1. Introduction
Many cancers are currently treated by a combination of local and systemic therapy. Local therapy often consists of a combination of surgery and radiotherapy. The latter requires a sophisticated planning process to guarantee a successful treatment. To improve the efficiency and quality of treatment planning in radiotherapy, methods for predicting possible dose distributions or dose volume histograms (DVH) curves have been introduced in the previous years. Since 2012, knowledge-based methods have been used to predict what is achievable in treatment planning. This was not only used for quality assurance in treatment planning [
1,
2], but also, to speed up the planning process by initiating the treatment plan based on the prediction [
3]. By reducing the number of inputs for the user, treatment planning becomes much more consistent, more resource-efficient and potentially beneficial for treatment quality.
In recent years, three-dimensional dose prediction through neural networks have shown to be a viable method for this purpose. In 2016 the first study using neural networks for dose predictions was published by Shiraishi et al. [
4]. In the following years, approximately 30 more studies were published that try to predict the 3D dose distribution with deep learning. Most of these studies are based on treatments with a relative standard target orientation, with minor anatomical variations from patient to patient, such as prostate and oro/nasopharyngeal cancers. However, there are also promising results for models in the brain, breast and lungs [
5,
6,
7]. In 2020 the open-access knowledge-based planning (OpenKBP) Challenge was organized providing an open-access data set of head and neck treatment plans to train prediction models and evaluate them on a set of standardized metrics [
8]. A total 195 participants competed in this challenge. The best-ranked team scored a mean absolute error (MAE) of 2.43 and 1.48 for the dose and DVH scores respectively (see methods section). Their methodology is publicly available and described as a technical note [
9].
Dose prediction models are mainly used for treatment planning. This means that in practice, in addition to the dose prediction, a second model is required to convert the predicted dose into an actual plan that is executable for the specific treatment technique. In this latter step, a final optimization incorporates individual case properties, physical constraints and dose delivery hardware. In our case, we want to use the dose prediction model for another purpose.
Contouring of targets and organs at risk (OAR), the step that takes place prior to planning, is also subject to automation to improve efficiency and consistency with respect to the current manual process. To ensure quality, an assessment of the contours is required. Usually, visual inspection is the go-to method; however, this is a time-consuming task. For each target and OAR, every slice needs to be visually inspected and if necessary, manually adjusted if deemed incorrect. Especially for deep learning based auto-segmentation models, a lack of robustness could result in unpredictable errors that can happen anywhere within the image volume [
10]. Although often such errors are small and might not have a critical effect on the treatment, in assessing the contours, we postulate that it would be beneficial to know the possible clinical impact of critical errors and of those where no further assessment is required. A deep learning model that can give an accurate prediction of the dose received by an OAR instantly, could provide the required information to assess the clinical impact of contour variations.
In this study, we aim for a deep learning model that can predict the dose for glioblastoma cases. Based on the network of Liu et al. [
9] that was used in the OpenKBP challenge, a model is trained on a set of curated glioblastoma (GBM) cases. Unlike current dose prediction algorithms, we want to verify the model’s performance for contouring quality assessment (QA). This means that specific accuracy and sensitivity are required as well as robustness for a broad range of situations. To do so, we test our trained model on specific sets of contour alterations to assess its sensitivity. Furthermore, we submit the model to a specific worst-case test-set, including rare cases where we expect it to fail. This enables us to determine the robustness of the model and understand where further improvements are required. Subsequently, based on the outcome observed on the worst-case test set, we improved the robustness of the model by augmenting the training set with synthetically generated cases characterizing the observed failure patterns.
