inhibit T cell activity in tumours through STAT1 with upregulation of IRF1, as well as degradation of tryptophan and NK cell suppression. It is considered that the T cell response is determined by the concentration of type II IFN–γ, within the TME, aside from Mϕ phenotypes and tumour–associated antigens (TAAs), as well as type I/III IFNs unknown to date [
43]. But also, that escape from type II IFN immune cell detection can regulate tumorigenesis [
43,
133,
135].
5. The Role of Interferon during SARS–CoV–2 induced COVID–19 in 2023
5.1. Introduction to SARS–CoV–2 Structure and Interferon Regulation
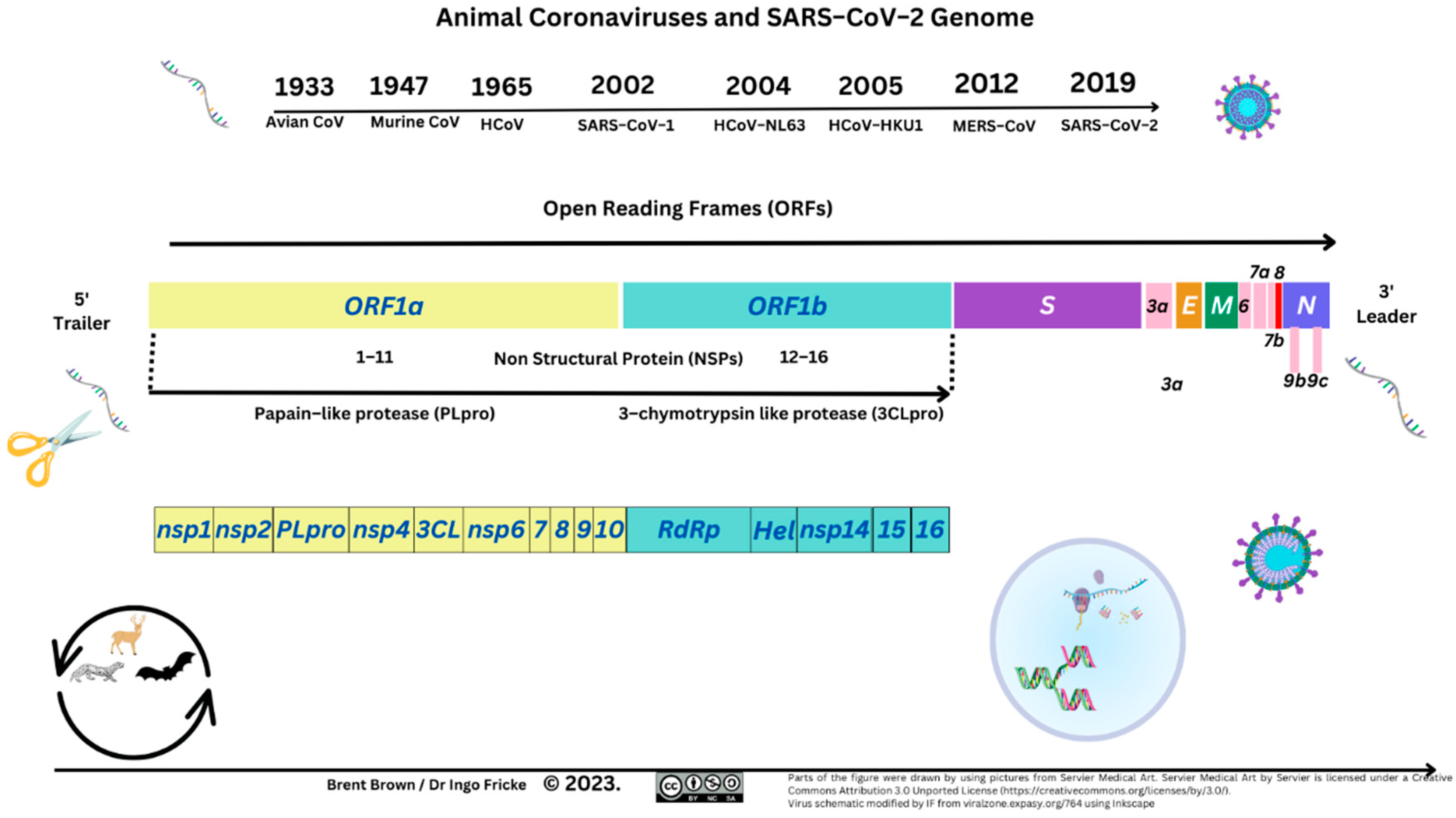
The SARS–CoV–2 viral protein domains consist of protein domains that include spike (S1/S2), membrane (M), nucleocapsid (N) and envelope (E) protein domains encoded by and 16 non–structural proteins (NSP1–NSP16) with differing detailed host roles. The first encompasses a receptor binding domain (RBD) that has an affinity to the human cell receptor angiotensin–converting enzyme (ACE2) receptor. These are depicted below (see
Figure 6).
Figure 6.
SARS–COV–2 Genome Structure.
Figure 6.
SARS–COV–2 Genome Structure.
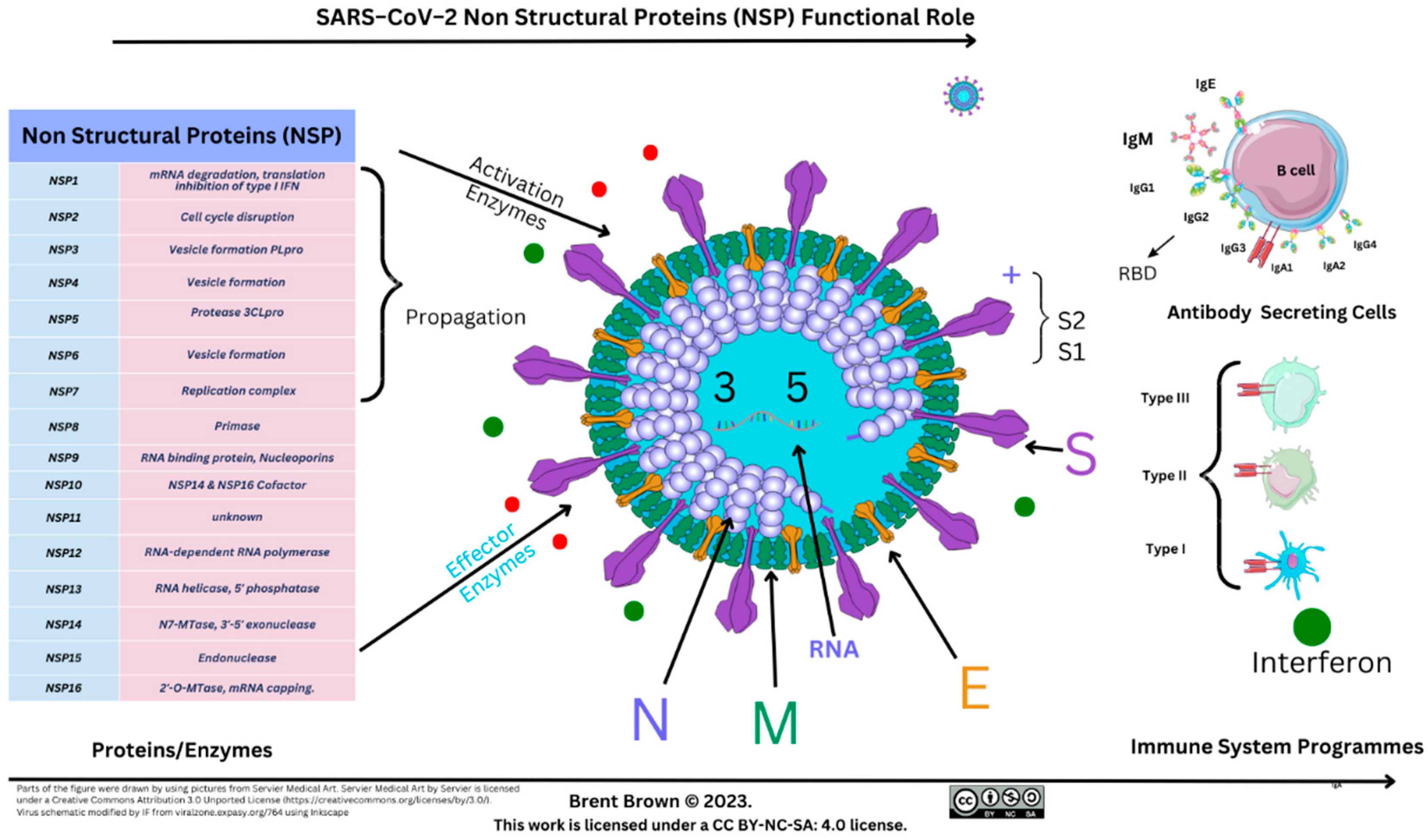
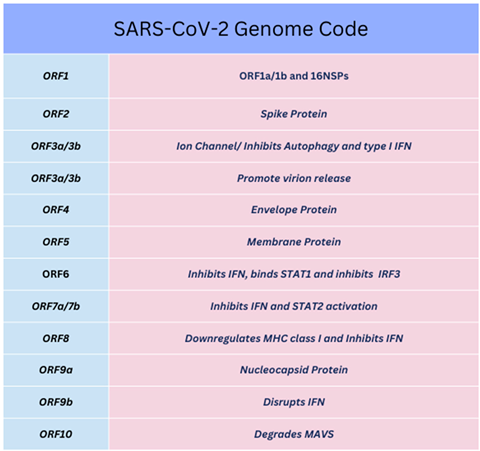
The first NSP (NSP1), lyses host mRNA affecting type I IFN signalling through STAT proteins. The last NSP (NSP16), towards the 3’ end of viral RNA is followed by a variety of structural/accessory proteins encoded by open reading frames (ORFs). Many NSPs are defined and end with a cleaving methyltransferase enzyme at NSP16 (2ʹ–O–MTase) [
136]. The latter enzyme is conserved in a viral pocket known as the S–Adenosyl–L–Methionine complex, activated by joining NSP10, forming the 2’–O–Methyltransferase enzyme complex. The resulting methyl group (–CH3) transfer occurs to the 5’ end of the +ssRNA VP upon activation. Moreover, specific ORFs (e.g., ORF6) encode other proteins affecting IFN signalling through STAT1/STAT2 and other membrane proteins as above [
137,
138,
139]. The –CH3 group is transported from S–adenosyl–L–methionine pocket formed after reacting with the S–adenosyl molecule and L–methionine amino–acids to NSP13/NSP14 before binding to an NSP16 effector protein. Each of NSP13–16 as well as ORF6 were indicated to suppress IFN production through STAT1 [
138,
140,
141,
142]. Below is shown the NSP and ORF function characterised for SARS–CoV–2 (see
Figure 7 and
Table 1).
Figure 7.
SARS–CoV–2 Non–Structural Protein (NSP) Function.
Figure 7.
SARS–CoV–2 Non–Structural Protein (NSP) Function.
Table 1.
SARS-CoV-2 Open Reading Frames (ORF).
Table 1.
SARS-CoV-2 Open Reading Frames (ORF).
These can have implications upon adaptive immune responses with lesser or greater incidence of moderate to severe disease [
143]. The effective regulation by IFN products play a major role in anti–viral defence [
144]. As above, SARS–CoV–2 proteins (e.g., ORF3a, ORF9b and ORF10) perform a role in modulating type I/III IFN mediated immune responses through inhibiting nuclear transport of IRF3, NF–κB and STAT polymers, leading to downregulated expression of a variety of IFI genes [
145]. Type I IFN is described as critical to regulating the activation and recruitment of neutrophils [
146]. Indeed, neutrophils may reduce
IFIT3 translation leading to a reduction in IRF3 phosphorylation [
147,
148]. Similar events occur in RSV and other infections [
149]. Each virus synthesises NSP1 inhibiting translation of type I and type III IFN–encoding genes leading to stimulation or inhibition of IFN receptor activity (e.g., IFNAR2); whilst RSV translates NSP2 to suppress responses as such [
150]. As above, RSV activates Rab5a host cell proteins to downregulate the activation of type III IFN signalling cascades [
151].
As above, SARS–CoV–2 ORF9b impairs type I IFN responses specifically through receptors but also intracellular enzymes (e.g., IκB kinase (IKK)–related kinases (IKKα/β/γ), as well as nuclear transcription factors (e.g., TFs, NF–κB). These can activate PRR proteins localised near mitochondria, namely, RIG–I and MAVS, usually resulting in robust IFN expression [
152,
153]. Other signalling pathways affected through type I IFN–α and IFNAR1/2 can result in homeostatic signal transduction through STAT proteins including cytokines (e.g., transforming growth factor (TGF–β) and TNF–α).
During chronic COVID–19, the rate of increase in cytokine synthesis can therefore affect cytolytic/cytotoxic NK/T cell function alongside T
H cell recognition of peptide antigens [
1,
154,
155,
156]. Moreover, chronic viral, bacterial, fungal or oncological pathology can involve the inhibition of both innate or adaptive type II IFN–γ synthesis together with significant inhibition of the lesser understood β2 integrin (CD18) by NK cells around vascular endothelial cells [
1,
43,
157]. Sixteen NSPs are translated that can directly suppress the immune–stimulatory action of type I IFN and type III IFNs through ISGs [
158,
159].
5.2. Interferon Research during SARS–CoV–2 induced COVID–19
Therefore, with regard to COVID–19 mechanisms, laboratory research examined genetic mutations (n=659) to estimate that 3.5% of mortality may occur from a deficiency in IRF7 proteins. Possible reasons include pDCs secreting less type I IFN or unknown IFN subtypes requiring further detail. Susceptibility in cell samples,
in vitro, established that fibroblast cells could also be susceptible to SARS–CoV–2 infection [
85]. In comparison, during HCoV–229E viral infection,
in vitro research indicated that IRF1/IRF3 and IRF7 were activated with IRF3 required for viral RNA transcription [
160]. As recently as 2023, Cao et al. showed the SARS–CoV–2 protease (PLpro), may inhibit phosphorylated IRF3 as a result of de–ubiquitination of STING at L289. This resulted in the inhibition of type I IFN gene transcript promoters (IFNB) alongside three other gene transcripts (
ISG56,
CXCL10, and
CCL5) [
161]. Whereas others examined respiratory responses examining the chemokines and immune cell changes (n=174) in individual health and heightened during community–acquired pneumonia (CAP), as well as COVID–19. It was found that IFN signalling gene transcripts were present [
162]. As above and below, part of the SARS–CoV–2 infection pathogenesis can be in part explained. It was predicted then there were more activated mast cells and neutrophils together with less activated DCs/M2ϕ but 3 chemokine gene transcripts and more chemokines were upregulated (
CXCL8,
CXCL17,
CCL2) together with one subunit of the type I IFN receptor,
IFNAR1 [
162].
It was further evidenced in serum blood samples, both type III IFN–λ1/ IFN–λ2 were observed at increased concentrations in recovering individuals (non–ICU vs ICU), independent of other cytokines measured by enzyme–linked immunosorbent assays (ELISA) [
163,
164]. Such assays may not have been widely available prior. Comprehensive IFN analysis of the lower respiratory tract occurred during chronic COVID–19 over the last 3 years through single–cell RNA (scRNA) sequencing. Specifically, it was observed that type III IFN gene transcripts (
IFN–λ2,
IFN–λ3 and
IFN–λ4), were upregulated alongside type I IFN gene transcripts, but type III IFN–λ2/3 proteins were translated and expressed in broncho–alveolar lavage fluid (BALF) samples [
31]. Moreover, it appears that epithelial cells do produce IFN–λ1 stimulated
in vitro, through TLR3, RIG–I, and MDA–5 [
31]. However, IFN–λ1 was evidenced as expressed in the upper respiratory tract with decreased production further down the respiratory tract with at least five gene transcripts upregulated appearing to correlate with SARS–CoV–2 viral load [
31]. This could therefore reflect cellular changes in IFNLR expression unknown so far. Each of the other type I/III IFN gene transcripts (
IFN–λ1,
IFN–λ2,
IFN–λ3,
IFN–β,
IFN–α4,) showed a lower correlation compared to acquired respiratory distress syndrome (ARDS) in the lower respiratory tract, although age and other pathological conditions are a factor [
31]. Interestingly, bioinformatic data suggests upregulation of TNFR/NFKB, IFN–λ2 and STAT3 pathways that usually stimulate immune cell phenotype differentiation [
31]. In a recent mixed age case (n=192) report, nasopharyngeal swabs were purported to measure that type I IFN (IFN–α, IFN–β, IFN–ε, IFN–ω), as well as type III IFN (IFN–λ1, IFN–λ2 and IFN–λ3) proteins were all synthesised and measured in children [
121,
165]. Indeed, increases in the synthesis of TNF/IL–6 through STAT1/2/3 does affect both metabolic and NF–κB causal in inflammatory processes that did not significantly restrict viral load increase [
166,
167,
168].
