Submitted:
14 August 2023
Posted:
14 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Structure and function of PRAME
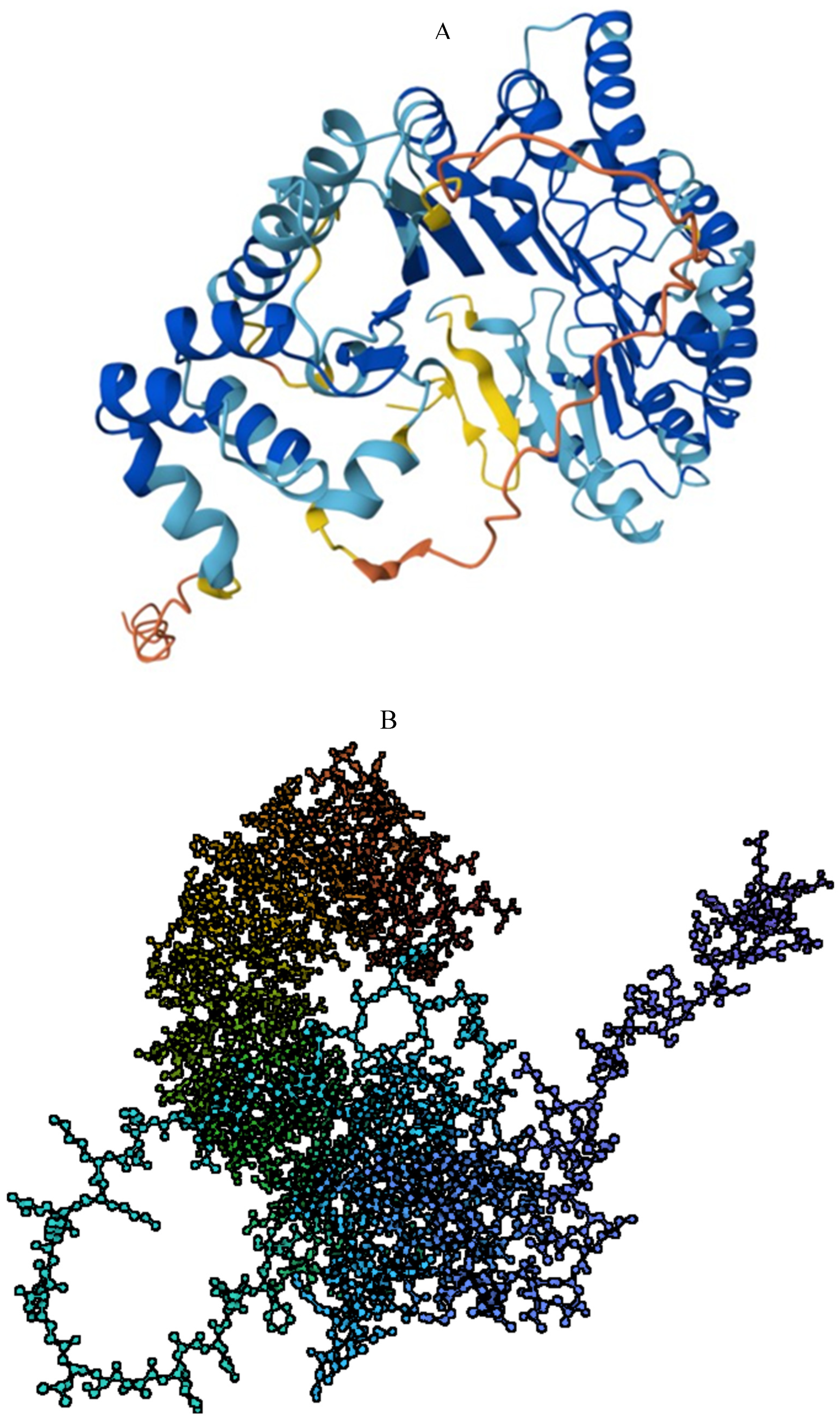
2.1. Structure
2.2. Structure Determines Function
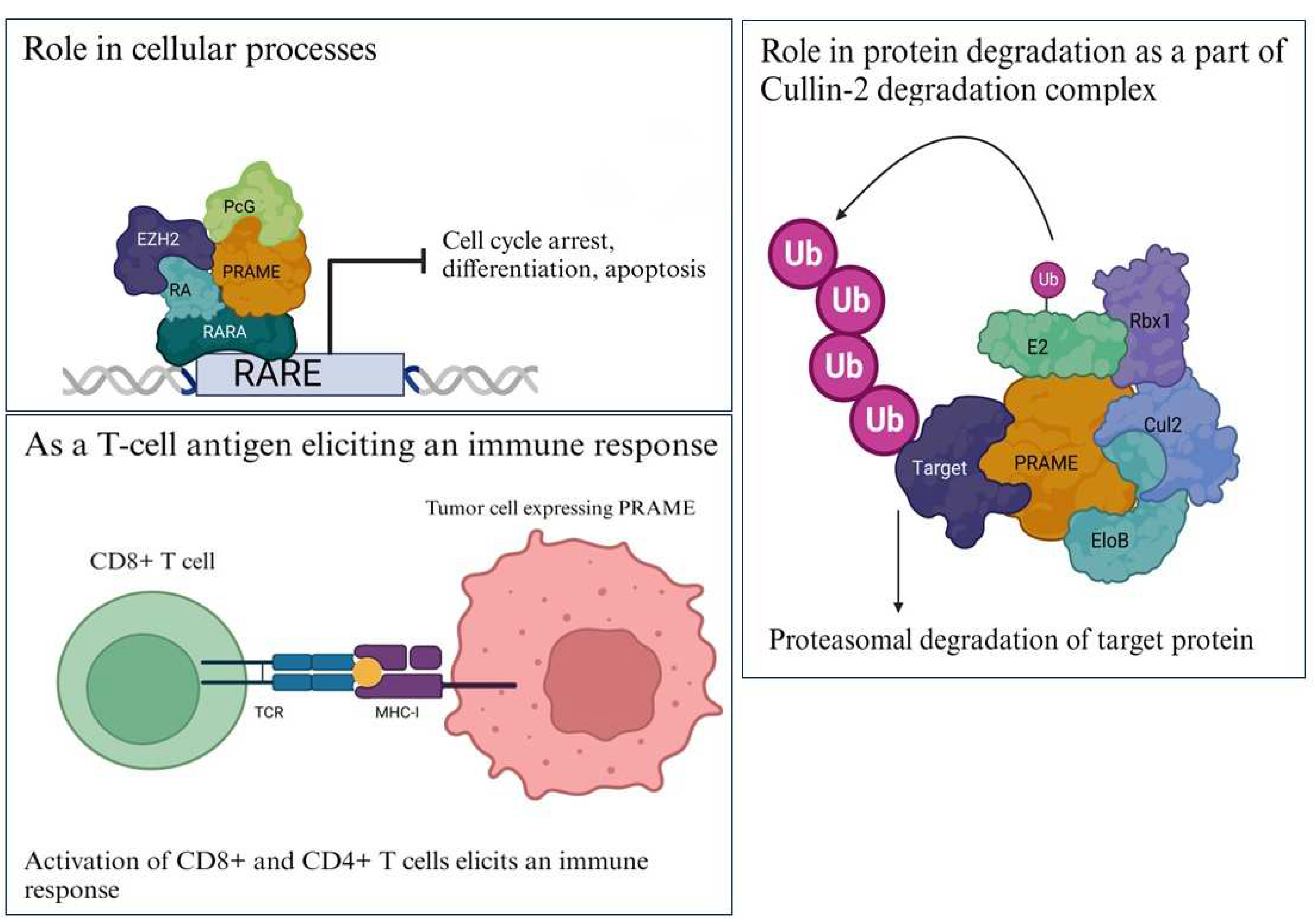
1.2.1. Differentiation