2. Materials and Methods
2.1. Data collection and preparation
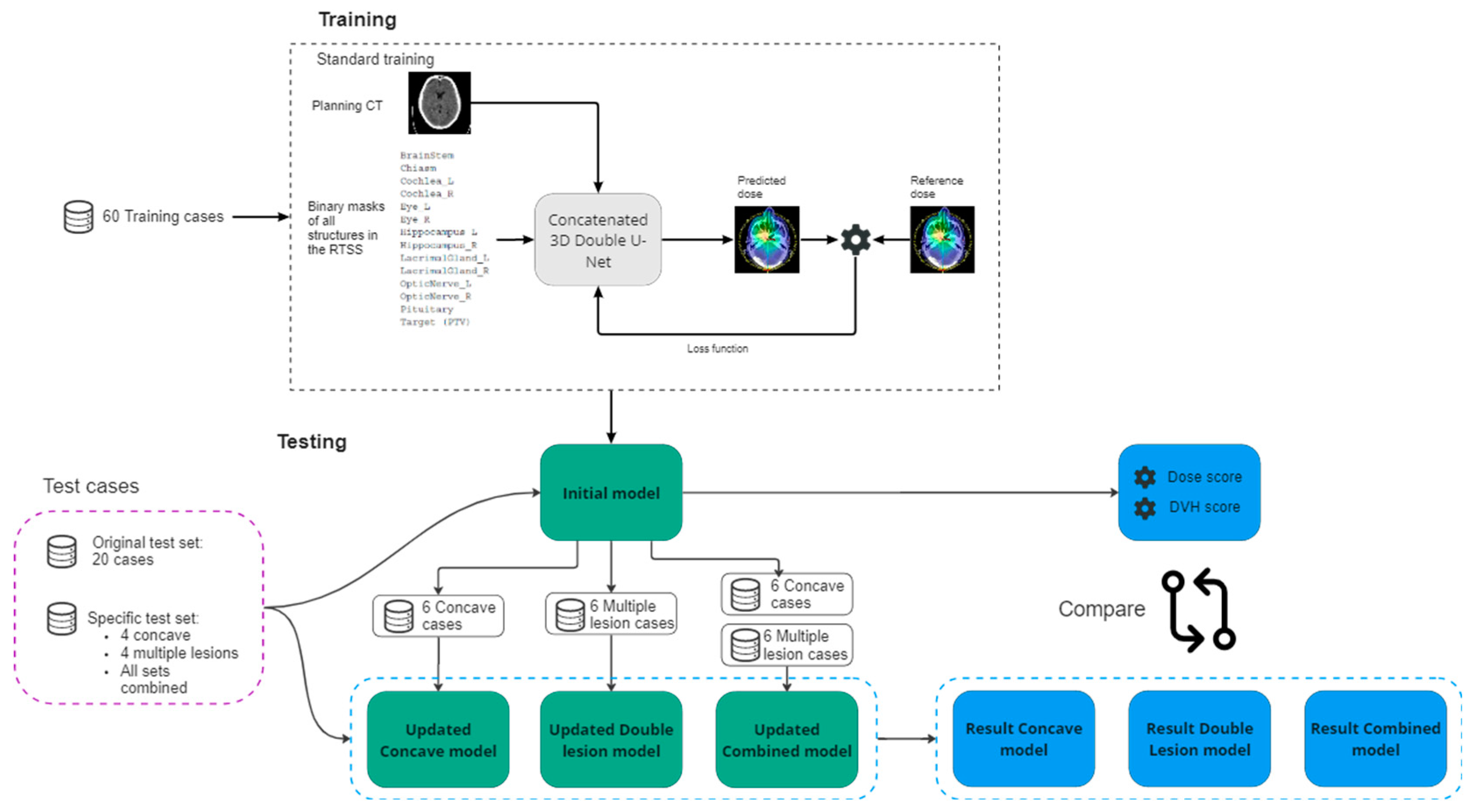
A cohort of 125 GBM patients treated with radiotherapy at the Inselspital University Hospital (Bern, Switzerland) was available. For all patients, the planning target volume (PTV) and the OARs were curated by mutual agreement of radiation oncology professionals. A plan was constructed consistently using a strict dose prescription and standard templates for planning setup and dose optimization initiation for all cases. Of the first 95 cases, 60 were randomly selected for model training, 15 were randomly chosen as validation, and 20 (also randomly chosen) were used as a test set. Of the remaining 30 cases, 10 were used to construct a “worst case” test set manually, and the other 20 for improving the training by adding specific out-of-distribution cases. The following sections further detail how the worst-case test set and the out-of-distribution cases were designed.
2.2. Dose planning
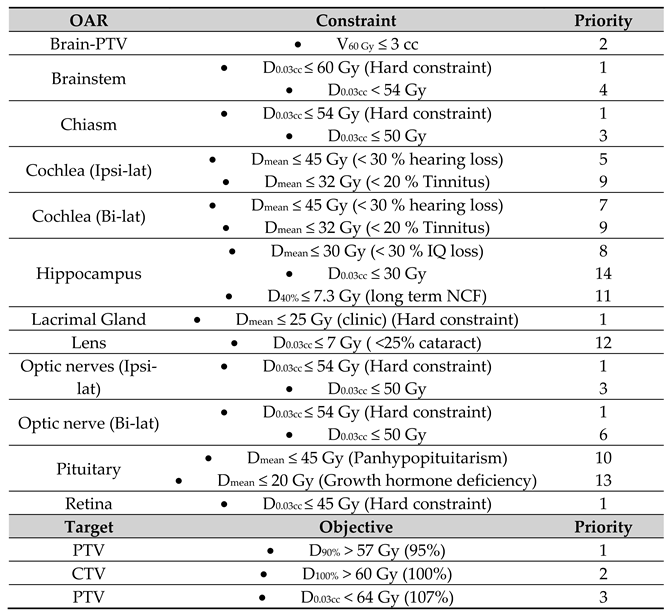
All cases were planned according to the clinical dose prescription of 60 Gy in 30 fractions in the Eclipse treatment planning system (TPS) V15.06.05 (Varian Medical Systems, Palo Alto). All OARs were subject to a dose constraint, which according to a priority list, could or could not be compromised (
Table 1). All plans used a volumetric arc technique (VMAT) with a double full co-planar arc with 6 MV beams containing a flattening filter. The plans were optimized with the photon optimizer, and doses where calculated with the Anisotropic Analytical Algorithm. After dose calculation, the dose was normalized so that 50% of the PTV was covered by 100% of the prescribed dose, according to the institutional clinical guidelines.