5.3. Two Types of Interferon during COVID–19
During 2020, many studies indicate that type I/III IFN homeostasis could be pertinent to COVID–19 regulation. An
in vitro study showed SARS–CoV–2 infection does not induce usual levels of type I IFN production by antigen presenting cells like myeloid DCs (mDC) or differentiated Mϕ [
169]. Specifically, it was suggested that there was no detectable IFN–α, with reduced synthesis and secretion of IFN–β1 and IFN–λ1, where normalised read counts were lower than 10 [
170]. Some observed in individual patients (n=101), decreased rate of type I/II IFN synthesis during COVID–19 disease by gene transcripts present (
IFN–α,
IFN–β,
IFN–λ1,
ISG), in the upper respiratory trace [
171]. Notably, measured assay scales indicated the rate of type I IFN–α protein concentrations (<2.1 pg/ml), together with type II IFN–γ (<15 IU/mL), could be independent of each other. Interestingly, one cytokine, IL–17 was indicated as not produced by T
H17 cells [
171]. Therefore, at least with the novel SARS–CoV–2, each of the STAT1/STAT2/STAT3 pathways could be affected. It was noted
in vitro that with the same cell samples stimulated, both DCs and NK cells were functionally exhausted [
171].
During 2020, results showed that out of 6 immune cell phenotypes (T cells, B cells, NK cells, and APCs), pDCs could respond to type I IFN–α2 and were more responsive to type I IFN–λ1 than B cells [
172,
173,
174]. This involves differential phosphorylation rates of more than 6 IFN signalling proteins (STAT1–STAT6), affecting T
REG cell differentiation [
172,
173,
174]. Notably, type III IFN–λ2 and IFN–λ3 induced STAT1 phosphorylation and activation in the B cells that produce antibodies [
44,
175]. Authors noted that naïve B cells more than plasmablasts could be blocked by JAK inhibitors [
44,
175]. The time point noted that antibody isotype class–switching did not occur within five days [
175]. There are 5 types of antibodies secreted in humans (IgG, IgA, IgM, IgE, IgD) with isotypes [
1]. One isotype, IgD was historically elusive [
176]. However, IgA1/IgA2 receptors (CD89/FcαRI) expressed by neutrophils in 2022 were indicated as possibly affected by type III IFN remaining unclear. In combination with CD16 (FcγRIII A/B), CD32 (FcγRII A/B/C) and CD64 (FcγRI), the predominant transduction pathways of IgG/IgA secretion occurs through antibody Ig Fc receptors to effect leukocyte type II IFN production in both the mucosal and respiratory tract [
1,
27,
177].
Shortly prior, significant details emerged (p<0.05) in a smaller cohort (n=54), that specific gene transcripts were expressed (
IFN–α,
IFN–β,
IFN–λ1,
IFN–λ2), in patients alongside IRF7 in comparison to health care workers (HCW) that would have been exposed to SARS–CoV–2 infection [
165]. One project set out to measure the time–points of host type I IFN–α2/IFN–β (n=65) expression that clearly showed type I IFN–α2 synthesised and reduced within 1 week in all but 25% of recovered patients between 2 timepoints, one week apart [
178,
179]. This was accompanied by a clear differential of reduced type I IFN–α2 concentrations during severe COVID–19. Albeit, assay scales may differ, it could therefore be seen then the dysregulated immune response with IFN may be affected by SARS–CoV–2 proteins [
178]. The homeostatic balance of type I IFN is usually maintained and largely unclear in different pathologies [
180]. At this juncture, homeostatic regulation of the rate of type I/III IFN synthesis could affect the immune cell phenotypes through STAT1/STAT3 signalling. During maturation other dominant peripheral blood γδ T cells as well as mucosal–associated invariant T (MAIT) cells producing tumour necrosis factor (TNF–α) can also stimulate STAT phosphorylation and immune cell maturation through IFN receptors [
1,
181]. Viral modulation of antigen presentation is largely unknown. However, during the COVID–19 pandemic, emerging reports appeared linking caspase activation and recruitment domains (CARD) to nucleotide–binding oligomerization domain (NOD) like receptor (NLRC5) central to the STAT1/IRF3 pathways [
159]. The significance of this was that NLRC5 seems to be central to nuclear transcription–producing MHC class I molecules that can be suppressed during infection affecting antigen presentation to T
C cells [
159]. Simultaneously MHC class II molecules regulation on DCs may affect NK cells [
159].
During COVID–19 disease severity, STAT6 in individuals (n=38) in pneumocytes and lymphocytes was measured by CD8/CD4 ratios evidenced and indicated at less than a ratio of 1 affecting survival. Specifically, higher STAT6 expression in pneumocyte cytoplasm and the nucleus was indicated [
182]. With regards to IFN signalling, some studies indicate that during infection of individuals (n=32), both TLR7/8 may potentially have been impaired unknown, ruling out type II IFN deficiency, with increases of relevant TLR gene transcripts (TLR3) during pulmonary infection [
183]. It was noted that IFN–λR1/IL10RB2 is required by NK cells for type II IFN signalling [
183]. Given the above, it is necessary to consider additional metabolic factors (e.g., pyruvate, hexanoate), where type IFN–λ significantly correlated with glycolytic metabolites potentially indicative of dysregulation of mitochondrial metabolism [
184]. Recent
in vitro and
in vivo research further indicates a link between type I IFN and long–interspersed–element 1 (LINE–1) retro–transposition. Specifically, suggestions were that type I IFN/ LINE–1 retrotransposons regulate each other, with exaggerated type I IFN expression potentially linked to a higher incidence of AI disease but also senescence [
185]. With regards to SARS–CoV–2 infection, it was indicated that 4 proteins (NSP1–3/NSP14) could suppress LINE–1 transposons. Therefore, it is possible that LINE–1 retrotransposon–encoding DNA representing around 17% of the human genome could be affected unknown to date.
5.4. Other Pattern Recognition Receptors
With regards to SARS–CoV–2 infection, viral host entry is sensed through PRRs and TLRs (e.g., TLR3, TLR7/8/9), as well as pathogen–associated molecular patterns (PAMPs). Endosomal PRRs like the four TLRs include cytoplasmic TLR4 which senses an array of pathogenic antigens. Activation of TLR3/TLR4 results in the phosphorylation of TRIFF and then the IRF3 dimer, whilst the activation of TLR7/8/9 causes phosphorylation of the IRF7 dimer [
153,
186,
187]. Unlike TLR3, activating TLR4 phosphorylates other proteins before the activation of the IRF3 dimer. The outcome is an expression of type I IFN–encoding genes with autocrine and paracrine signalling [
188]. Within these, IRF3 and IRF7, are considered to be cytokine and chemokine inducers inducing CXCL10, CCR5, ISG56, IL–12p35, IL–23, and IL–15 [
51,
189]. Other cytoplasmic PRRs include RIG–I and MDA5, which phosphorylate IRF3 and IRF7 dimers [
190]. Once the SARS–CoV–2 particle enters, TLR3 and TLR7 are activated through PAMPs, either at plasma or vesicular membranes during cellular infection localised with other RIG–I like proteins MDA5 affecting IFN regulatory factors (IRF3/IRF7) near mitochondrial and peroxisomal organelle membranes [
51,
76,
189,
191]. The overall cellular recognition activating IRF3 can induce both chemokines and cytokines like CXCL10, as well as IL–12p35 and IL–23 whilst inhibiting another subunit receptor IL–12β with all 3 required that influence DC phenotypes [
1].
Type I IFNs have been found not to recruit NK cells directly but through the activation of chemokines and monocyte maturation proteins (e.g., CCL2, CCL5, CXCL8, CXCL10, CXCL17) [
162,
192]. Interferon–stimulated genes result in producing chemokines, like CXCL1, CCL2 and CXCL10, that each have ligands and receptors on neutrophils, and T cells respectively [
193]. While, DCs expressing CD14 can reversibly differentiate into monocytes expressing CD163 and either M1ϕ/M2ϕ with recruitment of NK cells stimulating type II IFN–γ inducing lysis of infected cells [
36].
6. The Four Types of Type III Interferon during Health and Disease
6.1. Overview
Type III IFN–λ was first discovered in 2003 with subsequence confirmation there were 4 subtypes of type III IFN–λ1, IFN–λ2, IFN–λ3, as well as IFN–λ4. Upon discovery, early observations were that type III IFN–λ could influence immune cell (monocytes) development into DCs as several cytokines (e.g., IL–2) induce T
REG cell development, differentially through STAT protein and IFN signalling [
194]. It is considered that type III IFN–λ has overall anti–viral properties unclear to date through IFN–λR1/IL10RB2 binding affecting intracellular signal transduction. In 2010 this was clarified further when genomic analysis showed one gene for
IL10RA (
IFNLR1) was common to many animals including humans, monkeys, mice, horses and chickens. The gene transcript was subsequently found to be expressed by LNs, testis, but also by germinal centre B cells and in various types of cancer (lymphoma, acute lymphoblastic leukaemia, head and neck cancer); but expressed at high concentrations within tissue like the pancreas (thyroid, skeletal muscle, and heart tissues) indicating these could respond to type III IFN synthesis [
195]. Interestingly, authors postulated that the three important adaptive arms of the immune system responsive were NK cells, and T
C cells, through promoting the other phenotype by evoking a T
H1 cellular response [
195].
During early 2011 research, a gene regulator of B/T cell differentiation (LyF) was described as having a transcriptional binding site within the
IFNLR1 domain encoding for one part of the type III IFN receptor. Furthermore, the activator protein 2 (AP–2), c–Jun and a p53 binding site within 1kb of the start of the transcription sequence on IFNLR1 in humans was described [
195]. Conversely, the first report appeared after evidencing the other key gene transcript for type I IFN synthesis,
ISG56, as well as
RIG–I induced by synthetic IFN–λ2
in vitro in
P. alecto bats [
196]. As type III IFN research unfolded, it was in 2014 clarified that JAK2 was essential to regulating signal transduction for type III IFN–λ1, when
in vitro it was observed L. monocytogenes could potentiate type III IFN–λ1 around peroxisomes [
197].
The newer type III IFN–λ4 was investigated at the same time using transcriptome sequencing (RNA–seq) as to how liver hepatocytes and primary human airway epithelial cells (pHAE) could be affected [
198]. This comparison and others note that the type III
IFNL4 gene can be polymorphic and differentially expressed which could change the function after protein translation with frameshift mutation disrupting translation of the IFNL4 mRNA [
8,
183]. During 2016, no difference in IFN/ISG expression was observed according to clinical asthma severity (n=66) in individuals. It was observed that neutrophilic asthmatics overexpressed both type I/III IFN (IFN–β, IFN–λ1(IL–29)), rather than eosinophilic asthmatics, but not IFN–λ2/IFN–λ3 (IL–28) [
199]. Interestingly, in 2020, research on SLE further confirmation examined the unclear mechanisms of type III IFN that IFN–λR1 may correlate with B cell proliferation signalling through TLR7/8 PM receptors. It was plausibly suggested that increased IgM production could occur but outside the lymphatic follicular environment where B cell antibody clonal selection and isotype switching usually occurs [
1,
200]. The gap in these studies does concur with evolution in clarification of T cell phenotypes.
6.2. Early Type III Interferon Research During Viral Diseases
In comparison, supporting evidence with type III IFN–λ synthesis suppression during rotavirus infection and porcine epidemic diarrhoea virus (PEDV) similarly remains under investigation [
201,
202,
203,
204,
205]. Following on from above, in pigs, just before the recent pandemic (2019),
in vitro experiments compared the transcriptional profile of porcine epithelial cells to observe that type III IFN–λ3 was noted to upregulate at least 983 differentially expressed genes (
DEGs). This STRING analysis indicated 7x as many type III
DEGs in comparison to type I IFN could be upregulated illustrating the diversity of type III IFN. These observations remained pivotal to potential anti–viral inhibition of Porcine Epidemic Diarrhoea virus (PEDV) infection. In comparison, supporting evidence with IFN–λ synthesis suppression during rotavirus infection and porcine epidemic diarrhoea virus (PEDV) similarly remains under investigation [
201,
202,
203,
204,
205]. These can have implications upon adaptive immune responses with lesser or greater incidence of moderate to severe disease [
143]. Although a different viral infection, type III IFN is historically considered to affect mucosal epithelial cells. This was one of the initial projects that revealed relevant STAT protein gene transcripts affected with IFN–λ3 upregulation occurring could be STAT2/JAK2 of relevance given that STAT proteins and JAK enzymes affected can potentially be activation/inhibition therapeutic targets [
206]. Recently, in 2020 in between waves of SARS–CoV–2 infection, during a clinical trial (NCT04354259), it was observed that polyethylene glycol–conjugated (PEG) IFN–λ may potentially have a beneficial therapeutic effect in acute COVID–19 during a phase 2 clinical trial [
207].
Amidst 2015 and following the EBOV outbreaks, detail on the newly discovered type IIII IFN in immune cells emerged to show that gene transcripts during severity (EBOV) were observed within DCs that were
IL28A and
IL28B [
70]. Through 2016, investigating subtypes of type I IFN, above, during HCV infection, it was observed that STAT2 changed ISG15 synthesis through
MX1 transcription required for type I IFN synthesis in Mϕs with type III IFN phosphorylating JAK2 evocative that STAT2 may heterodimerize; Subsequently an increase of protein kinase R, IRF9, was seen in cells deficient in these proteins stimulated by type I IFN [
208]. These results confirmed that type I IFN–λ1 transduction was dependent on STAT1/STAT2. Interferon could also reduce replication and inhibit HCV, whilst STAT1 was essential for type II IFN synthesis [
208]. The paradoxical role of IFN–λ4 as the most studied polymorphic IFN indicates that during HCV infection, type III IFN–λ4 is secreted at lower concentrations from a stressed endoplasmic reticulum. This in effect could attenuate HCV–specific peptide presentation to T cells (CD8
+) through MHC class I peptide–dependant presentation, whilst type III IFN–λ3 was secreted at normal concentrations [
209].
Current 2022 investigations imply that the IFN–λR1 is also expressed within gingival keratinocytes, with
in vitro IFN–λ1 stimulation at low doses stimulating RIG–I/TLR3, with both PRRs recognising viral RNA without evoking high expression of pro–inflammatory cytokines, like IL–6, and therefore may be of consideration as an anti–viral [
210]. Whereas IFN–λ3 expression in a vector is being looked at to counter a variety of dog–affecting pathogens like canine coronavirus (CCoV), parvovirus (CPV), as well as distemper virus (CDV) [
211]. During 2020,
in vivo expression of a recombinant type III IFN (IFN–λ2, IFN–λ3), during Rabies virus infection (RABV) was shown to have an anti–viral response administered intranasally and reducing viral load in a neurotropic virus. The overall observations were that upregulation of SOCS1/SOCS3 occurred simultaneously with decreasing cytokine production and reduced viral pathogenicity when various type I IFN proteins (IFN–α4, IFN–α5, IFN–β, STAT1, IFIT2) were observed that could change vascular blood–brain barrier permeability [
212]. Recent kinetic reports indicate intracellular potency in hepatic cell lines
in vitro of the more polymorphic IFN–λ4, seemingly translated before 24 hours after cellular infection instigating STAT1/STAT2 phosphorylation earlier. This was characterised by gene transcripts (
MX1,
ISG15,
OAS2,
RIG–I,
STAT1) while IFN–λ3 was sustained in contradiction to other reports after 24 hours [
213]. Conversely, in other viruses (Human papillomavirus, HPV) that is implicated to cause cervical cancer, the differential expression of mucosal epithelial cell type III IFN (
λ1, λ2,
λ3) gene transcripts (n=56) was observed to be upregulated in low–risk HPV infection [
214]. Furthermore, utilising
in vitro HPV18 expression in cell lines it could be seen that type I IFN–β and type III IFN–λ1 in basal epithelial cells could be inhibited by DNA ligand stimulation and through suppression of the cGAS–STING pathway necessary for IFN synthesis [
215].
6.3. Immune System Production of Cytokines and Type I/III IFNs
Most immune cells produce type I IFNs, meaning that the glycoprotein is widely bioavailable within the human body [
216]. During immune cell characterisation before 2010 it was observed that T
REGS (T–bet
+Foxp3
–) could produce another cytokine, IL–10, regulating the majority of immune cell phenotypes including T
H cells (CD4
+) [
217]. The cytokine, IL–10, is an anti–inflammatory cytokine with an essential role in development and proliferation of T
REG cells, restricting myeloid and chronic inflammatory T cell responses. However, IL–10 or the expansion of antigen–activated, tumour–specific CD8
+ T cells remains unknown. In comparison, the role of IFN–λ is better described through the expression of IFN–λR1 through functional absence on fibroblasts, splenocytes, endothelial cells and some M1ϕ/M2ϕ phenotypes and thus these are non–responsive [
35,
36,
146,
186]. Whereas IFN–λR1 is constitutively expressed by pDCs and B cell phenotypes with IFN–λ activity on pDCs considered to occur through phosphorylation of STAT proteins (STAT1, STAT2, STAT3 as well as STAT5) [
218].
It has been hinted that IFN–λ could be regulated
by ISG15 transcription and encode the respective ISG15 protein at early/late stages after hepatocyte cell stimulation with different inhibitory regulation to type I IFNs. Studies using immortalized hepatocytes
in vitro ascertain that this occurs independently of IRF1, but IFN–β is maintained between 24–72 hours after viral infection [
186]. Contrasting with this, DCs are considered to produce more type I IFNs (IFN–α), within 24–48 hours after viral infection [
186].