2.3. Training
The planning CT and the structures where available in DICOM format for each case. All data is converted from DICOM to nifti files using the PyRaDise package [
11]. The RTSS files containing the PTV and the OARs are divided into 14, separate 3D binary masks each containing a single structure. The input files consisted of 16 3D volumes per case; the planning CT, the dose distribution, the PTV binary mask and 13 OAR binary masks (
Figure 1).
Training took place on a two-level, cascaded 3D (C3D) U-Net [
9] as the dose prediction network (i.e., the input to the second U-Net is the output of the first concatenated with the input to the first U-Net). The model input was a normalized CT volume and binary segmentation masks for each of the 13 OARs and target volume. As output, the model predicts a continuous-valued 3D dose (upscaled from [0,1] to [0, 70 Gy] ) of the same dimension as the input. The loss was computed as:
Where A and B are the outputs of the first and second U-Nets respectively. In this equation, reference indicates the reference dose and L1 refers to the L1 loss. All volumes were resampled to 1283 voxels, due to GPU memory constraints. The hyperparameters for training the C3D model were unchanged from the original implementation [
9], except the number of input binary masks was updated to 14, to match the number of structures in our data set. The model’s weights were randomly initialized using the ‘He’ method [
12]. The training process ran for 80 000 iterations and the model with the best validation dose score was saved. All experiments were run with PyTorch1.12 on an NVIDIA RTX A5000 graphics processing unit. We trained the model five times with the same hyperparameter set but different random seed initialization to ensure reliable convergence. Each training run took 24 hours.
2.4. Assessing the model’s sensitivity
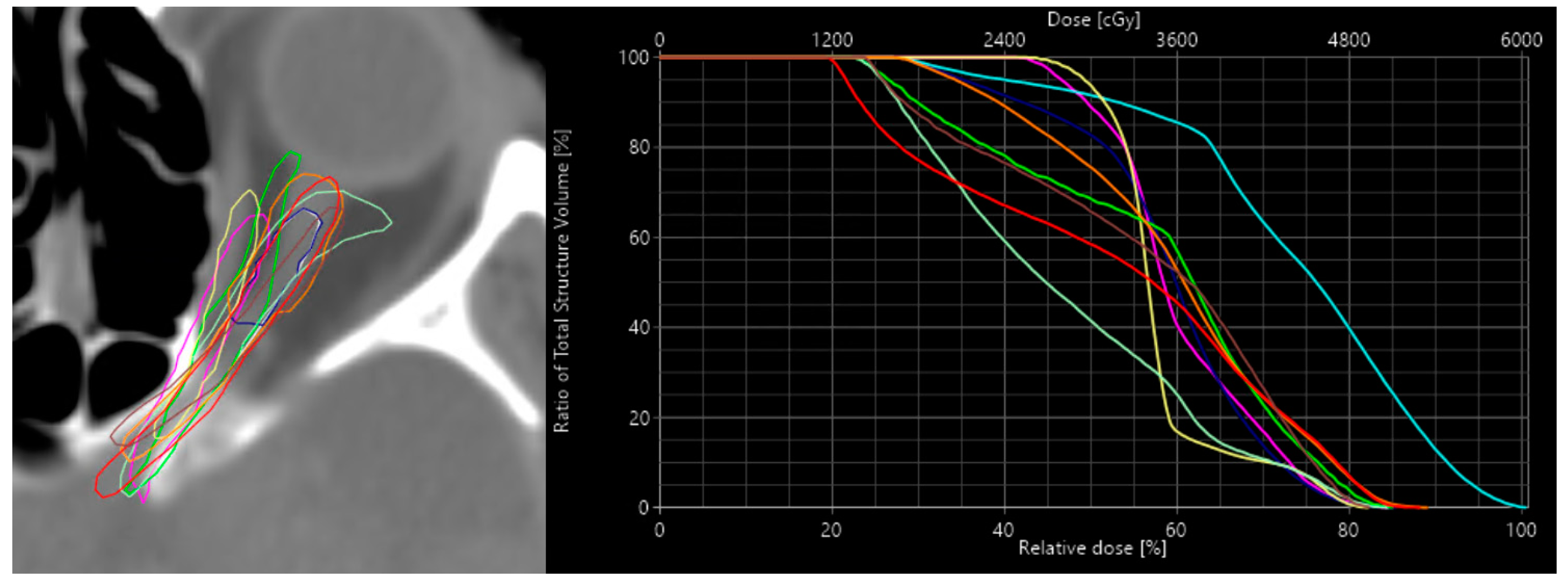
One of the goals of the dose predictor is to provide realistic dosimetric information based on given contours. It should be able to predict realistic dose changes produced by an organ’s small and realistic contour changes of an organ (i.e., inter-expert variability). To analyze the sensitivity of the dose prediction model to these changes, a specific case was chosen where the GBM target is near the left optic nerve (ONL). In practice the optic nerves are prone to variability in contours due to the inter and inter-fractional movement of the eyes which also affect the optic nerves. In this case, small changes in the contour of the ONL would lead to significant dose changes. Manually, 10 alternative contours of the ONL where drawn. The dose was re-optimized and recalculated on the TPS for each alternative contour, to serve as a reference dose. The reference doses were then compared qualitatively and quantitatively to the doses predicted by the model. Dice similarity coefficients (DSC) for the alternatives are calculated to correlate the dose differences to the geometric discrepancy of the ONL contours.
2.5. Improving the model – Worst case test set
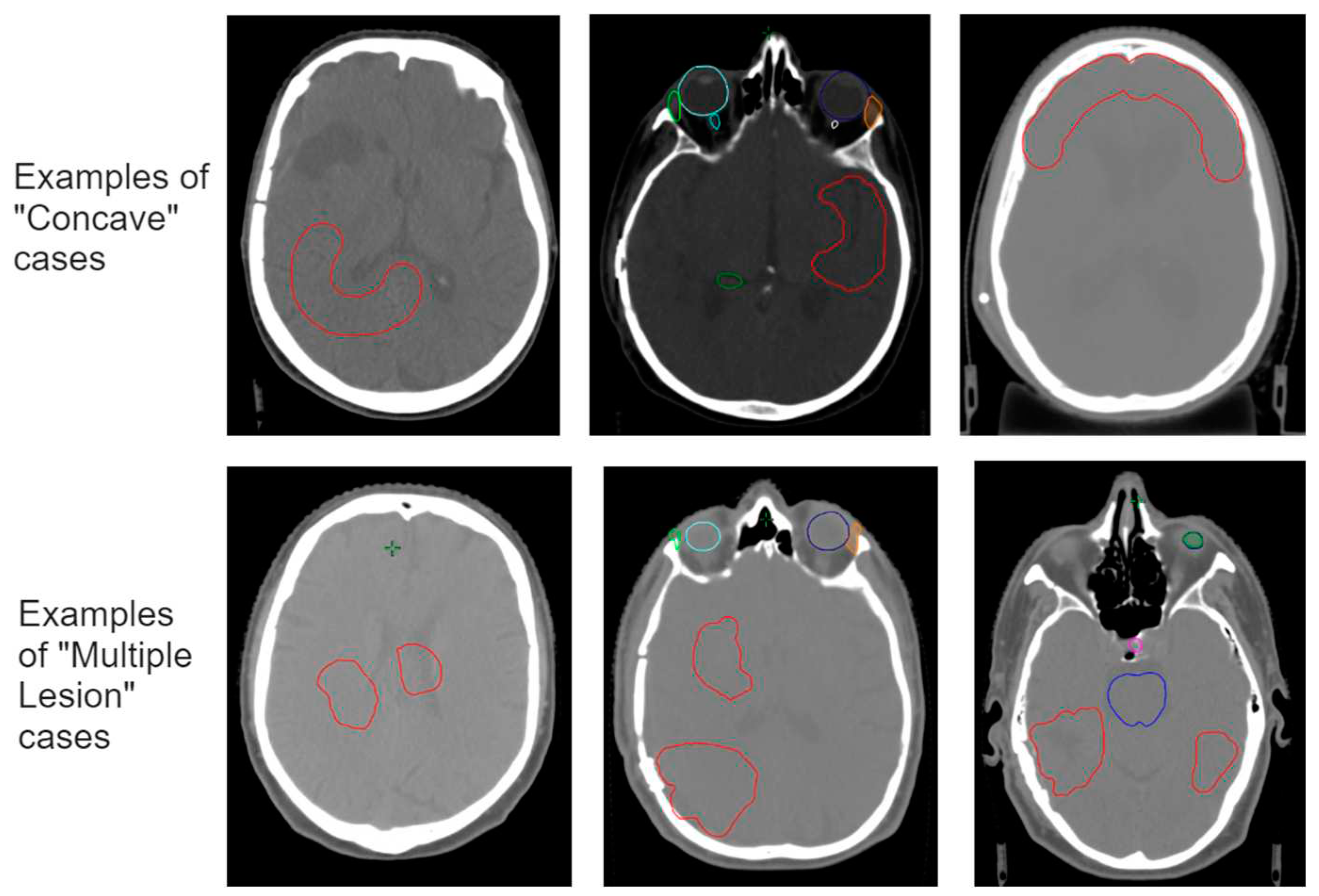
To assess the robustness of the model, and upon first analysis and evaluation of the standard test set of 20 cases, a specific “worst-case” test set was selected of cases where we expect the model to fail or have difficulties. The PTVs of these cases were manually manipulated to simulate rare cases not described by the training dataset (out-of-distribution cases), but also present a challenge in terms of the physical limitations of obtaining perfect dose conformity. Among these 10 cases, we included: (i) targets of larger and smaller size than those present in the training set, (ii) targets consisting of multiple lesions, (iii) irregular shapes such as elongated or concave targets, and (iv) present an overlap between the target and OARs.