The tolerogenic profile of DCs together with the development of weakened T lymphocyte–mediated adaptive immune responses requires IFN stimulation to stimulate type II IFN [
169]. Dendritic cells require other TFs and cellular activation factors like Runt–related transcription factor 2 (RUNX3) as well as IRFs to determine activation and cellular lineage of CD8
+T
C cells outside the lymphoid tissue and in tumours [
7,
81]
Shortly after 2012, employing cellular
in vitro stimulation, more detail arose of monocyte–derived phenotypes secreting variable type I/III IFN subtypes. It was then observed that differential phenotypes arose into the other APCs (DCs or M1ϕ/M2ϕ), that type I IFN–β and type III IFN–λ1 were synthesised by divergent phenotypes; but also this was described as followed by type III IFN–λ2/λ3 secretion in both monocyte/myeloid derived lineages [
219]. In 2013, transcriptional reports began to emerge on the role of TNF–α stimulation of monocytes and type I IFN–β synthesis. Specifically, these inferred that FOXP3/IFNAR2 expression were independent, both of which are required for IFN signalling; but also, that CD38/CD83 upregulation could be induced on monocytes also by type I IFN–β independent of T cells through differentially expressed genes (
DEGs) [
83]. In other laboratory analyses, observations in peripheral blood, in comparison to healthy individuals, suggestive of type IFN–β temporarily downregulated during COVID–19 with the measurement of two chemokines. Specifically, CCL2 along with a type II IFN–inducing protein (CXCL10), were significantly upregulated during COVID–19 (P < 0.01), but the receptor CCR2 was not [
220]. Further comparisons (n=17) observed that classical monocytes exhibited a type I IFN gene signature during disease severity. In relation to other respiratory viruses, like Influenza, it could be seen that T cells express more gene transcripts (
granzyme B+/IFN–γ+ ) than during COVID–19 alluding that there may be an imbalance in regulatory IFN production with
IFITM1, ISG15, and
JAK3 gene transcripts observed during severity [
221].
As above, DCs can upregulate the expression of two predominant type III IFNs, IFN–λ2 and IFN–λ3 [
31]. Type III IFN–λ1 was evidenced in pig epithelial cells to induce gene transcripts (
ISG15/
OASL) in intestinal cells up to 24 hours, historically considered to be the typical timepoint of type I IFN–α synthesis by DCs [
222]. Much remains unknown about the effects of type III IFN on CD16
+ monocytes or macrophages. More recently in 2021, PM profiling showed that type I IFN–α2 effects a greater than five–fold change in CD14
+ monocytes of CD38/ISG15 protein expression with minimal change in CD86/Fas expression, with both immune cell cycle regulation proteins controlling immune cell apoptosis [
223]. Notable observations in this study suggest maintenance of monocyte cell regulation with ISG15 as the type I IFN–inducing protein; but also a fifth, CD244, recognised as activating Mϕ during RA, Crohns disease (CD) and COVID–19 [
223]. Therefore, as CD244 was observed during lung inflammation and is a marker expressed by T
REG (CD8
+CD28
–CD57+) cells during myelomas, it has also been suggested as a potential cancer therapeutic target [
224,
225]. Concurrently, T cells significantly upregulated expression of endothelin converting enzyme–1 (ECE1), as well as the IL–1 receptor antagonist (IL–1RA), with the latter as known as a risk factor in arthritis [
223,
226]. In yet to be reviewed pre–prints type I/III IFN synthesis and signalling rates were decreased in primary human airway epithelial (pHAE) cells infected with SARS–CoV–2 [
227]. In addition, it is suggested that type I IFNs (IFN–α/IFN–β/IFN–ω), and type III IFNs (IFN–λ1, IFN–λ3) synthesis may remain when stimulated by SARS–CoV–2 with stimulated pDCs expressing chemokines (CXCL10, CXCL11, and CCL2) that contribute to effectively clear viral antigens by stimulating T cells [
227].
6.4. Other Factors in IFN Signalling
Above, many of the relevant protein factors that can be stimulated by IFN have been outlined. However, other proteins transcribed and translated also play a role. These include
IFI27, encoding IFI27 proteins (122 amino–acid hydrophobic protein, MW 12kDa), next to IFI6, both present within the mitochondrial membrane. These may contribute towards cellular IFN sensitization and organelle permeability through modulation of RIG–I. For example, IFI27 was observed elevated in peripheral serum (Influenza, SARS–CoV–2 and RSV) during infection [
228,
229]. The proposed mechanism is that IFI27 can impair RIG–I before binding to MAVS [
229]. Remarkably, the same research group showed an RNA analogue, poly (I:C), bound to IFI6 localising with RIG–I protein. In this case, it was shown in cell line deficiency
in vitro that two type III IFN gene transcripts (
IFN–λ2,
IFN–λ3) were upregulated with
STAT gene transcripts and
ISGs (
IFIT/
IFITM and
OAS). Each of these gene classes is required to be transcribed and translated for IFN synthesis and secretion to occur [
228]. Cell lines infected by either SARS–CoV–2/Influenza in cells without IFI6, in comparison to controls, could be observed to reduce viral replication with other type I IFN gene transcripts (
ISG15/
IFIT1) silenced [
228]. The role of ISG15 protein remains unclear. In ISG15 deficiency some individuals show enhanced anti–viral immune responses or similarities with other disorders. It can be considered that ISG15 is a regulatory protein [
58]. As an ubiquitin–like protein (Ubl), ISG15 can be covalently linked to substrates either intracellularly, via a cascade of E1, E2, and E3 enzymes, or secreted, so termed “ISGylation”. However, another endogenously produced metabolite, retinoic acid A metabolised from Vitamin A, and found at high levels in the gastrointestinal (GI) tract may play an as yet unknown role in IFN subtypes regulation. Retinoic acid was seen to induce IL–10 from T
REG cells in the gastrointestinal (GI) tract where it is present at higher concentrations [
230]. Research
in vivo in 2014, indicated that stimulation of naïve T cells (T
N) can produce IL–10; but also
in vivo, retinoic acid primes DCs in the lamina propria (LP) to express CD103 and synthesise retinoic acid [
230].
Table 2.
Genetic Regulation during Interferon Signalling.
Table 2.
Genetic Regulation during Interferon Signalling.
6.4. Interferon from Early Clinical Trials to Licensing as a Therapeutic
Early research, during and published towards the end of the 1969 Influenza (H2N2) pandemic investigated the efficacy of a nasal spray (n=14,000), based on a small concentration of human leukocyte–produced IFN, with a calculated efficacy rate range (56.3%–69.2%), with age–group variations, in prevention of symptomatic disease (see
Supplementary Materials). In the 20th century, other clinical trials evaluating the administration of PEG IFN–α therapies (n=168) indicate a restriction of HIV replication in individuals (n=168) with PEG thought to increase serum availability of IFN [
231].
Currently, synthetic type I IFN–β derivatives (IFN–1β, IFNβ–1a, peg–IFN–β1a) are approved and licensed by the FDA and some also the EMA. These are used, and monitored, as therapeutics for relapsing–remitting multiple sclerosis (MS), considered to reduce inflammation and promote the repair of damaged neurons [
232]. Type II IFN–γ is approved for the treatment of chronic granulomatous disease (CGD) [
233]. Below are the current synthetic IFN therapeutics licensed for therapeutic use (see
Table 3).
Fifty years later, during the 2009 Influenza (swine flu, H1N1) pandemic, the effects of type I IFN lozenges were examined (n=200) appearing to reduce severity and fever [
234]. It was assessed then that IFN–based therapeutics may reduce Influenza severity significantly. Furthermore, prior Influenza (H2N2) exhibited different strains that could be equally divided into IFN–positive and IFN–negative evoking strains (see
Supplementary Materials). Therefore, SARS–CoV–2 variants may also differentially affect type I IFN (IFN–α/IFN–β) expression [
235,
236].
The scientific rationale could plausibly be IFN subtype specificity, with as yet unknown effects of type III IFN subtypes or others. Some results recommended synthetic IFN–α2b therapy for the treatment of COVID–19 disease [
237]. Type I IFN–α2b was also observed to result in an early reduction of viral load in the upper respiratory tract [
237,
238]. Whilst a clinical trial in Iran indicated IFN–α treatment had encouraging results reducing COVID–19 mortality rate by more than 50% with early intervention reducing mortality [
239]. In 2020, the University of Western Australia signified low–dose oral type I IFN (nose drops/mouth spray/ lozenges) may prevent severe COVID–19 (see
Supplementary Materials). Furthermore, a clinical trial (n=2944) in China examined prophylactic administration of low–dose recombinant IFN–α2b also demonstrating similar protection of individuals during COVID–19 outbreaks [
240,
241]. With regards to type III IFN, an experiment in NHP involving a dosage of IFN–λ2
in vivo has so far denoted positive results in mitigating viral–induced disease [
242].
Table 3.
Current FDA licensed Interferon Therapeutics, adapted from, Deckers et al. [
13]. Accessed on 16
th August 2023
Table 3.
Current FDA licensed Interferon Therapeutics, adapted from, Deckers et al. [
13]. Accessed on 16
th August 2023
Discussion
Since Influenza pandemics of the 20th century, ongoing IFN research and receptor cloning has clarified the complexities of interferon thus far. Type I IFN–α synthesis can affect viral replication, whilst type II IFN is historically considered to be beneficially secreted from adaptive immune cells training the immune system response to pathologies including cancer. Overall, future examination of effects of IFN subtypes could be explored further. Malignant tumours and neurodegenerative disorders can be affected through type II IFN cellular regulation. The effects of a naturally produced human chemical remains a target for development subject to
in vitro/
in vivo methodology with toxicological profiling useful with recent developments of other cancer immunotherapeutics including cytokine modulators. Illustrated for type II IFN function could be considered to additionally vary depending on dosage. Type II IFN plays a key role and can be cytostatic, as well as apoptotic and may prevent cellular proliferation within the TME. Type II IFN is considered to be predominantly produced by CD4+ TH1 cells. In 3 cancer types (metastatic melanoma, head and neck squamous cell carcinoma, and gastric cancer patients), IFN gene signatures may also correlate with checkpoint inhibitor activity through M1ϕ selectively activated to overcome tumour progression within the TME [
43].
However, in 2022, other longitudinal studies of non–related acute/chronic inflammatory conditions observed gradual reduction of type I IFN–α, from a clear horizontal homeostatic line that may induce metabolic reprogramming changes through RIG–1/TLR7 in RA patients (n=19,1) independently of type II/III IFNs. In this cohort, the relevant IFN gene signatures show semblance of matching those discussed above relevant (ISG15, IFI44L, OAS1, IFI6, MxA) [
180]. Type I IFN synthesis and exocytosis from tumour cells also represent an essential step in the adequate signalling of tumour cells to immune cell components implicated in both angiogenesis and oncological diseases [
243,
244].
Furthermore, the rate of type I IFN synthesis can be affected during a number of other acute or chronic diseases [
185,
245,
246]. Questions arise as to when and how synthetic type I/IIII IFN proteins can be better utilised to mimic naturally synthesised proteins that can cause complete cancer remission. Type III IFN (encoded by IFNL1, IFNL2 and IFNL3), alongside polymorphic type III (IFNL4) gene transcripts may potentially be associated with less anti–viral activity [
247]. This has been observed during rotavirus and other infections [
201,
202,
203,
204,
205]. Observations were made that a type II IFN gene and sera transcript (IFNG/IL1A) were upregulated, together with key chemokines (CCL2/CCL3/CCL8) with type II IFN (IFNG) unchanged in lungs [
30]. This is further indicative that T cell production of type II IFN could be independent of type I/III interferon. Strikingly, type I/III IFN variation across lung cell types, and variable gene expression between myeloid cells (monocytes/Mϕs/neutrophils/cDC), could be affected by temporal reduction of specific immune cell lymphoid cells (B/T cells or pDCs) as well as other immune cell phenotypes [
181,
248]. One cytokine, TNF–α, can be reduced with variable other T cell phenotypes observed affected (T
N/T
EM/T
H17/T
REG cells), as well NK cells and cytotoxic T cells. Interferon pathways can affect STAT1/STAT3 signalling and immune cell differentiation through type I IFN regulation but the other IFN signalling pathway STAT5 was not [
30].
Interestingly, it would thus appear that if IFN gene transcripts are present that unidentified T cell phenotypes could affect this balance and rate of IFN/TNF synthesis [
1,
249,
250,
251]. However, research shows that specific type I IFN (α2/α4/β) as well as type III IFN (λ2/3) gene transcripts can be upregulated in the respiratory tract during COVID–19, but not IL1B or IL–6 (in nasopharyngeal swabs ) [
31]. These results would therefore imply that neutrophils may not be causal during chronic COVID–19 remaining unknown [
31]. Therefore, the type III IFN receptors that are expressed could be researched further. Albeit, the gene transcripts of type III IFN (λ1, λ2, λ3) may correlate with viral load, type I IFN (IFN–β, IFN–α2, IFN–α4) gene transcripts correlated with COVID–19 individual human immune responses described above. This is reflective that nuclear IFN subtype transcription occurs with viral pathogens affecting metabolic, cytosolic pathways or nuclear pathways protein translocation.
Such other factors are becoming evident with the investigations into MIS–C, another pathology not yet understood. Although various theories circulate regarding the origins of SARS–CoV–2, it is important to note that there is a semblance of superantigen–like–properties. Notable other studies indicate that a combined diagnostic approach could be utilised exemplified by a combined CD64/CD169 diagnostic to differentiate between bacterial/viral infections where both neutrophils and monocytes are affected [
252]. The role of cellular membrane Fc receptors requires further clarity that bind Abs to effector cell function [
177].
Prior to the pandemic, CD64 was suggested as a diagnostic marker of sepsis whilst CD169 could be considered an activation marker in other viral pathologies [
253,
254]. Yet the IFN gene signatures evoked during type I IFN homeostatic responses in the early onset of arthritis appear to be similar and mirror human type I IFN gene signatures and are observed to inhibit lyssaviruses [
180,
255]. Further details on other AI conditions and therapeutic development as above are detailed and in development [
256,
257,
258,
259,
260,
261].
Characterised this century, bat gene transcripts evoked by a type I IFN response
in vivo appear to be
Mx1,
ISG15, IFIT3, and
ISG56 that could be individually unique to type I IFN-ω as well as unknown so far [
253]. Type I IFN–ω was described as antigenically distinct, understandably as different species genetic regulation is metabolically different. Yet, type I IFN–ω along with both IFN–α8 and IFN–β, were the most potent inhibitors of SARS–CoV–2 viral load
in vitro according to published research last year using human alveolar type II epithelial cells (A549) transduced with the ACE2 receptor [
236]. Few studies document which species specific pDC produce IFN subtypes [
70]. Type III IFN–λ is likely a potential major player in DC–mediated immune responses downstream of the STING, also known as transmembrane protein 173 (TMEM173) activation. It would be interesting to see the mechanisms of how type III IFN regulates and induces STING in DCs alongside DC apoptosis [
70,
79,
210,
262].
Limitations
Assay scales vary measuring host type I/III IFN with regards to the early stage of research concerning potential further prophylactic/early therapeutic effects highlighted in 2019 [
263]. A common problem of clinical trial completion is a lack of funding with insufficient participants. Likewise, the bioavailability of natural or recombinant IFNs has an impact upon the severity of multiple diseases. Adverse effects of recombinant IFN were noted in some pharmacokinetic studies [
264]; however other studies are indicative that the production vectors used could be effective in delivering the appropriate pharmacokinetic profile further using other synthetic IFN derivatives.
Conclusion
In conclusion, pathogenic microbes and human organisms have co–existed and will evolve regardless of first–line human immunity. While SARS–CoV–2 and other viral pathogens as well as protein mutations discovered during development affect the homeostatic immune system balance. Some disorders may impair the IFN system through subsequent recognition by host cell pattern recognition receptors. Given the above, and the results from NCT data, type I/III IFN therapeutic research is worthy of further investigation as a potential prophylactic treatment. It can be seen in small cohort studies that type I IFN was seen to be reduced during severe COVID–19. Developments in single–cell RNA (scRNA) sequencing gene transcripts have occurred and above is discussed where type III IFN could be a factor during the host immune response. Therefore, though the prophylactic uses of IFN small MW proteins continue to be investigated, the outline above should serve as a complete analysis of IFN subtypes in health and disease. The IFN gene regulatory pathways have been detailed.
Type I IFN was researched heavily before the SARS–CoV–2 pandemic, and also during oncogenic pathologies. It can be considered that SARS–CoV–2 viral proteins affect complexities of type I/II/III IFN subtype regulation. Therefore, this added layer of immune cell regulation remains subject to further research. Furthermore, studies appearing since 2022, although small cohorts, do consistently show type I IFN reduction in patients during the COVID–19 pandemic. While vaccines have prevented severe COVID–19, cellular factors require detail. Examining further administration of type I IFN from NCTs clarifies that other type I IFNs like IFNα–2b, IFN–ω, as well as type III IFN (λ1/2/3) in hosts may counteract act cellular infection to stimulate a robust and natural immune response against viral/neoplastic or other pathologies. Likewise, this intervention fits the definition of a traditional immunogen. However, because this requires further research, the potential immunisation process can be considered a long–term process.