According to the “worst case test set” results, we gained insight into which situations the model performs poorly and where it could benefit from additional training. Our observations showed that the prediction model mainly struggled with the physical limitations of conforming the dose according to the targets outlines for specific shapes. Where conformity decreases with concave shapes or multiple targets close to each other, the dose predictor overestimates the dose fall-off in these regions. To increase the overall robustness of the model but specifically to improve the model for these situations, we updated the trained model by including a set of concave-shaped target cases and a set of cases where the target consists of multiple lesions.
The respective new training sets were constructed by means of manually adjusting the target volume (
Figure 2). The 10 cases used for both sets are from different patients and have not been used in any previous model training. All new cases received a dose planning according to the same protocol described above, to serve as the reference dose.
First, we trained an updated “Concave Model” with 60 standard cases + 6 concave cases. The remaining 4 concave cases were used as test-set. Second, we trained an updated “Multiple Lesion Model”, with the 60 standard cases + 6 concave cases. Again, 4 cases were used as a test-set. Finally, we retrained the initial model with 60 standard cases + 6 concave cases + 6 multiple lesion cases. We tested the latter model on the standard test set as well as on the specific test cases, and compared this with the results of the initial model. An overview of the experimental setup is given in
Figure 1.
2.6. Evaluation
The trained models were tested on the test set and the prediction of the dose was compared to the actual planned dose by means of the standardized metrics used by the OpenKBP challenge [
8]: The dose score and the DVH score. The dose score measures the mean error over all the voxels between the two 3D volumes. In this case, we used the whole brain to measure the dose score instead of the whole CT volume or whole body volume. Taking a larger volume will dilute the results to a more positive outcome. The DVH score is the mean error over a set of criteria specific to the given volume. For OARs, these criteria are the mean dose (Dmean) and the maximum dose to 0.1cc (D0.1cc). For the target volume the criteria are the dose received by 1%, 95% and 99% of the voxels within the volume (D1, D95 and D99). The DVH score is calculated for all OARs used in training (lens and retina are combined within the eye, since overlapping masks were not possible). We report the mean results over the 5 trained models in a five-fold split for the evaluation.
Additionally, the initially trained model and the updated models were tested on a set of concave target cases, a set of multiple lesion cases and a combined test set that includes both plus the standard test set.
3. Results
Based on the initial training set of 60 cases, the performance of the initial model was determined on the standard test set of 20 cases. The mean results over 5 independently trained models showed a dose score, measured over the whole brain volume of 0.94 (standard deviation [SD] = 0.36). The mean DVH score over all OARs and the target was 1.95 (SD = 0.95).
3.1. Reults on sensitivity
An overview of the 9 alternative left optic nerve contours is shown in
Figure 3. The mean dose to the ONL based on the treatment planning system and the mean dose based on the prediction model for the reference and the 9 alternative contours are shown in
Table 2. There is a reasonable variation in mean dose among the different alternative contours with respect to the reference contour. In some cases, only minor changes to the mean dose occur, even though the DSC metric shows a significant difference in contour similarity. In other cases, the mean dose change with respect to the reference contour can be up to 5 or 7 Gy difference. The difference between the calculated dose and the predicted dose seems to follow a trend and varies between a maximum of 3.50 Gy with a mean of 1.38 Gy. This states that the predicted dose is more often overestimated.
The average difference of the calculated mean dose for the alternatives with respect to the calculated reference mean dose was 2.44 Gy. (i.e., the difference between the alternatives to the reference dose). For the predicted dose, this difference was 2.32 Gy. This means the correlation coefficient between reference and predicted dose differences across the contour alternatives was 0.89, while the correlation coefficient with the DSC was only −0.42 [
13].
3.2. Improving the model – Worst case test set
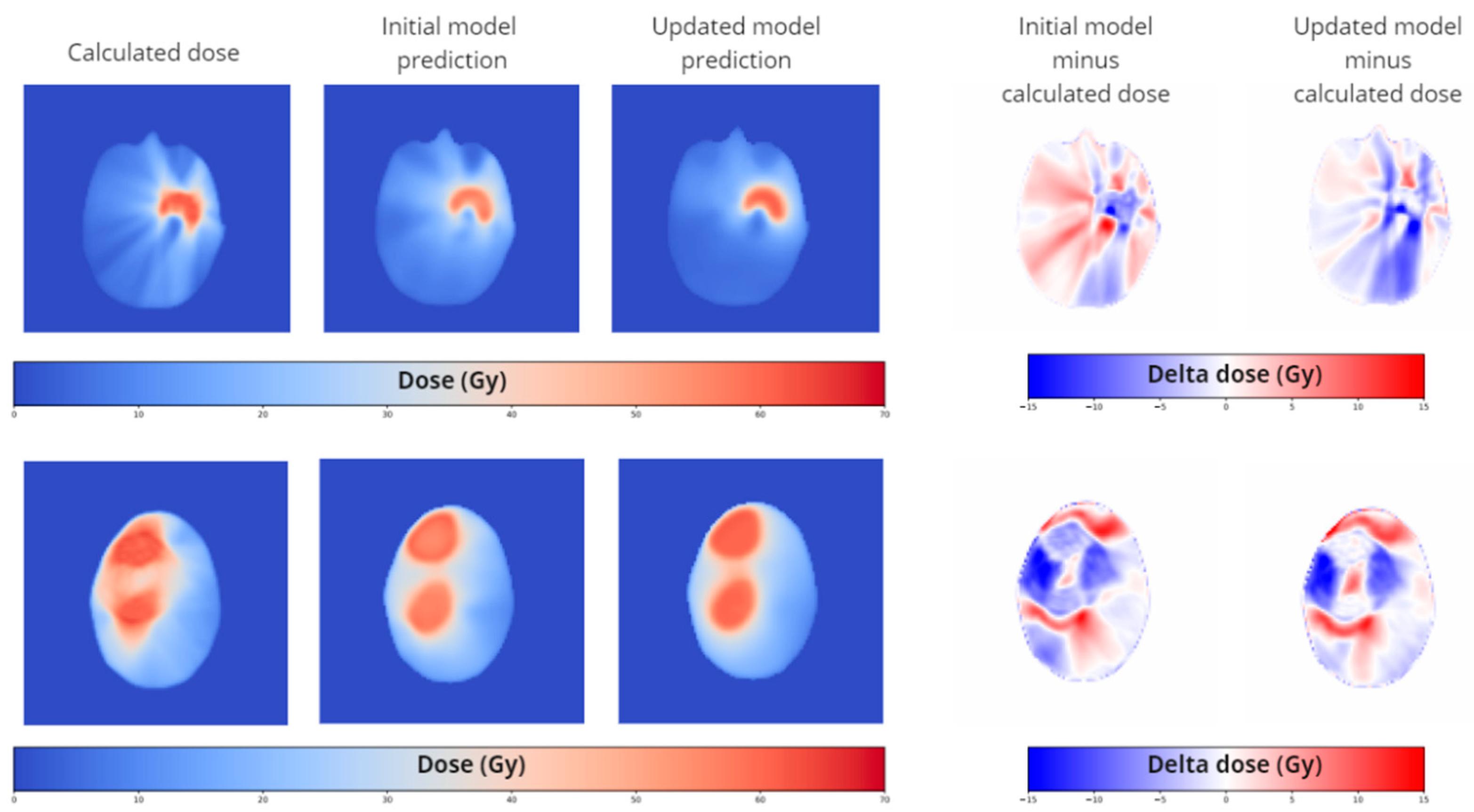
While analyzing the results of the worst-case test set, in particular we saw some flaws, particularly in cases where targets have concave shapes and where target consists of multiple lesions (see
Figure 4). In such cases the prediction model overestimated the ability to conform the dose to the targets. This is mainly reflected in higher dose scores and less so in the DVH scores of the target since the dose discrepancy occurs just outside of the target structure. We updated our training data with 6 concave target cases and 6 multiple lesion target cases as well as a combination of both. The results for different test sets are given for the dose score, the DVH score for the OARs and the DVH score for the Targets in
Table 3.