Above is discussed the variability in IFN synthesis in both immunodeficiency as well as current knowledge of IFN subtypes that may yet have unknown biological roles. The complexities of STAT proteins have been compared throughout pathologies, some of which were only considered in the 21st century during information technology (IT) development. This report should therefore serve academics, clinicians and researchers as a holistic overview of interferon roles in health and disease.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org,
Figure S1: Type I Interferon Receptor/Ligand Binding.;
Figure S2: Systemic Interferon Signalling;
Figure S3: STAT3 Protein role in immune system and cancer;
Figure S4 Protein and Cytokine STAT Protein Interactions Summary known today;
Figure S5: The Role of Type I Interferon in the Immune System;
Figure S6: SARS–COV–2 Genome Structure;
Table S1: SARS–CoV–2 Open Reading Frames (ORF);
Table S2: Genetic Regulation during Interferon Signalling;
Table S3: Current FDA licensed Interferon Therapeutics; UWA Expert Series – A few daily drops may have the potential to guard against COVID–19 : Archive Page : The University of Western Australia; Welcome to Interferome (monash.edu.au); The results of controlled observations on the prophylaxis of influenza with interferon – PubMed (nih.gov); PMC2427762.pdf (who.int); Home - ClinicalTrials.gov; European Medicines Agency | (europa.eu); U.S. Food and Drug Administration (fda.gov); MHRA Products | Home; Haemophilus influenzae: guidance, data and analysis - GOV.UK (
www.gov.uk);
https://www.wma.net/what-we-do/medical-ethics/declaration-of-helsinki//
Author Contributions
Original manuscript draft written by, B.B.; conceptualisation by: B.B., formal analysis by: B.B.; data curation by: B.B.; writing—reviewing and editing by: B.B., C.I., I.F., P.M.: software: B.B.: visualization: B.B., I.F.; supervision by B.B.; validation: B.B., All authors have signed, read and agreed to the published version of the manuscript according to IJCME guidelines.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not–for–profit sectors.
Institutional Review Board Statement
Not applicable.
Ethical Approval Statement and/or Informed Consent Statement
See originating citations and
Supplementary Materials. The author (s) have given signed consent. Individual citations are subject to the Helsinki Declaration as above and individual regulatory authorities.
Trial registration details
Not applicable contained within citations.
Conflict or Competing Interest
The author(s) declares no conflict of interest in the production of this manuscript.
Use of Artificial Intelligence (AI) Disclaimer
During the preparation of this work the author(s) did not use AI tools or service. Citations are referenced using Mendeleey. After using the above tool/service, each author(s) reviewed and edited the content.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Brown, B.; Ojha, V.; Fricke, I.; Al-Sheboul, S.A.; Imarogbe, C.; Gravier, T.; Green, M.; Peterson, L.; Koutsaroff, I.P.; Demir, A.; et al. Innate and Adaptive Immunity during SARS-CoV-2 Infection: Biomolecular Cellular Markers and Mechanisms. Vaccines (Basel) 2023, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- Brown, B. Immunopathogenesis of Orthopoxviridae: Insights into Immunology from Smallpox to Monkeypox (Mpox). 2023. [Google Scholar] [CrossRef]
- Brown, B. Filoviridae: Insights into Immune Responses to Ebolavirus. 2023. [Google Scholar] [CrossRef]
- Sheng, L.; Chen, X.; Wang, Q.; Lyu, S.; Li, P. Interferon-A2b Enhances Survival and Modulates Transcriptional Profiles and the Immune Response in Melanoma Patients Treated with Dendritic Cell Vaccines. Biomedicine & Pharmacotherapy 2020, 125, 109966. [Google Scholar] [CrossRef] [PubMed]
- Mesev, E. V.; LeDesma, R.A.; Ploss, A. Decoding Type I and III Interferon Signalling during Viral Infection. Nat Microbiol 2019, 4, 914–924. [Google Scholar] [CrossRef]
- Högner, K.; Wolff, T.; Pleschka, S.; Plog, S.; Gruber, A.D.; Kalinke, U.; Walmrath, H.-D.; Bodner, J.; Gattenlöhner, S.; Lewe-Schlosser, P.; et al. Macrophage-Expressed IFN-β Contributes to Apoptotic Alveolar Epithelial Cell Injury in Severe Influenza Virus Pneumonia. PLoS Pathog 2013, 9, e1003188. [Google Scholar] [CrossRef]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef]
- O’Brien, T.R.; Prokunina-Olsson, L.; Donnelly, R.P. IFN-Λ4: The Paradoxical New Member of the Interferon Lambda Family. Journal of Interferon & Cytokine Research 2014, 34, 829–838. [Google Scholar] [CrossRef]
- Shields, L.E.; Jennings, J.; Liu, Q.; Lee, J.; Ma, W.; Blecha, F.; Miller, L.C.; Sang, Y. Cross-Species Genome-Wide Analysis Reveals Molecular and Functional Diversity of the Unconventional Interferon-ω Subtype. Front Immunol 2019, 10. [Google Scholar] [CrossRef]
- Wittling, M.C.; Cahalan, S.R.; Levenson, E.A.; Rabin, R.L. Shared and Unique Features of Human Interferon-Beta and Interferon-Alpha Subtypes. Front Immunol 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Tachedjian, M.; Wynne, J.W.; Boyd, V.; Cui, J.; Smith, I.; Cowled, C.; Ng, J.H.J.; Mok, L.; Michalski, W.P.; et al. Contraction of the Type I IFN Locus and Unusual Constitutive Expression of IFN-α in Bats. Proceedings of the National Academy of Sciences 2016, 113, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Mattapallil, J.J. Interferon-α Subtypes As an Adjunct Therapeutic Approach for Human Immunodeficiency Virus Functional Cure. Front Immunol 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Deckers, J.; Anbergen, T.; Hokke, A.M.; de Dreu, A.; Schrijver, D.P.; de Bruin, K.; Toner, Y.C.; Beldman, T.J.; Spangler, J.B.; de Greef, T.F.A.; et al. Engineering Cytokine Therapeutics. Nature Reviews Bioengineering 2023, 1, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Paul, F.; Pellegrini, S.; Uzé, G. IFNA2: The Prototypic Human Alpha Interferon. Gene 2015, 567, 132–137. [Google Scholar] [CrossRef]
- Giron, L.B.; Colomb, F.; Papasavvas, E.; Azzoni, L.; Yin, X.; Fair, M.; Anzurez, A.; Damra, M.; Mounzer, K.; Kostman, J.R.; et al. Interferon-α Alters Host Glycosylation Machinery during Treated HIV Infection. EBioMedicine 2020, 59, 102945. [Google Scholar] [CrossRef]
- Liu, S.; Liu, X. IgG N-Glycans. In; 2021; pp. 1–47.
- Katla, S.; Yoganand, K.N.R.; Hingane, S.; Ranjith Kumar, C.T.; Anand, B.; Sivaprakasam, S. Novel Glycosylated Human Interferon Alpha 2b Expressed in Glycoengineered Pichia Pastoris and Its Biological Activity: N-Linked Glycoengineering Approach. Enzyme Microb Technol 2019, 128, 49–58. [Google Scholar] [CrossRef]
- How, J.; Hobbs, G. Use of Interferon Alfa in the Treatment of Myeloproliferative Neoplasms: Perspectives and Review of the Literature. Cancers (Basel) 2020, 12, 1954. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia Pastoris : A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J Cell Physiol 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Chung, J.-H.; Hong, S.-H.; Seo, N.; Kim, T.-S.; An, H.J.; Lee, P.; Shin, E.-C.; Kim, H.M. Structure-Based Glycoengineering of Interferon Lambda 4 Enhances Its Productivity and Anti-Viral Potency. Cytokine 2020, 125, 154833. [Google Scholar] [CrossRef]
- Li, S.; Gong, M.; Zhao, F.; Shao, J.; Xie, Y.; Zhang, Y.; Chang, H. Type I Interferons: Distinct Biological Activities and Current Applications for Viral Infection. Cellular Physiology and Biochemistry 2018, 51, 2377–2396. [Google Scholar] [CrossRef]
- Zanin, N.; Viaris de Lesegno, C.; Lamaze, C.; Blouin, C.M. Interferon Receptor Trafficking and Signaling: Journey to the Cross Roads. Front Immunol 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Lochte, S.; Piehler, J.; Schreiber, G. IFNAR1 and IFNAR2 Play Distinct Roles in Initiating Type I Interferon–Induced JAK-STAT Signaling and Activating STATs. Sci Signal 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, T.B.; Kalie, E.; Crisafulli-Cabatu, S.; Abramovich, R.; DiGioia, G.; Moolchan, K.; Pestka, S.; Schreiber, G. Binding and Activity of All Human Alpha Interferon Subtypes. Cytokine 2011, 56, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Shin, E.-C. Type I and III Interferon Responses in SARS-CoV-2 Infection. Exp Mol Med 2021, 53, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mann-Nüttel, R.; Schulze, A.; Richter, L.; Alferink, J.; Scheu, S. Sources of Type I Interferons in Infectious Immunity: Plasmacytoid Dendritic Cells Not Always in the Driver’s Seat. Front Immunol 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, T.J.; Gonye, A.L.K.; Freeman, S.S.; Kaplonek, P.; Gushterova, I.; Kays, K.R.; Manakongtreecheep, K.; Tantivit, J.; Rojas-Lopez, M.; Russo, B.C.; et al. Longitudinal Characterization of Circulating Neutrophils Uncovers Phenotypes Associated with Severity in Hospitalized COVID-19 Patients. Cell Rep Med 2022, 3, 100779. [Google Scholar] [CrossRef]
- de Weerd, N.A.; Samarajiwa, S.A.; Hertzog, P.J. Type I Interferon Receptors: Biochemistry and Biological Functions. Journal of Biological Chemistry 2007, 282, 20053–20057. [Google Scholar] [CrossRef]
- Lam, J.-Y.; Yuen, C.-K.; Ip, J.D.; Wong, W.-M.; To, K.K.-W.; Yuen, K.-Y.; Kok, K.-H. Loss of Orf3b in the Circulating SARS-CoV-2 Strains. Emerg Microbes Infect 2020, 9, 2685–2696. [Google Scholar] [CrossRef]
- Overholt, K.J.; Krog, J.R.; Zanoni, I.; Bryson, B.D. Dissecting the Common and Compartment-Specific Features of COVID-19 Severity in the Lung and Periphery with Single-Cell Resolution. iScience 2021, 24, 102738. [Google Scholar] [CrossRef]
- Sposito, B.; Broggi, A.; Pandolfi, L.; Crotta, S.; Clementi, N.; Ferrarese, R.; Sisti, S.; Criscuolo, E.; Spreafico, R.; Long, J.M.; et al. The Interferon Landscape along the Respiratory Tract Impacts the Severity of COVID-19. Cell 2021, 184, 4953–4968. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral Activities of Type I Interferons to SARS-CoV-2 Infection. Antiviral Res 2020, 179, 104811. [Google Scholar] [CrossRef]
- Brzoska, J.; von Eick, H.; Hündgen, M. Interferons in COVID-19: Missed Opportunities to Prove Efficacy in Clinical Phase III Trials? Front Med (Lausanne) 2023, 10. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I Interferons in Infectious Disease. Nat Rev Immunol 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Villani, A.-C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S.; et al. Single-Cell RNA-Seq Reveals New Types of Human Blood Dendritic Cells, Monocytes, and Progenitors. Science (1979) 2017, 356. [Google Scholar] [CrossRef]
- Di Blasio, S.; van Wigcheren, G.F.; Becker, A.; van Duffelen, A.; Gorris, M.; Verrijp, K.; Stefanini, I.; Bakker, G.J.; Bloemendal, M.; Halilovic, A.; et al. The Tumour Microenvironment Shapes Dendritic Cell Plasticity in a Human Organotypic Melanoma Culture. Nat Commun 2020, 11, 2749. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Yau, D.; Holbrook, C.; Reder, A.T. Type I Interferons Inhibit Interleukin-10 Production in Activated Human Monocytes and Stimulate IL-10 in T Cells: Implications for Th1-Mediated Diseases. Journal of Interferon & Cytokine Research 2002, 22, 311–319. [Google Scholar] [CrossRef]
- Blumer, T.; Coto-Llerena, M.; Duong, F.H.T.; Heim, M.H. SOCS1 Is an Inducible Negative Regulator of Interferon λ (IFN-λ)–Induced Gene Expression in Vivo. Journal of Biological Chemistry 2017, 292, 17928–17938. [Google Scholar] [CrossRef]
- Klopfenstein, N.; Brandt, S.L.; Castellanos, S.; Gunzer, M.; Blackman, A.; Serezani, C.H. SOCS-1 Inhibition of Type I Interferon Restrains Staphylococcus Aureus Skin Host Defense. PLoS Pathog 2021, 17, e1009387. [Google Scholar] [CrossRef]
- Piganis, R.A.R.; De Weerd, N.A.; Gould, J.A.; Schindler, C.W.; Mansell, A.; Nicholson, S.E.; Hertzog, P.J. Suppressor of Cytokine Signaling (SOCS) 1 Inhibits Type I Interferon (IFN) Signaling via the Interferon α Receptor (IFNAR1)-Associated Tyrosine Kinase Tyk2. Journal of Biological Chemistry 2011, 286, 33811–33818. [Google Scholar] [CrossRef] [PubMed]
- Medrano, R.F.V.; Hunger, A.; Mendonça, S.A.; Barbuto, J.A.M.; Strauss, B.E. Immunomodulatory and Antitumor Effects of Type I Interferons and Their Application in Cancer Therapy. Oncotarget 2017, 8, 71249–71284. [Google Scholar] [CrossRef]
- Belardelli, F.; Ferrantini, M.; Proietti, E.; Kirkwood, J.M. Interferon-Alpha in Tumor Immunity and Immunotherapy. Cytokine Growth Factor Rev 2002, 13, 119–134. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in Tumor Progression and Regression: A Review. Biomark Res 2020, 8, 49. [Google Scholar] [CrossRef]
- Syedbasha, M.; Egli, A. Interferon Lambda: Modulating Immunity in Infectious Diseases. Front Immunol 2017, 8. [Google Scholar] [CrossRef]
- Miknis, Z.J.; Magracheva, E.; Li, W.; Zdanov, A.; Kotenko, S. V.; Wlodawer, A. Crystal Structure of Human Interferon-Λ1 in Complex with Its High-Affinity Receptor Interferon-ΛR1. J Mol Biol 2010, 404, 650–664. [Google Scholar] [CrossRef]
- Egli, A.; Santer, D.M.; O’Shea, D.; Barakat, K.; Syedbasha, M.; Vollmer, M.; Baluch, A.; Bhat, R.; Groenendyk, J.; Joyce, M.A.; et al. IL-28B Is a Key Regulator of B- and T-Cell Vaccine Responses against Influenza. PLoS Pathog 2014, 10, e1004556. [Google Scholar] [CrossRef] [PubMed]
- Sherer, N.M.; Mothes, W. Cytonemes and Tunneling Nanotubules in Cell-Cell Communication and Viral Pathogenesis. Trends Cell Biol 2008, 18, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, N.; Liongue, C.; Ward, A.C. STAT Proteins: A Kaleidoscope of Canonical and Non-Canonical Functions in Immunity and Cancer. J Hematol Oncol 2021, 14, 198. [Google Scholar] [CrossRef]