Based on the standard test set, the updated models scored similar to the initial model on the dose score and the DVH score for targets. The DVH score for OARs improved for all updated models. The updated model with the concave target cases shows by far the best results of all trained models.
Focusing on the concave updated model we observed improvements in dose and DVH score on the concave test set, but also improved results on the multiple test set. This means that on this small set of specific cases the concave updated model scores significantly better than the initial model, as expected. For the multiple-lesion updated model we also observed an improvement, but to a lesser extent. The combined updated model, containing both concave and multiple target lesions scored the worst of all updated models. For the standard test set, the combined updated model did not show improved scores with respect to the initial model.
For the combined test set, which is a combination of the 3 previous test sets, the updated models score consistently better than the initial model.
Qualitatively we can see an improvement in the spatial distribution of the predicted dose in the updated models at exactly the locations of concern, the concave parts of the target and the space between multiple lesions especially in the axial direction.
3. Discussion
By means of an existing dose prediction model that was trained for head and neck cases, we obtained good results for translating the dose prediction model to glioblastoma cases in the brain. For these initial results, only 60 cases where used for training. Compared to the results of the OpenKBP challenge, using their proposed metrics, our initial trained model shows better scores than the top ranked participants. However, we are aware that the anatomies are different. It must be noted that different 3D volumes are used, as well as different OAR structures, which will have an influence on the scores. We do not yet have a benchmark for these metrics in the brain region. On the other hand, whereas head and neck targets are much more similar from case to case, GBM targets vary much more in size, shape and location. Nonetheless, the model was able to achieve good results on a relatively small training set. We conclude from this that a cascade 3D U-Net is capable of predicting dose if high-quality and curated data is used to train it.
Although some other groups have published on dose prediction in the brain [
4,
5,
14], for both VMAT and conventional intensity modulated radiotherapy (IMRT), they did not specify the nature of the brain tumors. Furthermore, different treatment prescriptions were included in these works, certain tumor locations were excluded and additional planning parameters were used in the training model. This is the first prediction model specified for GBM treatment.
It seems to be that 3D dose distribution prediction is a great application for deep learning models. Although the predictions are not perfect, they are useful in the initiation processes of automatic planning. It also provides a near-instant estimation of the dose distribution that outperforms any currently available analytical or mathematical prediction method. Our work shows that a dose prediction model for a specific purpose, i.e. treatment area and modality, is relatively easy to obtain. For our specific purpose, we wanted to predict the dose to OARs. We were able to improve the accuracy of the DVH score for the OARs with some minor additions to the training set.
In order to obtain the training data we used plans that were made according to a strict and standardized protocol. This makes the resulting dose distribution better to predict. It might therefore be one of the reasons for the good results. On the other hand, using such a strict protocol makes the model only valid for treatments following this strict protocol. However, in clinical reality different approaches are used depending on case specifics but also on the individual preference of doctors, planners and the availability of specific hardware. In this case, we used a VMAT technique. The dose distribution of such plans is easy to predict since in every case two full 360-degree co-planar arcs are used. In other techniques such as conventional IMRT, using a set number of beam angles, or more sophisticated VMAT techniques making use of non-coplanar arcs, the dose might be more difficult to predict. Solving this issue really comes down to making a prediction model for the different treatment strategies. Although this seems cumbersome, this can probably be singled down to a few treatment strategies per treatment site. Given the results of our model, only a limited amount of data is required to obtain a viable model.
The main contribution of this paper was to show the feasibility of training an accurate deep learning-based dose predictor for GBM treatment. If data is limited for a particular scenario, but a demarcated treatment protocol exists, even for non-homogeneous anatomies (I.e., not prostate or head and neck, where most of the dose prediction methods are proposed for) good results can be obtained compared to currently reported outcomes. Although the main drive behind dose prediction models is the purpose of automation in dose planning, dose prediction models can also be important for many other purposes. Our hypothesis is that it can be useful in the quality management of the steps prior to planning to make the (auto) contouring, the review and the quality assurance more relevant towards the final goal; good dose coverage of targets while sparing the OARs.
We tested the obtained dose prediction model for some specific criteria that are important for a dose predictor in terms of quality management, sensitivity and robustness. We have shown that the initially trained model is sensitive enough to detect dose trends on realistic contour variability in a critical organ such as the optic nerve. We also tested the initial model against robustness. Although we found that the model did lack a certain accuracy in specific situations, we showed that with a simple strategy of adding specific cases to the training set, the robustness and the overall accuracy of the model increased. We anticipate that dose prediction models can be improved even more when using larger data sets of carefully curated data. In addition, the models can be tailored to have specific characteristics to fulfil the needs of different tasks in radiotherapy management.