- Hu, X.; li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT Signaling Pathway: From Bench to Clinic. Signal Transduct Target Ther 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.; Prince, A. Participation of the IL-10RB Related Cytokines, IL-22 and IFN-λ in Defense of the Airway Mucosal Barrier. Front Cell Infect Microbiol 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, C.A. Regulating IRFs in IFN Driven Disease. Front Immunol 2019, 10. [Google Scholar] [CrossRef]
- Gal-Ben-Ari, S.; Barrera, I.; Ehrlich, M.; Rosenblum, K. PKR: A Kinase to Remember. Front Mol Neurosci 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Adenosine Deaminase Acting on RNA (ADAR1), a Suppressor of Double-Stranded RNA–Triggered Innate Immune Responses. Journal of Biological Chemistry 2019, 294, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Xue, Y. ADAR1: A Mast Regulator of Aging and Immunity. Signal Transduct Target Ther 2023, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A Virus NS1 Protein Activates the PI3K/Akt Pathway to Mediate Antiapoptotic Signaling Responses. J Virol 2007, 81, 3058–3067. [Google Scholar] [CrossRef]
- Zhang, M.; Meng, Y.; Ying, Y.; Zhou, P.; Zhang, S.; Fang, Y.; Yao, Y.; Li, D. Selective Activation of STAT3 and STAT5 Dictates the Fate of Myeloid Progenitor Cells. Cell Death Discov 2023, 9, 274. [Google Scholar] [CrossRef]
- Deimel, L.P.; Li, Z.; Roy, S.; Ranasinghe, C. STAT3 Determines IL-4 Signalling Outcomes in Naïve T Cells. Sci Rep 2021, 11, 10495. [Google Scholar] [CrossRef]
- Meyer zu Horste, G.; Przybylski, D.; Schramm, M.A.; Wang, C.; Schnell, A.; Lee, Y.; Sobel, R.; Regev, A.; Kuchroo, V.K. Fas Promotes T Helper 17 Cell Differentiation and Inhibits T Helper 1 Cell Development by Binding and Sequestering Transcription Factor STAT1. Immunity 2018, 48, 556–569. [Google Scholar] [CrossRef]
- Lee, D.; Le Pen, J.; Yatim, A.; Dong, B.; Aquino, Y.; Ogishi, M.; Pescarmona, R.; Talouarn, E.; Rinchai, D.; Zhang, P.; et al. Inborn Errors of OAS–RNase L in SARS-CoV-2–Related Multisystem Inflammatory Syndrome in Children. Science (1979) 2023, 379. [Google Scholar] [CrossRef]
- Gurlevik, S.L.; Ozsurekci, Y.; Sağ, E.; Derin Oygar, P.; Kesici, S.; Akca, Ü.K.; Cuceoglu, M.K.; Basaran, O.; Göncü, S.; Karakaya, J.; et al. The Difference of the Inflammatory Milieu in MIS-C and Severe COVID-19. Pediatr Res 2022, 92, 1805–1814. [Google Scholar] [CrossRef]
- Kang, J.A.; Kim, Y.J.; Jeon, Y.J. The Diverse Repertoire of ISG15: More Intricate than Initially Thought. Exp Mol Med 2022, 54, 1779–1792. [Google Scholar] [CrossRef]
- Jiménez Fernández, D.; Hess, S.; Knobeloch, K.-P. Strategies to Target ISG15 and USP18 Toward Therapeutic Applications. Front Chem 2020, 7. [Google Scholar] [CrossRef]
- Dubois, A.; François, C.; Descamps, V.; Fournier, C.; Wychowski, C.; Dubuisson, J.; Castelain, S.; Duverlie, G. Enhanced Anti-HCV Activity of Interferon Alpha 17 Subtype. Virol J 2009, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Scagnolari, C.; Trombetti, S.; Selvaggi, C.; Carbone, T.; Monteleone, K.; Spano, L.; Di Marco, P.; Pierangeli, A.; Maggi, F.; Riva, E.; et al. In Vitro Sensitivity of Human Metapneumovirus to Type I Interferons. Viral Immunol 2011, 24, 159–164. [Google Scholar] [CrossRef]
- Markušić, M.; Šantak, M.; Košutić-Gulija, T.; Jergović, M.; Jug, R.; Forčić, D. Induction of IFN- α Subtypes and Their Antiviral Activity in Mumps Virus Infection. Viral Immunol 2014, 27, 497–505. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Wang, Y.; Zhao, P.; Cui, C.; Tu, L.; Li, X.; Yu, Y.; Li, H.; Wang, L. Diversity of Locally Produced IFN-α Subtypes in Human Nasopharyngeal Epithelial Cells and Mouse Lung Tissues during Influenza Virus Infection. Appl Microbiol Biotechnol 2020, 104, 6351–6361. [Google Scholar] [CrossRef]
- Safadi, D. El; Lebeau, G.; Lagrave, A.; Mélade, J.; Grondin, L.; Rosanaly, S.; Begue, F.; Hoareau, M.; Veeren, B.; Roche, M.; et al. Extracellular Vesicles Are Conveyors of the NS1 Toxin during Dengue Virus and Zika Virus Infection. Viruses 2023, 15, 364. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.; Gravier, T.; Fricke, I.; Al-Sheboul, S.A.; Carp, T.-N.; Leow, C.Y.; Imarogbe, C.; Arabpour, J. Immunopathogenesis of Nipah Virus Infection and Associated Immune Responses. Immuno 2023, 3, 160–181. [Google Scholar] [CrossRef]
- Mateo, M.; Reid, S.P.; Leung, L.W.; Basler, C.F.; Volchkov, V.E. Ebolavirus VP24 Binding to Karyopherins Is Required for Inhibition of Interferon Signaling. J Virol 2010, 84, 1169–1175. [Google Scholar] [CrossRef]
- Ilinykh, P.A.; Lubaki, N.M.; Widen, S.G.; Renn, L.A.; Theisen, T.C.; Rabin, R.L.; Wood, T.G.; Bukreyev, A. Different Temporal Effects of Ebola Virus VP35 and VP24 Proteins on Global Gene Expression in Human Dendritic Cells. J Virol 2015, 89, 7567–7583. [Google Scholar] [CrossRef]
- Leung, L.W.; Park, M.-S.; Martinez, O.; Valmas, C.; López, C.B.; Basler, C.F. Ebolavirus VP35 Suppresses IFN Production from Conventional but Not Plasmacytoid Dendritic Cells. Immunol Cell Biol 2011, 89, 792–802. [Google Scholar] [CrossRef]
- Kuroda, M.; Halfmann, P.J.; Hill-Batorski, L.; Ozawa, M.; Lopes, T.J.S.; Neumann, G.; Schoggins, J.W.; Rice, C.M.; Kawaoka, Y. Identification of Interferon-Stimulated Genes That Attenuate Ebola Virus Infection. Nat Commun 2020, 11, 2953. [Google Scholar] [CrossRef] [PubMed]
- Woolsey, C.; Menicucci, A.R.; Cross, R.W.; Luthra, P.; Agans, K.N.; Borisevich, V.; Geisbert, J.B.; Mire, C.E.; Fenton, K.A.; Jankeel, A.; et al. A VP35 Mutant Ebola Virus Lacks Virulence but Can Elicit Protective Immunity to Wild-Type Virus Challenge. Cell Rep 2019, 28, 3032–3046. [Google Scholar] [CrossRef] [PubMed]
- Coldbeck-Shackley, R.C.; Romeo, O.; Rosli, S.; Gearing, L.J.; Gould, J.A.; Lim, S.S.; Van der Hoek, K.H.; Eyre, N.S.; Shue, B.; Robertson, S.A.; et al. Constitutive Expression and Distinct Properties of IFN-Epsilon Protect the Female Reproductive Tract from Zika Virus Infection. PLoS Pathog 2023, 19, e1010843. [Google Scholar] [CrossRef]
- Toledo Pinto, T.G.; Batista-Silva, L.R.; Medeiros, R.C.A.; Lara, F.A.; Moraes, M.O. Type I Interferons, Autophagy and Host Metabolism in Leprosy. Front Immunol 2018, 9. [Google Scholar] [CrossRef]
- Irving, A.T.; Zhang, Q.; Kong, P.-S.; Luko, K.; Rozario, P.; Wen, M.; Zhu, F.; Zhou, P.; Ng, J.H.J.; Sobota, R.M.; et al. Interferon Regulatory Factors IRF1 and IRF7 Directly Regulate Gene Expression in Bats in Response to Viral Infection. Cell Rep 2020, 33, 108345. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, G.; Zhu, L.; Zhao, Z.; Liu, Y.; Han, W.; Zhang, X.; Zhang, Y.; Xiong, T.; Zeng, H.; et al. HIV-1 Vif Suppresses Antiviral Immunity by Targeting STING. Cell Mol Immunol 2022, 19, 108–121. [Google Scholar] [CrossRef]
- Khatamzas, E.; Hipp, M.M.; Gaughan, D.; Pichulik, T.; Leslie, A.; Fernandes, R.A.; Muraro, D.; Booth, S.; Zausmer, K.; Sun, M.-Y.; et al. Snapin Promotes HIV-1 Transmission from Dendritic Cells by Dampening TLR8 Signaling. EMBO J 2017, 36, 2998–3011. [Google Scholar] [CrossRef]
- Guo, K.; Barrett, B.S.; Morrison, J.H.; Mickens, K.L.; Vladar, E.K.; Hasenkrug, K.J.; Poeschla, E.M.; Santiago, M.L. Interferon Resistance of Emerging SARS-CoV-2 Variants. Proceedings of the National Academy of Sciences 2022, 119. [Google Scholar] [CrossRef]
- Bucciol, G.; Moens, L.; Ogishi, M.; Rinchai, D.; Matuozzo, D.; Momenilandi, M.; Kerrouche, N.; Cale, C.M.; Treffeisen, E.R.; Al Salamah, M.; et al. Human Inherited Complete STAT2 Deficiency Underlies Inflammatory Viral Diseases. Journal of Clinical Investigation 2023, 133. [Google Scholar] [CrossRef] [PubMed]
- Milner, J.J.; Toma, C.; Yu, B.; Zhang, K.; Omilusik, K.; Phan, A.T.; Wang, D.; Getzler, A.J.; Nguyen, T.; Crotty, S.; et al. Runx3 Programs CD8+ T Cell Residency in Non-Lymphoid Tissues and Tumours. Nature 2017, 552, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, L.; Estecha, A.; Samaniego, R.; Sánchez-Ramón, S.; Vega, M.Á.; Sánchez-Mateos, P. The Chemokine CXCL12 Regulates Monocyte-Macrophage Differentiation and RUNX3 Expression. Blood 2011, 117, 88–97. [Google Scholar] [CrossRef]
- Henig, N.; Avidan, N.; Mandel, I.; Staun-Ram, E.; Ginzburg, E.; Paperna, T.; Pinter, R.Y.; Miller, A. Interferon-Beta Induces Distinct Gene Expression Response Patterns in Human Monocytes versus T Cells. PLoS One 2013, 8, e62366. [Google Scholar] [CrossRef]
- Kerner, G.; Rosain, J.; Guérin, A.; Al-Khabaz, A.; Oleaga-Quintas, C.; Rapaport, F.; Massaad, M.J.; Ding, J.-Y.; Khan, T.; Ali, F. Al; et al. Inherited Human IFN-γ Deficiency Underlies Mycobacterial Disease. Journal of Clinical Investigation 2020, 130, 3158–3171. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dupuis, S.; Kong, X.-F.; Okada, S.; Cypowyj, S.; Puel, A.; Abel, L.; Casanova, J.-L. Inborn Errors of Human STAT1: Allelic Heterogeneity Governs the Diversity of Immunological and Infectious Phenotypes. Curr Opin Immunol 2012, 24, 364–378. [Google Scholar] [CrossRef]
- Bogunovic, D.; Byun, M.; Durfee, L.A.; Abhyankar, A.; Sanal, O.; Mansouri, D.; Salem, S.; Radovanovic, I.; Grant, A. V.; Adimi, P.; et al. Mycobacterial Disease and Impaired IFN-γ Immunity in Humans with Inherited ISG15 Deficiency. Science (1979) 2012, 337, 1684–1688. [Google Scholar] [CrossRef]
- Crow, Y.J.; Stetson, D.B. The Type I Interferonopathies: 10 Years On. Nat Rev Immunol 2022, 22, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.-P.; Ding, J.-Y.; Yeh, C.-F.; Chi, C.-Y.; Ku, C.-L. Anti-Interferon-γ Autoantibody-Associated Immunodeficiency. Curr Opin Immunol 2021, 72, 206–214. [Google Scholar] [CrossRef]
- Dupuis, S.; Döffinger, R.; Picard, C.; Fieschi, C.; Altare, F.; Jouanguy, E.; Abel, L.; Casanova, J.-L. Human Interferon-g-Mediated Immunity Is a Genetically Controlled Continuous Trait That Determines the Outcome of Mycobacterial Invasion. Immunol Rev 2000, 178, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Kreins, A.Y.; Ciancanelli, M.J.; Okada, S.; Kong, X.-F.; Ramírez-Alejo, N.; Kilic, S.S.; El Baghdadi, J.; Nonoyama, S.; Mahdaviani, S.A.; Ailal, F.; et al. Human TYK2 Deficiency: Mycobacterial and Viral Infections without Hyper-IgE Syndrome. Journal of Experimental Medicine 2015, 212, 1641–1662. [Google Scholar] [CrossRef]
- van de Vosse, E.; Haverkamp, M.H.; Ramirez-Alejo, N.; Martinez-Gallo, M.; Blancas-Galicia, L.; Metin, A.; Garty, B.Z.; Sun-Tan, Ç.; Broides, A.; de Paus, R.A.; et al. IL-12Rβ1 Deficiency: Mutation Update and Description of the IL12RB1 Variation Database. Hum Mutat 2013, 34, 1329–1339. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; van de Veerdonk, F.L.; Crossland, K.L.; Smeekens, S.P.; Chan, C.M.; Al Shehri, T.; Abinun, M.; Gennery, A.R.; Mann, J.; Lendrem, D.W.; et al. Gain-of-Function STAT1 Mutations Impair STAT3 Activity in Patients with Chronic Mucocutaneous Candidiasis (CMC). Eur J Immunol 2015, 45, 2834–2846. [Google Scholar] [CrossRef] [PubMed]
- Passarelli, C.; Civino, A.; Rossi, M.N.; Cifaldi, L.; Lanari, V.; Moneta, G.M.; Caiello, I.; Bracaglia, C.; Montinaro, R.; Novelli, A.; et al. IFNAR2 Deficiency Causing Dysregulation of NK Cell Functions and Presenting With Hemophagocytic Lymphohistiocytosis. Front Genet 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Sakata, S.; Tsumura, M.; Matsubayashi, T.; Karakawa, S.; Kimura, S.; Tamaura, M.; Okano, T.; Naruto, T.; Mizoguchi, Y.; Kagawa, R.; et al. Autosomal Recessive Complete STAT1 Deficiency Caused by Compound Heterozygous Intronic Mutations. Int Immunol 2020, 32, 663–671. [Google Scholar] [CrossRef]
- Lim, H.K.; Huang, S.X.L.; Chen, J.; Kerner, G.; Gilliaux, O.; Bastard, P.; Dobbs, K.; Hernandez, N.; Goudin, N.; Hasek, M.L.; et al. Severe Influenza Pneumonitis in Children with Inherited TLR3 Deficiency. Journal of Experimental Medicine 2019, 216, 2038–2056. [Google Scholar] [CrossRef]
- Sakata, S.; Tsumura, M.; Matsubayashi, T.; Karakawa, S.; Kimura, S.; Tamaura, M.; Okano, T.; Naruto, T.; Mizoguchi, Y.; Kagawa, R.; et al. Autosomal Recessive Complete STAT1 Deficiency Caused by Compound Heterozygous Intronic Mutations. Int Immunol 2020, 32, 663–671. [Google Scholar] [CrossRef]
- Alphonse, N.; Wanford, J.J.; Voak, A.A.; Gay, J.; Venkhaya, S.; Burroughs, O.; Mathew, S.; Lee, T.; Evans, S.L.; Zhao, W.; et al. A Family of Conserved Bacterial Virulence Factors Dampens Interferon Responses by Blocking Calcium Signaling. Cell 2022, 185, 2354–2369. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jing, H.; Martin-Nalda, A.; Bastard, P.; Rivière, J.G.; Liu, Z.; Colobran, R.; Lee, D.; Tung, W.; Manry, J.; et al. Inborn Errors of TLR3- or MDA5-Dependent Type I IFN Immunity in Children with Enterovirus Rhombencephalitis. Journal of Experimental Medicine 2021, 218. [Google Scholar] [CrossRef]
- Duncan, C.J.A.; Hambleton, S. Human Disease Phenotypes Associated with Loss and Gain of Function Mutations in STAT2: Viral Susceptibility and Type I Interferonopathy. J Clin Immunol 2021, 41, 1446–1456. [Google Scholar] [CrossRef]
- Holland, S.M.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.; et al. STAT3 Mutations in the Hyper-IgE Syndrome. New England Journal of Medicine 2007, 357, 1608–1619. [Google Scholar] [CrossRef]
- Takenaka, M.C.; Quintana, F.J. Tolerogenic Dendritic Cells. Semin Immunopathol 2017, 39, 113–120. [Google Scholar] [CrossRef]
- Parackova, Z.; Zentsova, I.; Vrabcova, P.; Sediva, A.; Bloomfield, M. Aberrant Tolerogenic Functions and Proinflammatory Skew of Dendritic Cells in STAT1 Gain-of-Function Patients May Contribute to Autoimmunity and Fungal Susceptibility. Clinical Immunology 2023, 246, 109174. [Google Scholar] [CrossRef]
- Wolf, Y.; Anderson, A.C.; Kuchroo, V.K. TIM3 Comes of Age as an Inhibitory Receptor. Nat Rev Immunol 2020, 20, 173–185. [Google Scholar] [CrossRef]
- Lupov, I.P.; Voiles, L.; Han, L.; Schwartz, A.; De La Rosa, M.; Oza, K.; Pelloso, D.; Sahu, R.P.; Travers, J.B.; Robertson, M.J.; et al. Acquired STAT4 Deficiency as a Consequence of Cancer Chemotherapy. Blood 2011, 118, 6097–6106. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Mai, H.; Peng, J.; Zhou, B.; Hou, J.; Jiang, D. STAT4: An Immunoregulator Contributing to Diverse Human Diseases. Int J Biol Sci 2020, 16, 1575–1585. [Google Scholar] [CrossRef]
- Meng, Z.-Z.; Liu, W.; Xia, Y.; Yin, H.-M.; Zhang, C.-Y.; Su, D.; Yan, L.-F.; Gu, A.-H.; Zhou, Y. The Pro-Inflammatory Signalling Regulator Stat4 Promotes Vasculogenesis of Great Vessels Derived from Endothelial Precursors. Nat Commun 2017, 8, 14640. [Google Scholar] [CrossRef]
- Torpey, N.; Maher, S.E.; Bothwell, A.L.M.; Pober, J.S. Interferon α but Not Interleukin 12 Activates STAT4 Signaling in Human Vascular Endothelial Cells. Journal of Biological Chemistry 2004, 279, 26789–26796. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, G.M.; Grassmann, S.; Lau, C.M.; Rapp, M.; Villarino, A. V.; Friedrich, C.; Gasteiger, G.; O’Shea, J.J.; Sun, J.C. Divergent Role for STAT5 in the Adaptive Responses of Natural Killer Cells. Cell Rep 2020, 33, 108498. [Google Scholar] [CrossRef]
- Hatami, M.; Salmani, T.; Arsang-Jang, S.; Davood Omrani, M.; Mazdeh, M.; Ghafouri-Fard, S.; Sayad, A.; Taheri, M. STAT5a and STAT6 Gene Expression Levels in Multiple Sclerosis Patients. Cytokine 2018, 106, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Sumida, T.S.; Dulberg, S.; Schupp, J.C.; Lincoln, M.R.; Stillwell, H.A.; Axisa, P.-P.; Comi, M.; Unterman, A.; Kaminski, N.; Madi, A.; et al. Type I Interferon Transcriptional Network Regulates Expression of Coinhibitory Receptors in Human T Cells. Nat Immunol 2022, 23, 632–642. [Google Scholar] [CrossRef]
- Kanai, T.; Jenks, J.; Nadeau, K.C. The STAT5b Pathway Defect and Autoimmunity. Front Immunol 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Karpathiou, G.; Papoudou-Bai, A.; Ferrand, E.; Dumollard, J.M.; Peoc’h, M. STAT6: A Review of a Signaling Pathway Implicated in Various Diseases with a Special Emphasis in Its Usefulness in Pathology. Pathol Res Pract 2021, 223, 153477. [Google Scholar] [CrossRef]
- Meloni, A.; Furcas, M.; Cetani, F.; Marcocci, C.; Falorni, A.; Perniola, R.; Pura, M.; Bøe Wolff, A.S.; Husebye, E.S.; Lilic, D.; et al. Autoantibodies against Type I Interferons as an Additional Diagnostic Criterion for Autoimmune Polyendocrine Syndrome Type I. J Clin Endocrinol Metab 2008, 93, 4389–4397. [Google Scholar] [CrossRef]
- von Stemann, J.H.; Rigas, A.S.; Thørner, L.W.; Rasmussen, D.G.K.; Pedersen, O.B.; Rostgaard, K.; Erikstrup, C.; Ullum, H.; Hansen, M.B. Prevalence and Correlation of Cytokine-Specific Autoantibodies with Epidemiological Factors and C-Reactive Protein in 8,972 Healthy Individuals: Results from the Danish Blood Donor Study. PLoS One 2017, 12, e0179981. [Google Scholar] [CrossRef] [PubMed]
- Ghale, R.; Spottiswoode, N.; Anderson, M.S.; Mitchell, A.; Wang, G.; Calfee, C.S.; DeRisi, J.L.; Langelier, C.R. Prevalence of Type-1 Interferon Autoantibodies in Adults with Non-COVID-19 Acute Respiratory Failure. Respir Res 2022, 23, 354. [Google Scholar] [CrossRef]
- Vanker, M.; Särekannu, K.; Fekkar, A.; Jørgensen, S.E.; Haljasmägi, L.; Kallaste, A.; Kisand, K.; Lember, M.; Peterson, P.; Menon, M.; et al. Autoantibodies Neutralizing Type III Interferons Are Uncommon in Patients with Severe Coronavirus Disease 2019 Pneumonia. Journal of Interferon & Cytokine Research 2023, 43, 379–393. [Google Scholar] [CrossRef]
- Hong, G.H.; Ortega-Villa, A.M.; Hunsberger, S.; Chetchotisakd, P.; Anunnatsiri, S.; Mootsikapun, P.; Rosen, L.B.; Zerbe, C.S.; Holland, S.M. Natural History and Evolution of Anti-Interferon-γ Autoantibody-Associated Immunodeficiency Syndrome in Thailand and the United States. Clinical Infectious Diseases 2020, 71, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against Type I IFNs in Patients with Life-Threatening COVID-19. Science (1979) 2020, 370. [Google Scholar] [CrossRef]
- Goncalves, D.; Mezidi, M.; Bastard, P.; Perret, M.; Saker, K.; Fabien, N.; Pescarmona, R.; Lombard, C.; Walzer, T.; Casanova, J.; et al. Antibodies against Type I Interferon: Detection and Association with Severe Clinical Outcome in COVID-19 Patients. Clin Transl Immunology 2021, 10. [Google Scholar] [CrossRef]
- Gervais, A.; Rovida, F.; Avanzini, M.A.; Croce, S.; Marchal, A.; Lin, S.-C.; Ferrari, A.; Thorball, C.W.; Constant, O.; Le Voyer, T.; et al. Autoantibodies Neutralizing Type I IFNs Underlie West Nile Virus Encephalitis in ∼40% of Patients. Journal of Experimental Medicine 2023, 220. [Google Scholar] [CrossRef]
- Pierangeli, A.; Gentile, M.; Oliveto, G.; Frasca, F.; Sorrentino, L.; Matera, L.; Nenna, R.; Viscido, A.; Fracella, M.; Petrarca, L.; et al. Comparison by Age of the Local Interferon Response to SARS-CoV-2 Suggests a Role for IFN-ε and -ω. Front Immunol 2022, 13. [Google Scholar] [CrossRef]
- Zhu, H.; Zheng, F.; Li, L.; Jin, Y.; Luo, Y.; Li, Z.; Zeng, J.; Tang, L.; Li, Z.; Xia, N.; et al. A Chinese Host Genetic Study Discovered IFNs and Causality of Laboratory Traits on COVID-19 Severity. iScience 2021, 24, 103186. [Google Scholar] [CrossRef]
- Martin-Sancho, L.; Lewinski, M.K.; Pache, L.; Stoneham, C.A.; Yin, X.; Becker, M.E.; Pratt, D.; Churas, C.; Rosenthal, S.B.; Liu, S.; et al. Functional Landscape of SARS-CoV-2 Cellular Restriction. Mol Cell 2021, 81, 2656–2668. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.-C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef]
- Bedford, J.G.; O’Keeffe, M.; Reading, P.C.; Wakim, L.M. Rapid Interferon Independent Expression of IFITM3 Following T Cell Activation Protects Cells from Influenza Virus Infection. PLoS One 2019, 14, e0210132. [Google Scholar] [CrossRef]
- Yánez, D.C.; Ross, S.; Crompton, T. The IFITM Protein Family in Adaptive Immunity. Immunology 2020, 159, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Compton, A.A.; Roy, N.; Porrot, F.; Billet, A.; Casartelli, N.; Yount, J.S.; Liang, C.; Schwartz, O. Natural Mutations in IFITM3 Modulate Post-translational Regulation and Toggle Antiviral Specificity. EMBO Rep 2016, 17, 1657–1671. [Google Scholar] [CrossRef]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.H.; Everitt, A.R.; Vora, M.; et al. The CD225 Domain of IFITM3 Is Required for Both IFITM Protein Association and Inhibition of Influenza A Virus and Dengue Virus Replication. J Virol 2013, 87, 7837–7852. [Google Scholar] [CrossRef]
- Klein, S.; Golani, G.; Lolicato, F.; Lahr, C.; Beyer, D.; Herrmann, A.; Wachsmuth-Melm, M.; Reddmann, N.; Brecht, R.; Hosseinzadeh, M.; et al. IFITM3 Blocks Influenza Virus Entry by Sorting Lipids and Stabilizing Hemifusion. Cell Host Microbe 2023, 31, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Kenney, A.D.; Zani, A.; Kawahara, J.; Eddy, A.C.; Wang, X.; Mahesh, K.; Lu, M.; Thomas, J.; Kohlmeier, J.E.; Suthar, M.S.; et al. Interferon-induced Transmembrane Protein 3 (IFITM3) Limits Lethality of SARS-CoV-2 in Mice. EMBO Rep 2023, 24. [Google Scholar] [CrossRef]
- Hur, J.-Y.; Frost, G.R.; Wu, X.; Crump, C.; Pan, S.J.; Wong, E.; Barros, M.; Li, T.; Nie, P.; Zhai, Y.; et al. The Innate Immunity Protein IFITM3 Modulates γ-Secretase in Alzheimer’s Disease. Nature 2020, 586, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Pidugu, V.K.; Pidugu, H.B.; Wu, M.-M.; Liu, C.-J.; Lee, T.-C. Emerging Functions of Human IFIT Proteins in Cancer. Front Mol Biosci 2019, 6. [Google Scholar] [CrossRef]
- Yu, R.; Zhu, B.; Chen, D. Type I Interferon-Mediated Tumor Immunity and Its Role in Immunotherapy. Cellular and Molecular Life Sciences 2022, 79, 191. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, D.; Cui, X.; Dai, Y.; Wang, C.; Feng, W.; Lv, X.; Li, Y.; Wang, L.; Ru, Y.; et al. Loss of IRF7 Accelerates Acute Myeloid Leukemia Progression and Induces VCAM1-VLA-4 Mediated Intracerebral Invasion. Oncogene 2022, 41, 2303–2314. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-F.; Lin, C.-M.; Lee, K.-Y.; Wu, S.-Y.; Feng, P.-H.; Chen, K.-Y.; Chuang, H.-C.; Chen, C.-L.; Wang, Y.-C.; Tseng, P.-C.; et al. Escape from IFN-γ-Dependent Immunosurveillance in Tumorigenesis. J Biomed Sci 2017, 24, 10. [Google Scholar] [CrossRef]
- Mahalapbutr, P.; Kongtaworn, N.; Rungrotmongkol, T. Structural Insight into the Recognition of S-Adenosyl-L-Homocysteine and Sinefungin in SARS-CoV-2 Nsp16/Nsp10 RNA Cap 2′-O-Methyltransferase. Comput Struct Biotechnol J 2020, 18, 2757–2765. [Google Scholar] [CrossRef] [PubMed]
- Setaro, A.C.; Gaglia, M.M. All Hands on Deck: SARS-CoV-2 Proteins That Block Early Anti-Viral Interferon Responses. Curr Res Virol Sci 2021, 2, 100015. [Google Scholar] [CrossRef]
- Yuen, C.-K.; Lam, J.-Y.; Wong, W.-M.; Mak, L.-F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 Nsp13, Nsp14, Nsp15 and Orf6 Function as Potent Interferon Antagonists. Emerg Microbes Infect 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 Hijacks Nup98 to Block STAT Nuclear Import and Antagonize Interferon Signaling. Proceedings of the National Academy of Sciences 2020, 117, 28344–28354. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, C.; Swanson, S.E.; Negatu, S.G.; Dittmar, M.; Miller, J.; Ramage, H.R.; Cherry, S.; Jurado, K.A. SARS-CoV-2 Viral Proteins NSP1 and NSP13 Inhibit Interferon Activation through Distinct Mechanisms. PLoS One 2021, 16, e0253089. [Google Scholar] [CrossRef]
- Kumar, A.; Ishida, R.; Strilets, T.; Cole, J.; Lopez-Orozco, J.; Fayad, N.; Felix-Lopez, A.; Elaish, M.; Evseev, D.; Magor, K.E.; et al. SARS-CoV-2 Nonstructural Protein 1 Inhibits the Interferon Response by Causing Depletion of Key Host Signaling Factors. J Virol 2021, 95. [Google Scholar] [CrossRef]
- Rincon-Arevalo, H.; Aue, A.; Ritter, J.; Szelinski, F.; Khadzhynov, D.; Zickler, D.; Stefanski, L.; Lino, A.C.; Körper, S.; Eckardt, K.; et al. Altered Increase in STAT1 Expression and Phosphorylation in Severe COVID-19. Eur J Immunol 2022, 52, 138–148. [Google Scholar] [CrossRef]
- Fisher, T.; Gluck, A.; Narayanan, K.; Kuroda, M.; Nachshon, A.; Hsu, J.C.; Halfmann, P.J.; Yahalom-Ronen, Y.; Tamir, H.; Finkel, Y.; et al. Parsing the Role of NSP1 in SARS-CoV-2 Infection. Cell Rep 2022, 39, 110954. [Google Scholar] [CrossRef]
- McKellar, J.; Rebendenne, A.; Wencker, M.; Moncorgé, O.; Goujon, C. Mammalian and Avian Host Cell Influenza A Restriction Factors. Viruses 2021, 13, 522. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and Functions of SARS-CoV-2 Proteins in Host Immune Evasion. Front Immunol 2022, 13, 940756. [Google Scholar] [CrossRef]
- Zanoni, I.; Granucci, F.; Broggi, A. Interferon (IFN)-λ Takes the Helm: Immunomodulatory Roles of Type III IFNs. Front Immunol 2017, 8, 1661. [Google Scholar] [CrossRef]
- Margaroli, C.; Fram, T.R.; Sharma, N.S.; Patel, S.B.; Tipper, J.; Robison, S.W.; Russell, D.W.; Fortmann, S.D.; Banday, M.M.; Soto-Vazquez, Y.M.; et al. Interferon-Dependent Signaling Is Critical for Viral Clearance in Airway Neutrophils. JCI Insight 2023. [Google Scholar] [CrossRef] [PubMed]
- Glennon-Alty, L.; Moots, R.J.; Edwards, S.W.; Wright, H.L. Type I Interferon Regulates Cytokine-Delayed Neutrophil Apoptosis, Reactive Oxygen Species Production and Chemokine Expression. Clin Exp Immunol 2021, 203, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Hijano, D.R.; Vu, L.D.; Kauvar, L.M.; Tripp, R.A.; Polack, F.P.; Cormier, S.A. Role of Type I Interferon (IFN) in the Respiratory Syncytial Virus (RSV) Immune Response and Disease Severity. Front Immunol 2019, 10, 566. [Google Scholar] [CrossRef]
- Munir, S.; Hillyer, P.; Le Nouën, C.; Buchholz, U.J.; Rabin, R.L.; Collins, P.L.; Bukreyev, A. Respiratory Syncytial Virus Interferon Antagonist NS1 Protein Suppresses and Skews the Human T Lymphocyte Response. PLoS Pathog 2011, 7, e1001336–e1001336. [Google Scholar] [CrossRef]
- Mo, S.; Tang, W.; Xie, J.; Chen, S.; Ren, L.; Zang, N.; Xie, X.; Deng, Y.; Gao, L.; Liu, E. Respiratory Syncytial Virus Activates Rab5a to Suppress IRF1-Dependent IFN-λ Production, Subverting the Antiviral Defense of Airway Epithelial Cells. J Virol 2021, 95, e02333–20. [Google Scholar] [CrossRef]
- Wu, J.; Shi, Y.; Pan, X.; Wu, S.; Hou, R.; Zhang, Y.; Zhong, T.; Tang, H.; Du, W.; Wang, L.; et al. SARS-CoV-2 ORF9b Inhibits RIG-I-MAVS Antiviral Signaling by Interrupting K63-Linked Ubiquitination of NEMO. Cell Rep 2021, 34, 108761. [Google Scholar] [CrossRef]
- Jefferies, C.A. Regulating IRFs in IFN Driven Disease. Front Immunol 2019, 10. [Google Scholar] [CrossRef]
- Ferreira-Gomes, M.; Kruglov, A.; Durek, P.; Heinrich, F.; Tizian, C.; Heinz, G.A.; Pascual-Reguant, A.; Du, W.; Mothes, R.; Fan, C.; et al. SARS-CoV-2 in Severe COVID-19 Induces a TGF-β-Dominated Chronic Immune Response That Does Not Target Itself. Nat Commun 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Barros-Martins, J.; Förster, R.; Bošnjak, B. NK Cell Dysfunction in Severe COVID-19: TGF-β-Induced Downregulation of Integrin Beta-2 Restricts NK Cell Cytotoxicity. Signal Transduct Target Ther 2022, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Tizian, C.; Ferreira-Gomes, M.; Niemeyer, D.; Jones, T.C.; Heinrich, F.; Frischbutter, S.; Angermair, S.; Hohnstein, T.; Mattiola, I.; et al. Untimely TGFβ Responses in COVID-19 Limit Antiviral Functions of NK Cells. Nature 2021, 600, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Bi, J. NK Cell Dysfunction in Patients with COVID-19. Cellular & Molecular Immunology 2022, 19, 127–129. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.-S.; Sasaki, M.; Cho, S.X.; Kasuga, Y.; Zhu, B.; Ouda, R.; Orba, Y.; de Figueiredo, P.; Sawa, H.; Kobayashi, K.S. SARS-CoV-2 Inhibits Induction of the MHC Class I Pathway by Targeting the STAT1-IRF1-NLRC5 Axis. Nat Commun 2021, 12, 6602. [Google Scholar] [CrossRef]