5. Conclusion
With currently available deep learning networks, making a 3 dimensional dose prediction model with a dataset of fewer than 100 cases is possible, provided they are carefully curated. The prediction model shows good sensitivity to realistic minor changes in structure segmentations. It is also possible to improve the model for specific cases by updating the training dataset based on analyzing its failure patterns. Due to the near-instant results that 3D dose predictors can provide, it is a useful technique for dose awareness in radiotherapy treatment steps, prior to the actual dose planning.
Author Contributions
RP: Conceptualization, methodology, data curation, data analysis, writing original draft. AK: Conceptualization, methodology, training framework, model training, data analysis, writing review and editing. JW: Conceptualization, writing review and editing. NA: Conceptualization, writing review and editing. EE: Data curation, writing review and editing. DA: Supervision, writing review and editing, PM: Methodology, supervision, writing review and editing. MR: Conceptualization, methodology, supervision, writing review and editing.
Funding
This work is supported by the Innosuisse Grant 31274.1 IP-LS.
Informed Consent Statement
All subjects in this study have approved the use of their clinical data in a written statement. This work is in accordance with the Declaration of Helsinki in its most recent version.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data on which the models are trained is not able to be shared due to privacy and ethical considerations. The data of the networks that were trained are available from third party sources which is referred to in the main text of the manuscript.
Conflicts of Interest
The authors of this manuscript declare no conflict of interest.
References
- L. M. Appenzoller, J. M. Michalski, W. L. Thorstad, S. Mutic, and K. L. Moore, “Predicting dose-volume histograms for organs-at-risk in IMRT planning,” Med. Phys., vol. 39, no. 12, pp. 7446–7461, 2012. [CrossRef]
- J. P. Tol, M. Dahele, A. R. Delaney, B. J. Slotman, and W. F. A. R. Verbakel, “Can knowledge-based DVH predictions be used for automated, individualized quality assurance of radiotherapy treatment plans?,” Radiat. Oncol., vol. 10, no. 234, pp. 1–14, 2015. [CrossRef]
- C. McIntosh and T. G. Purdie, “Contextual Atlas Regression Forests : Multiple-Atlas-Based Automated Dose Prediction in Radiation Therapy,” IEEE Trans. Med. Imaging, vol. 35, no. 4, pp. 1000–1012, 2016. [CrossRef]
- S. Shiraishi and K. L. Moore, “Knowledge-based prediction of three-dimensional dose distributions for external beam radiotherapy,” Med. Phys., vol. 43, no. 1, pp. 378–387, 2016. [CrossRef]
- C. McIntosh and T. G. Purdie, “Voxel-based dose prediction with multi-patient atlas selection for automated radiotherapy treatment planning,” Phys. Med. Biol., vol. 62, no. 2, pp. 415–431, 2017. [CrossRef]
- N. Bakx, H. Bluemink, E. Hagelaar, M. Van Der Sangen, and J. Theuws, “Development and evaluation of radiotherapy deep learning dose prediction models for breast cancer,” Phys. Imaging Radiat. Oncol., vol. 17, no. August 2020, pp. 65–70, 2021. [CrossRef]
- N. Dahiya, G. Jhanwar, A. Yezzi, M. Zarepisheh, and S. Nadeem, “Deep Learning 3D Dose Prediction for Conventional Lung IMRT Using Consistent / Unbiased Automated Plans,” arXiv, no. 2, pp. 1–16, 2021.
- Babier et al., “OpenKBP: The open-access knowledge-based planning grand challenge and dataset,” Med. Phys., vol. 48, no. 9, pp. 5549–5561, 2021.
- J. Wang et al., “Technical Note : A deep learning-based autosegmentation of rectal tumors in MR images,” 2018. [CrossRef]
- R. Poel et al., “Impact of random outliers in auto-segmented targets on radiotherapy treatment plans for glioblastoma,” Radiat. Oncol., vol. 17, no. 1, p. 170, 2022. [CrossRef]
- E. Rüfenacht et al., “PyRaDiSe: A Python package for DICOM-RT-based auto-segmentation pipeline construction and DICOM-RT data conversion,” Comput. Methods Programs Biomed., vol. 231, 2023. [CrossRef]
- K. He, X. Zhang, S. Ren, and J. Sun, “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2015 International Conference on Computer Vision, ICCV 2015, pp. 1026–1034, 2015.
- R. Poel et al., “The predictive value of segmentation metrics on dosimetry in organs at risk of the brain,” Med. Image Anal., vol. 73, p. 102161, 2021.
- J. Yang, Y. Zhao, F. Zhang, M. Liao, and X. Yang, “Deep learning architecture with transformer and semantic field alignment for voxel-level dose prediction on brain tumors,” Med. Phys., pp. 1–13, 2022. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).