- Duncan, J.K.S.; Xu, D.; Licursi, M.; Joyce, M.A.; Saffran, H.A.; Liu, K.; Gohda, J.; Tyrrell, D.L.; Kawaguchi, Y.; Hirasawa, K. Interferon Regulatory Factor 3 Mediates Effective Antiviral Responses to Human Coronavirus 229E and OC43 Infection. Front Immunol 2023, 14. [Google Scholar] [CrossRef]
- Cao, D.; Duan, L.; Huang, B.; Xiong, Y.; Zhang, G.; Huang, H. The SARS-CoV-2 Papain-like Protease Suppresses Type I Interferon Responses by Deubiquitinating STING. Sci Signal 2023, 16. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890. [Google Scholar] [CrossRef]
- Shahbazi, M.; Amri Maleh, P.; Bagherzadeh, M.; Moulana, Z.; Sepidarkish, M.; Rezanejad, M.; Mirzakhani, M.; Ebrahimpour, S.; Ghorbani, H.; Ahmadnia, Z.; et al. Linkage of Lambda Interferons in Protection Against Severe COVID-19. Journal of Interferon & Cytokine Research 2021, 41, 149–152. [Google Scholar] [CrossRef]
- Syedbasha, M.; Linnik, J.; Santer, D.; O’Shea, D.; Barakat, K.; Joyce, M.; Khanna, N.; Tyrrell, D.L.; Houghton, M.; Egli, A. An ELISA Based Binding and Competition Method to Rapidly Determine Ligand-Receptor Interactions. Journal of Visualized Experiments 2016. [Google Scholar] [CrossRef]
- Scagnolari, C.; Pierangeli, A.; Frasca, F.; Bitossi, C.; Viscido, A.; Oliveto, G.; Scordio, M.; Mazzuti, L.; Di Carlo, D.; Gentile, M.; et al. Differential Induction of Type I and III Interferon Genes in the Upper Respiratory Tract of Patients with Coronavirus Disease 2019 (COVID-19). Virus Res 2021, 295, 198283. [Google Scholar] [CrossRef]
- Lee-Kirsch, M.A. The Type I Interferonopathies. Annu Rev Med 2017, 68, 297–315. [Google Scholar] [CrossRef]
- Lee, A.C.; Jeong, Y.; Lee, S.; Jang, H.; Zheng, A.; Kwon, S.; Repine, J.E. Nasopharyngeal Type-I Interferon for Immediately Available Prophylaxis Against Emerging Respiratory Viral Infections. Front Immunol 2021, 12, 660298. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Shin, E.-C. The Type I Interferon Response in COVID-19: Implications for Treatment. Nat Rev Immunol 2020, 20, 585–586. [Google Scholar] [CrossRef] [PubMed]
- Niles, M.A.; Gogesch, P.; Kronhart, S.; Ortega Iannazzo, S.; Kochs, G.; Waibler, Z.; Anzaghe, M. Macrophages and Dendritic Cells Are Not the Major Source of Pro-Inflammatory Cytokines Upon SARS-CoV-2 Infection. Front Immunol 2021, 12. [Google Scholar] [CrossRef]
- Trouillet-Assant, S.; Viel, S.; Gaymard, A.; Pons, S.; Richard, J.-C.; Perret, M.; Villard, M.; Brengel-Pesce, K.; Lina, B.; Mezidi, M.; et al. Type I IFN Immunoprofiling in COVID-19 Patients. Journal of Allergy and Clinical Immunology 2020, 146, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Ruetsch, C.; Brglez, V.; Crémoni, M.; Zorzi, K.; Fernandez, C.; Boyer-Suavet, S.; Benzaken, S.; Demonchy, E.; Risso, K.; Courjon, J.; et al. Functional Exhaustion of Type I and II Interferons Production in Severe COVID-19 Patients. Front Med (Lausanne) 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Seepathomnarong, P.; Ongarj, J.; Sophonmanee, R.; Seeyankem, B.; Chusri, S.; Surasombatpattana, S.; Pinpathomrat, N. Regulatory T Cells Decreased during Recovery from Mild COVID-19. Viruses 2022, 14, 1688. [Google Scholar] [CrossRef] [PubMed]
- Gocher-Demske, A.M.; Cui, J.; Szymczak-Workman, A.L.; Vignali, K.M.; Latini, J.N.; Pieklo, G.P.; Kimball, J.C.; Avery, L.; Cipolla, E.M.; Huckestein, B.R.; et al. IFNγ-Induction of TH1-like Regulatory T Cells Controls Antiviral Responses. Nat Immunol 2023, 24, 841–854. [Google Scholar] [CrossRef]
- Larange, A.; Takazawa, I.; Kakugawa, K.; Thiault, N.; Ngoi, S.; Olive, M.E.; Iwaya, H.; Seguin, L.; Vicente-Suarez, I.; Becart, S.; et al. A Regulatory Circuit Controlled by Extranuclear and Nuclear Retinoic Acid Receptor α Determines T Cell Activation and Function. Immunity 2023. [Google Scholar] [CrossRef] [PubMed]
- Syedbasha, M.; Bonfiglio, F.; Linnik, J.; Stuehler, C.; Wüthrich, D.; Egli, A. Interferon-λ Enhances the Differentiation of Naive B Cells into Plasmablasts via the MTORC1 Pathway. Cell Rep 2020, 33, 108211. [Google Scholar] [CrossRef]
- Flemming, A. Uncovering the Mystery of Secreted IgD. Nat Rev Immunol 2018, 18, 668–669. [Google Scholar] [CrossRef]
- Junker, F.; Gordon, J.; Qureshi, O. Fc Gamma Receptors and Their Role in Antigen Uptake, Presentation, and T Cell Activation. Front Immunol 2020, 11. [Google Scholar] [CrossRef]
- Contoli, M.; Papi, A.; Tomassetti, L.; Rizzo, P.; Vieceli Dalla Sega, F.; Fortini, F.; Torsani, F.; Morandi, L.; Ronzoni, L.; Zucchetti, O.; et al. Blood Interferon-α Levels and Severity, Outcomes, and Inflammatory Profiles in Hospitalized COVID-19 Patients. Front Immunol 2021, 12. [Google Scholar] [CrossRef]
- Contoli, M.; Message, S.D.; Laza-Stanca, V.; Edwards, M.R.; Wark, P.A.B.; Bartlett, N.W.; Kebadze, T.; Mallia, P.; Stanciu, L.A.; Parker, H.L.; et al. Role of Deficient Type III Interferon-λ Production in Asthma Exacerbations. Nat Med 2006, 12, 1023–1026. [Google Scholar] [CrossRef]
- Cooles, F.A.H.; Tarn, J.; Lendrem, D.W.; Naamane, N.; Lin, C.M.; Millar, B.; Maney, N.J.; Anderson, A.E.; Thalayasingam, N.; Diboll, J.; et al. Interferon-α-Mediated Therapeutic Resistance in Early Rheumatoid Arthritis Implicates Epigenetic Reprogramming. Ann Rheum Dis 2022, 81, 1214–1223. [Google Scholar] [CrossRef]
- López-Rodríguez, J.C.; Hancock, S.J.; Li, K.; Crotta, S.; Barrington, C.; Suárez-Bonnet, A.; Priestnall, S.L.; Aubé, J.; Wack, A.; Klenerman, P.; et al. Type I Interferons Drive MAIT Cell Functions against Bacterial Pneumonia. Journal of Experimental Medicine 2023, 220. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Birkenbach, M.; Chen, S. Patterns of Inflammatory Cell Infiltration and Expression of STAT6 in the Lungs of Patients With COVID-19: An Autopsy Study. Applied Immunohistochemistry & Molecular Morphology 2022, 30, 350–357. [Google Scholar] [CrossRef]
- Sorrentino, L.; Fracella, M.; Frasca, F.; D’Auria, A.; Santinelli, L.; Maddaloni, L.; Bugani, G.; Bitossi, C.; Gentile, M.; Ceccarelli, G.; et al. Alterations in the Expression of IFN Lambda, IFN Gamma and Toll-like Receptors in Severe COVID-19 Patients. Microorganisms 2023, 11, 689. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, M.D.; Kinning, K.T.; Sullivan, K.D.; Araya, P.; Smith, K.P.; Granrath, R.E.; Shaw, J.R.; Baxter, R.; Jordan, K.R.; Russell, S.; et al. Specialized Interferon Action in COVID-19. Proceedings of the National Academy of Sciences 2022, 119. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 Drives IFN in Senescent Cells and Promotes Age-Associated Inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Forero, A.; Ozarkar, S.; Li, H.; Lee, C.H.; Hemann, E.A.; Nadjsombati, M.S.; Hendricks, M.R.; So, L.; Green, R.; Roy, C.N.; et al. Differential Activation of the Transcription Factor IRF1 Underlies the Distinct Immune Responses Elicited by Type I and Type III Interferons. Immunity 2019, 51, 451–464. [Google Scholar] [CrossRef]
- Cytlak, U.; Resteu, A.; Pagan, S.; Green, K.; Milne, P.; Maisuria, S.; McDonald, D.; Hulme, G.; Filby, A.; Carpenter, B.; et al. Differential IRF8 Transcription Factor Requirement Defines Two Pathways of Dendritic Cell Development in Humans. Immunity 2020, 53, 353–370. [Google Scholar] [CrossRef]
- Cervantes, J.L.; Weinerman, B.; Basole, C.; Salazar, J.C. TLR8: The Forgotten Relative Revindicated. Cell Mol Immunol 2012, 9, 434–438. [Google Scholar] [CrossRef] [PubMed]
- AL Hamrashdi, M.; Brady, G. Regulation of IRF3 Activation in Human Antiviral Signaling Pathways. Biochem Pharmacol 2022, 200, 115026. [Google Scholar] [CrossRef]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front Immunol 2019, 10, 1586. [Google Scholar] [CrossRef] [PubMed]
- Izaguirre, A.; Barnes, B.J.; Amrute, S.; Yeow, W.-S.; Megjugorac, N.; Dai, J.; Feng, D.; Chung, E.; Pitha, P.M.; Fitzgerald-Bocarsly, P. Comparative Analysis of IRF and IFN-Alpha Expression in Human Plasmacytoid and Monocyte-Derived Dendritic Cells. J Leukoc Biol 2003, 74, 1125–1138. [Google Scholar] [CrossRef]
- Lee, A.J.; Ashkar, A.A. The Dual Nature of Type I and Type II Interferons. Front Immunol 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Capucetti, A.; Albano, F.; Bonecchi, R. Multiple Roles for Chemokines in Neutrophil Biology. Front Immunol 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Mennechet, F.J.D.; Uzé, G. Interferon-λ–Treated Dendritic Cells Specifically Induce Proliferation of FOXP3-Expressing Suppressor T Cells. Blood 2006, 107, 4417–4423. [Google Scholar] [CrossRef]
- Wei Integrative Genomic Analyses on IL28RA, the Common Receptor of Interferon-Λ1, -Λ2 and -Λ3. Int J Mol Med 2010, 25. [CrossRef]
- Zhou, P.; Cowled, C.; Todd, S.; Crameri, G.; Virtue, E.R.; Marsh, G.A.; Klein, R.; Shi, Z.; Wang, L.-F.; Baker, M.L. Type III IFNs in Pteropid Bats: Differential Expression Patterns Provide Evidence for Distinct Roles in Antiviral Immunity. The Journal of Immunology 2011, 186, 3138–3147. [Google Scholar] [CrossRef]
- Odendall, C.; Dixit, E.; Stavru, F.; Bierne, H.; Franz, K.M.; Durbin, A.F.; Boulant, S.; Gehrke, L.; Cossart, P.; Kagan, J.C. Diverse Intracellular Pathogens Activate Type III Interferon Expression from Peroxisomes. Nat Immunol 2014, 15, 717–726. [Google Scholar] [CrossRef]
- Lauber, C.; Vieyres, G.; Terczyńska-Dyla, E.; Anggakusuma; Dijkman, R. ; Gad, H.H.; Akhtar, H.; Geffers, R.; Vondran, F.W.R.; Thiel, V.; et al. Transcriptome Analysis Reveals a Classical Interferon Signature Induced by IFNλ4 in Human Primary Cells. Genes Immun 2015, 16, 414–421. [Google Scholar] [CrossRef]
- da Silva, J.; Hilzendeger, C.; Moermans, C.; Schleich, F.; Henket, M.; Kebadze, T.; Mallia, P.; Edwards, M.R.; Johnston, S.L.; Louis, R. Raised Interferon-β, Type 3 Interferon and Interferon-Stimulated Genes - Evidence of Innate Immune Activation in Neutrophilic Asthma. Clinical & Experimental Allergy 2017, 47, 313–323. [Google Scholar] [CrossRef]
- Barnas, J.L.; Albrecht, J.; Meednu, N.; Alzamareh, D.F.; Baker, C.; McDavid, A.; Looney, R.J.; Anolik, J.H. B Cell Activation and Plasma Cell Differentiation Are Promoted by IFN-λ in Systemic Lupus Erythematosus. The Journal of Immunology 2021, 207, 2660–2672. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, J.; Feng, H.; Wei, Q.; Zhao, S.; Yang, S.; Ma, H.; Liu, D.; et al. Antiviral Activity of Porcine Interferon Delta 8 against Pesudorabies Virus in Vitro. Int J Biol Macromol 2021, 177, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Liu, X.; Hu, D.; Luo, Y.; Zhang, G.; Liu, P. Swine Enteric Coronaviruses (PEDV, TGEV, and PDCoV) Induce Divergent Interferon-Stimulated Gene Responses and Antigen Presentation in Porcine Intestinal Enteroids. Front Immunol 2022, 12, 826882. [Google Scholar] [CrossRef]
- Zhang, Q.; Ke, H.; Blikslager, A.; Fujita, T.; Yoo, D. Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein Nsp1 in IRF1 Signaling. J Virol 2018, 92, e01677–17. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoo, D. Immune Evasion of Porcine Enteric Coronaviruses and Viral Modulation of Antiviral Innate Signaling. Virus Res 2016, 226, 128–141. [Google Scholar] [CrossRef]
- Iaconis, G.; Jackson, B.; Childs, K.; Boyce, M.; Goodbourn, S.; Blake, N.; Iturriza-Gomara, M.; Seago, J. Rotavirus NSP1 Inhibits Type I and Type III Interferon Induction. Viruses 2021, 13, 589. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xue, M.; Fu, F.; Yin, L.; Feng, L.; Liu, P. IFN-Lambda 3 Mediates Antiviral Protection Against Porcine Epidemic Diarrhea Virus by Inducing a Distinct Antiviral Transcript Profile in Porcine Intestinal Epithelia. Front Immunol 2019, 10. [Google Scholar] [CrossRef]
- Feld, J.J.; Kandel, C.; Biondi, M.J.; Kozak, R.A.; Zahoor, M.A.; Lemieux, C.; Borgia, S.M.; Boggild, A.K.; Powis, J.; McCready, J.; et al. Peginterferon Lambda for the Treatment of Outpatients with COVID-19: A Phase 2, Placebo-Controlled Randomised Trial. Lancet Respir Med 2021, 9, 498–510. [Google Scholar] [CrossRef]
- Yamauchi, S.; Takeuchi, K.; Chihara, K.; Honjoh, C.; Kato, Y.; Yoshiki, H.; Hotta, H.; Sada, K. STAT1 Is Essential for the Inhibition of Hepatitis C Virus Replication by Interferon-λ but Not by Interferon-α. Sci Rep 2016, 6, 38336. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Coto-Llerena, M.; Suslov, A.; Teixeira, R.D.; Fofana, I.; Nuciforo, S.; Hofmann, M.; Thimme, R.; Hensel, N.; Lohmann, V.; et al. Interferon Lambda 4 Impairs Hepatitis C Viral Antigen Presentation and Attenuates T Cell Responses. Nat Commun 2021, 12, 4882. [Google Scholar] [CrossRef] [PubMed]
- Shikama, Y.; Kurosawa, M.; Furukawa, M.; Kudo, Y.; Ishimaru, N.; Matsushita, K. The Priming Potential of Interferon Lambda-1 for Antiviral Defense in the Oral Mucosa. Inflammation 2022, 45, 1348–1361. [Google Scholar] [CrossRef]
- Kim, D.-H.; Han, S.-H.; Go, H.-J.; Kim, D.-Y.; Kim, J.-H.; Lee, J.-B.; Park, S.-Y.; Song, C.-S.; Lee, S.-W.; Choi, I.-S. Antiviral Activity of Canine Interferon Lambda 3 Expressed Using a Recombinant Adenovirus against Canine Coronavirus, Canine Parvovirus, and Canine Distemper Virus. Vet Res Commun 2022, 46, 1363–1368. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Luo, Z.; Zhang, Y.; Lv, L.; Zhao, J.; Sui, B.; Huang, F.; Cui, M.; Fu, Z.F.; et al. Interferon-λ Attenuates Rabies Virus Infection by Inducing Interferon-Stimulated Genes and Alleviating Neurological Inflammation. Viruses 2020, 12, 405. [Google Scholar] [CrossRef]
- Obajemu, A.A.; Rao, N.; Dilley, K.A.; Vargas, J.M.; Sheikh, F.; Donnelly, R.P.; Shabman, R.S.; Meissner, E.G.; Prokunina-Olsson, L.; Onabajo, O.O. IFN-Λ4 Attenuates Antiviral Responses by Enhancing Negative Regulation of IFN Signaling. The Journal of Immunology 2017, 199, 3808–3820. [Google Scholar] [CrossRef] [PubMed]
- Cannella, F.; Scagnolari, C.; Selvaggi, C.; Stentella, P.; Recine, N.; Antonelli, G.; Pierangeli, A. Interferon Lambda 1 Expression in Cervical Cells Differs between Low-Risk and High-Risk Human Papillomavirus-Positive Women. Med Microbiol Immunol 2014, 203, 177–184. [Google Scholar] [CrossRef]
- Albertini, S.; Lo Cigno, I.; Calati, F.; De Andrea, M.; Borgogna, C.; Dell’Oste, V.; Landolfo, S.; Gariglio, M. HPV18 Persistence Impairs Basal and DNA Ligand–Mediated IFN-β and IFN-Λ1 Production through Transcriptional Repression of Multiple Downstream Effectors of Pattern Recognition Receptor Signaling. The Journal of Immunology 2018, 200, 2076–2089. [Google Scholar] [CrossRef]
- De Maeyer, E.; De Maeyer-Guignard, J. Type I Interferons. Int Rev Immunol 1998, 17, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Jankovic, D.; Kugler, D.G.; Sher, A. IL-10 Production by CD4+ Effector T Cells: A Mechanism for Self-Regulation. Mucosal Immunol 2010, 3, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Finotti, G.; Tamassia, N.; Cassatella, M.A. Interferon-Λs and Plasmacytoid Dendritic Cells: A Close Relationship. Front Immunol 2017, 8. [Google Scholar] [CrossRef]
- Hillyer, P.; Mane, V.P.; Schramm, L.M.; Puig, M.; Verthelyi, D.; Chen, A.; Zhao, Z.; Navarro, M.B.; Kirschman, K.D.; Bykadi, S.; et al. Expression Profiles of Human Interferon-alpha and Interferon-lambda Subtypes Are Ligand- and Cell-dependent. Immunol Cell Biol 2012, 90, 774–783. [Google Scholar] [CrossRef]
- Xi, X.; Guo, Y.; Zhu, M.; Wei, Y.; Li, G.; Du, B.; Wang, Y. Higher Expression of Monocyte Chemotactic Protein 1 in Mild COVID-19 Patients Might Be Correlated with Inhibition of Type I IFN Signaling. Virol J 2021, 18, 12. [Google Scholar] [CrossRef]
- Seok Lee, J.; Park, S.; Won Jeong, H.; Young Ahn, J.; Jin Choi, S.; Lee, H.; Choi, B.; Kyung Nam, S.; Sa, M.; Kwon, J.-S.; et al. Immunophenotyping of COVID-19 and Influenza Highlights the Role of Type I Interferons in Development of Severe COVID-19; Vol. 5; 2020. [Google Scholar]
- Zhao, M.; Li, L.; Zhai, L.; Yue, Q.; Liu, H.; Ren, S.; Jiang, X.; Gao, F.; Bai, S.; Li, H.; et al. Comparative Transcriptomic and Proteomic Analyses Prove That IFN-Λ1 Is a More Potent Inducer of ISGs than IFN-α against Porcine Epidemic Diarrhea Virus in Porcine Intestinal Epithelial Cells. J Proteome Res 2020, 19, 3697–3707. [Google Scholar] [CrossRef]
- Soday, L.; Potts, M.; Hunter, L.M.; Ravenhill, B.J.; Houghton, J.W.; Williamson, J.C.; Antrobus, R.; Wills, M.R.; Matheson, N.J.; Weekes, M.P. Comparative Cell Surface Proteomic Analysis of the Primary Human T Cell and Monocyte Responses to Type I Interferon. Front Immunol 2021, 12. [Google Scholar] [CrossRef]
- Awwad, M.H.S.; Mahmoud, A.; Bruns, H.; Echchannaoui, H.; Kriegsmann, K.; Lutz, R.; Raab, M.S.; Bertsch, U.; Munder, M.; Jauch, A.; et al. Selective Elimination of Immunosuppressive T Cells in Patients with Multiple Myeloma. Leukemia 2021, 35, 2602–2615. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.P.; Nguyen, H.N.; Gomez-Rivas, E.; Jeong, Y.; Jonsson, A.H.; Chen, A.F.; Lange, J.K.; Dyer, G.S.; Blazar, P.; Earp, B.E.; et al. SLAMF7 Engagement Superactivates Macrophages in Acute and Chronic Inflammation. Sci Immunol 2022, 7. [Google Scholar] [CrossRef]
- Attur, M.; Zhou, H.; Samuels, J.; Krasnokutsky, S.; Yau, M.; Scher, J.U.; Doherty, M.; Wilson, A.G.; Bencardino, J.; Hochberg, M.; et al. Interleukin 1 Receptor Antagonist ( IL1RN) Gene Variants Predict Radiographic Severity of Knee Osteoarthritis and Risk of Incident Disease. Ann Rheum Dis 2020, 79, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Cervantes-Barragan, L.; Vanderheiden, A.; Royer, C.J.; Davis-, M.E.; Ralfs, P.; Chirkova, T.; Anderson, L.J.; Grakoui, A.; Suthar, S. Plasmacytoid Dendritic Cells Produce Type I Interferon and Reduce Viral Replication in 1 Airway Epithelial Cells after SARS-CoV-2 Infection 2 3 4. [CrossRef]
- Villamayor, L.; Rivero, V.; López-García, D.; Topham, D.J.; Martínez-Sobrido, L.; Nogales, A.; DeDiego, M.L. Interferon Alpha Inducible Protein 6 Is a Negative Regulator of Innate Immune Responses by Modulating RIG-I Activation. Front Immunol 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Villamayor, L.; López-García, D.; Rivero, V.; Martínez-Sobrido, L.; Nogales, A.; DeDiego, M.L. The IFN-Stimulated Gene IFI27 Counteracts Innate Immune Responses after Viral Infections by Interfering with RIG-I Signaling. Front Microbiol 2023, 14. [Google Scholar] [CrossRef]
- Bakdash, G.; Vogelpoel, L.T.; van Capel, T.M.; Kapsenberg, M.L.; de Jong, E.C. Retinoic Acid Primes Human Dendritic Cells to Induce Gut-Homing, IL-10-Producing Regulatory T Cells. Mucosal Immunol 2015, 8, 265–278. [Google Scholar] [CrossRef]
- Emilie, D.; Burgard, M.; Lascoux-Combe, C.; Laughlin, M.; Krzysiek, R.; Pignon, C.; Rudent, A.; Molina, J.-M.; Livrozet, J.-M.; Souala, F.; et al. Early Control of HIV Replication in Primary HIV-1 Infection Treated with Antiretroviral Drugs and Pegylated IFNα: Results from the Primoferon A (ANRS 086) Study. AIDS 2001, 15, 1435–1437. [Google Scholar] [CrossRef] [PubMed]
- Filipi, M.; Jack, S. Interferons in the Treatment of Multiple Sclerosis. Int J MS Care 2020, 22, 165–172. [Google Scholar] [CrossRef]
- Errante, P.; Frazao, J.; Condino-Neto, A. The Use of Interferon-Gamma Therapy in Chronic Granulomatous Disease. Recent Pat Antiinfect Drug Discov 2008, 3, 225–230. [Google Scholar] [CrossRef]
- Bennett, A.L.; Smith, D.W.; Cummins, M.J.; Jacoby, P.A.; Cummins, J.M.; Beilharz, M.W. Low-Dose Oral Interferon Alpha as Prophylaxis against Viral Respiratory Illness: A Double-Blind, Parallel Controlled Trial during an Influenza Pandemic Year. Influenza Other Respir Viruses 2013, 7, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Shalamova, L.; Felgenhauer, U.; Wilhelm, J.; Schaubmar, A.R.; Büttner, K.; Schoen, A.; Widera, M.; Ciesek, S.; Weber, F. Omicron Variant of SARS-CoV-2 Exhibits an Increased Resilience to the Antiviral Type I Interferon Response. PNAS Nexus 2022, 1. [Google Scholar] [CrossRef]
- Guo, K.; Barrett, B.S.; Morrison, J.H.; Mickens, K.L.; Vladar, E.K.; Hasenkrug, K.J.; Poeschla, E.M.; Santiago, M.L. Interferon Resistance of Emerging SARS-CoV-2 Variants. Proceedings of the National Academy of Sciences 2022, 119. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.-S.; Xiang, X.; Wang, X.; Wang, Z.-H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Interferon-A2b Treatment for COVID-19. Front Immunol 2020, 11. [Google Scholar] [CrossRef]
- Zhou, Q.; MacArthur, M.R.; He, X.; Wei, X.; Zarin, P.; Hanna, B.S.; Wang, Z.-H.; Xiang, X.; Fish, E.N. Interferon-A2b Treatment for COVID-19 Is Associated with Improvements in Lung Abnormalities. Viruses 2020, 13, 44. [Google Scholar] [CrossRef]
- Davoudi-Monfared, E.; Rahmani, H.; Khalili, H.; Hajiabdolbaghi, M.; Salehi, M.; Abbasian, L.; Kazemzadeh, H.; Yekaninejad, M.S. A Randomized Clinical Trial of the Efficacy and Safety of Interferon β-1a in Treatment of Severe COVID-19. Antimicrob Agents Chemother 2020, 64, e01061–20. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Wang, T.; Chen, L.; Chen, X.; Li, L.; Qin, X.; Li, H.; Luo, J. The Effect of Recombinant Human Interferon Alpha Nasal Drops to Prevent COVID-19 Pneumonia for Medical Staff in an Epidemic Area. Curr Top Med Chem 2021, 21, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lu, X.; Tong, L.; Shi, X.; Ma, J.; Lv, F.; Wu, J.; Pan, Q.; Yang, J.; Cao, H.; et al. Interferon-α-2b Aerosol Inhalation Is Associated with Improved Clinical Outcomes in Patients with Coronavirus Disease-2019. Br J Clin Pharmacol 2021, 87, 4737–4746. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.; Karl, C.E.; Halfmann, P.J.; Kawaoka, Y.; Winkler, E.S.; Keeler, S.P.; Holtzman, M.J.; Yu, J.; Diamond, M.S. Nasally Delivered Interferon-λ Protects Mice against Infection by SARS-CoV-2 Variants Including Omicron. Cell Rep 2022, 39, 110799. [Google Scholar] [CrossRef]
- Borden, E.C. Interferons α and β in Cancer: Therapeutic Opportunities from New Insights. Nat Rev Drug Discov 2019, 18, 219–234. [Google Scholar] [CrossRef]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at Age 50: Past, Current and Future Impact on Biomedicine. Nat Rev Drug Discov 2007, 6, 975–990. [Google Scholar] [CrossRef]
- Li, Y.; Yang, J.; Shen, S.; Wang, W.; Liu, N.; Guo, H.; Wei, W. SARS-CoV-2-encoded Inhibitors of Human LINE-1 Retrotransposition. J Med Virol 2023, 95. [Google Scholar] [CrossRef]
- Crow, M.K. Long Interspersed Nuclear Elements (LINE-1): Potential Triggers of Systemic Autoimmune Disease. Autoimmunity 2010, 43, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Møhlenberg, M.; Terczyńska-Dyla, E.; Winther, K.G.; Hansen, N.H.; Vad-Nielsen, J.; Laloli, L.; Dijkman, R.; Nielsen, A.L.; Gad, H.H.; et al. The IFNL4 Gene Is a Noncanonical Interferon Gene with a Unique but Evolutionarily Conserved Regulation. J Virol 2020, 94. [Google Scholar] [CrossRef]
- Marongiu, L.; Protti, G.; Facchini, F.A.; Valache, M.; Mingozzi, F.; Ranzani, V.; Putignano, A.R.; Salviati, L.; Bevilacqua, V.; Curti, S.; et al. Maturation Signatures of Conventional Dendritic Cell Subtypes in COVID-19 Suggest Direct Viral Sensing. Eur J Immunol 2022, 52, 109–122. [Google Scholar] [CrossRef]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front Cell Dev Biol 2019, 7. [Google Scholar] [CrossRef]
- Psarras, A.; Antanaviciute, A.; Alase, A.; Carr, I.; Wittmann, M.; Emery, P.; Tsokos, G.C.; Vital, E.M. TNF-α Regulates Human Plasmacytoid Dendritic Cells by Suppressing IFN-α Production and Enhancing T Cell Activation. The Journal of Immunology 2021, 206, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, D.I.; Koay, H.-F.; McCluskey, J.; Gherardin, N.A. The Biology and Functional Importance of MAIT Cells. Nat Immunol 2019, 20, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Bourgoin, P.; Soliveres, T.; Barbaresi, A.; Loundou, A.; Belkacem, I.A.; Arnoux, I.; Bernot, D.; Loosveld, M.; Morange, P.; Michelet, P.; et al. <scp>CD169</Scp> and <scp>CD64</Scp> Could Help Differentiate Bacterial from <scp>CoVID</Scp> -19 or Other Viral Infections in the Emergency Department. Cytometry Part A 2021, 99, 435–445. [Google Scholar] [CrossRef]
- He, X.; Korytář, T.; Schatz, J.; Freuling, C.M.; Müller, T.; Köllner, B. Anti-Lyssaviral Activity of Interferons κ and ω from the Serotine Bat, Eptesicus Serotinus. J Virol 2014, 88, 5444–5454. [Google Scholar] [CrossRef]
- Pavlovich, S.S.; Darling, T.; Hume, A.J.; Davey, R.A.; Feng, F.; Mühlberger, E.; Kepler, T.B. Egyptian Rousette IFN-ω Subtypes Elicit Distinct Antiviral Effects and Transcriptional Responses in Conspecific Cells. Front Immunol 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, F.; Shao, J.; Xie, Y.; Chang, H.; Zhang, Y. Interferon-Omega: Current Status in Clinical Applications. Int Immunopharmacol 2017, 52, 253–260. [Google Scholar] [CrossRef]
- Goebel, G.L.; Qiu, X.; Wu, P. Kinase-Targeting Small-Molecule Inhibitors and Emerging Bifunctional Molecules. Trends Pharmacol Sci 2022, 43, 866–881. [Google Scholar] [CrossRef]
- Volkov, M.; Coppola, M.; Huizinga, R.; Eftimov, F.; Huizinga, T.W.J.; van der Kooi, A.J.; Oosten, L.E.M.; Raaphorst, J.; Rispens, T.; Sciarrillo, R.; et al. Comprehensive Overview of Autoantibody Isotype and Subclass Distribution. Journal of Allergy and Clinical Immunology 2022, 150, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.R.; Kotenko, S. V.; Kaplan, M.J. Interferon Lambda in Inflammation and Autoimmune Rheumatic Diseases. Nat Rev Rheumatol 2021, 17, 349–362. [Google Scholar] [CrossRef]
- Papadopoulos, V.E.; Skarlis, C.; Evangelopoulos, M.-E.; Mavragani, C.P. Type I Interferon Detection in Autoimmune Diseases: Challenges and Clinical Applications. Expert Rev Clin Immunol 2021, 17, 883–903. [Google Scholar] [CrossRef]
- Volkov, M.; Coppola, M.; Huizinga, R.; Eftimov, F.; Huizinga, T.W.J.; van der Kooi, A.J.; Oosten, L.E.M.; Raaphorst, J.; Rispens, T.; Sciarrillo, R.; et al. Comprehensive Overview of Autoantibody Isotype and Subclass Distribution. Journal of Allergy and Clinical Immunology 2022, 150, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- Leonard, W.J.; Lin, J.-X. Strategies to Therapeutically Modulate Cytokine Action. Nat Rev Drug Discov 2023. [Google Scholar] [CrossRef]
- Pang, E.S.; Daraj, G.; Macri, C.; Balka, K.; de Nardo, D.; Hochrein, H.; Masterman, K.-A.; Zhan, Y.; Kile, B.; Lawlor, K.; et al. Activation of Stimulator of Interferon Genes in Dendritic Cells Induces Interferon-Lambda and Subset- and Species-Specific Dendritic Cell Death. The Journal of Immunology 2021, 206, 111–28. [Google Scholar] [CrossRef]
- Burska, A.; Rodríguez-Carrio, J.; Biesen, R.; Dik, W.A.; Eloranta, M.-L.; Cavalli, G.; Visser, M.; Boumpas, D.T.; Bertsias, G.; Wahren-Herlenius, M.; et al. Type I Interferon Pathway Assays in Studies of Rheumatic and Musculoskeletal Diseases: A Systematic Literature Review Informing EULAR Points to Consider. RMD Open 2023, 9, e002876. [Google Scholar] [CrossRef]
- Hauschild, A.; Gogas, H.; Tarhini, A.; Middleton, M.R.; Testori, A.; Dréno, B.; Kirkwood, J.M. Practical Guidelines for the Management of Interferon-α-2b Side Effects in Patients Receiving Adjuvant Treatment for Melanoma. Cancer 2008, 112, 982–994. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).