1.2.2. Protein Degradation
1.2.3. Immune Target
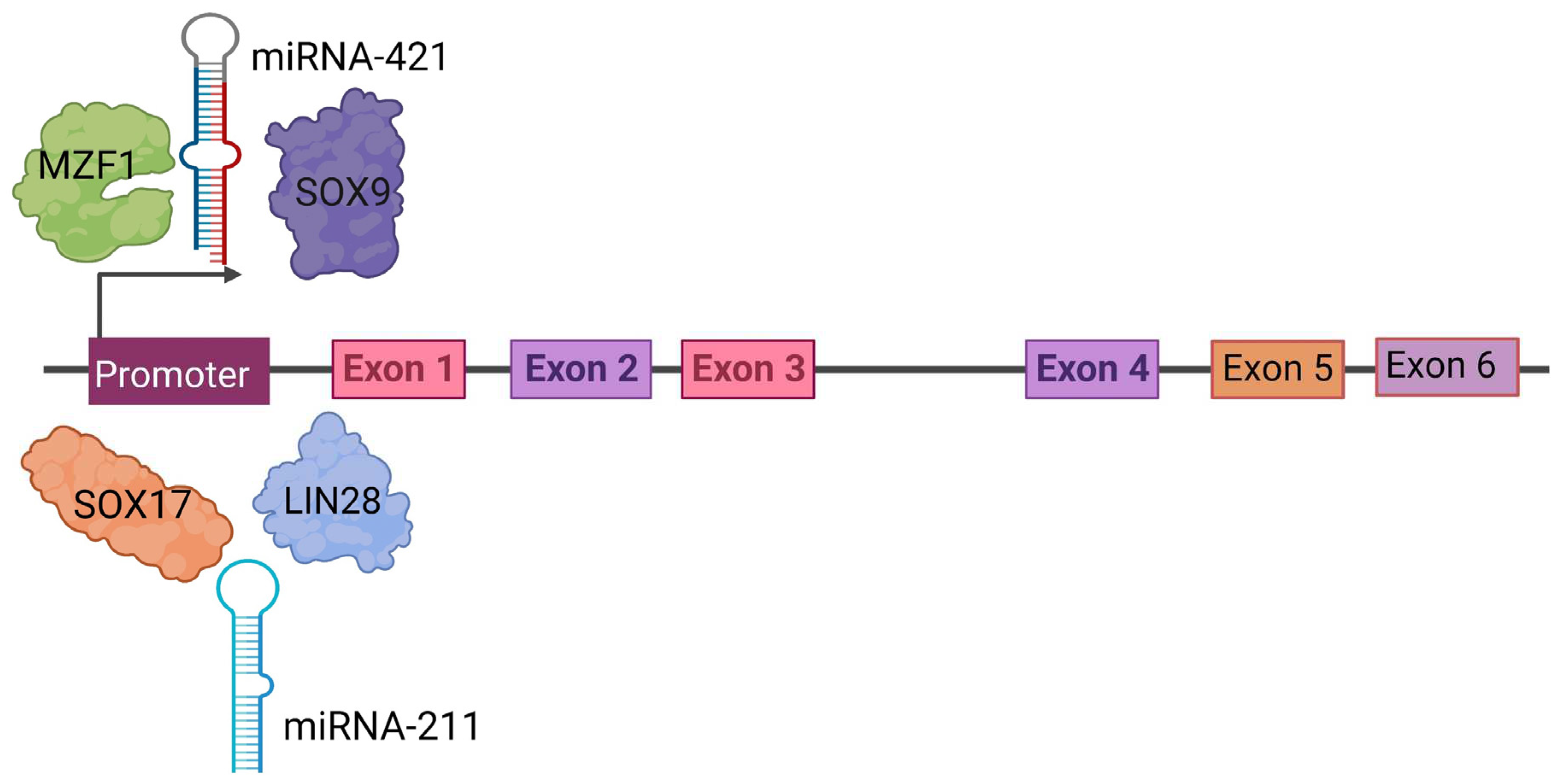
3. Regulation of PRAME Expression
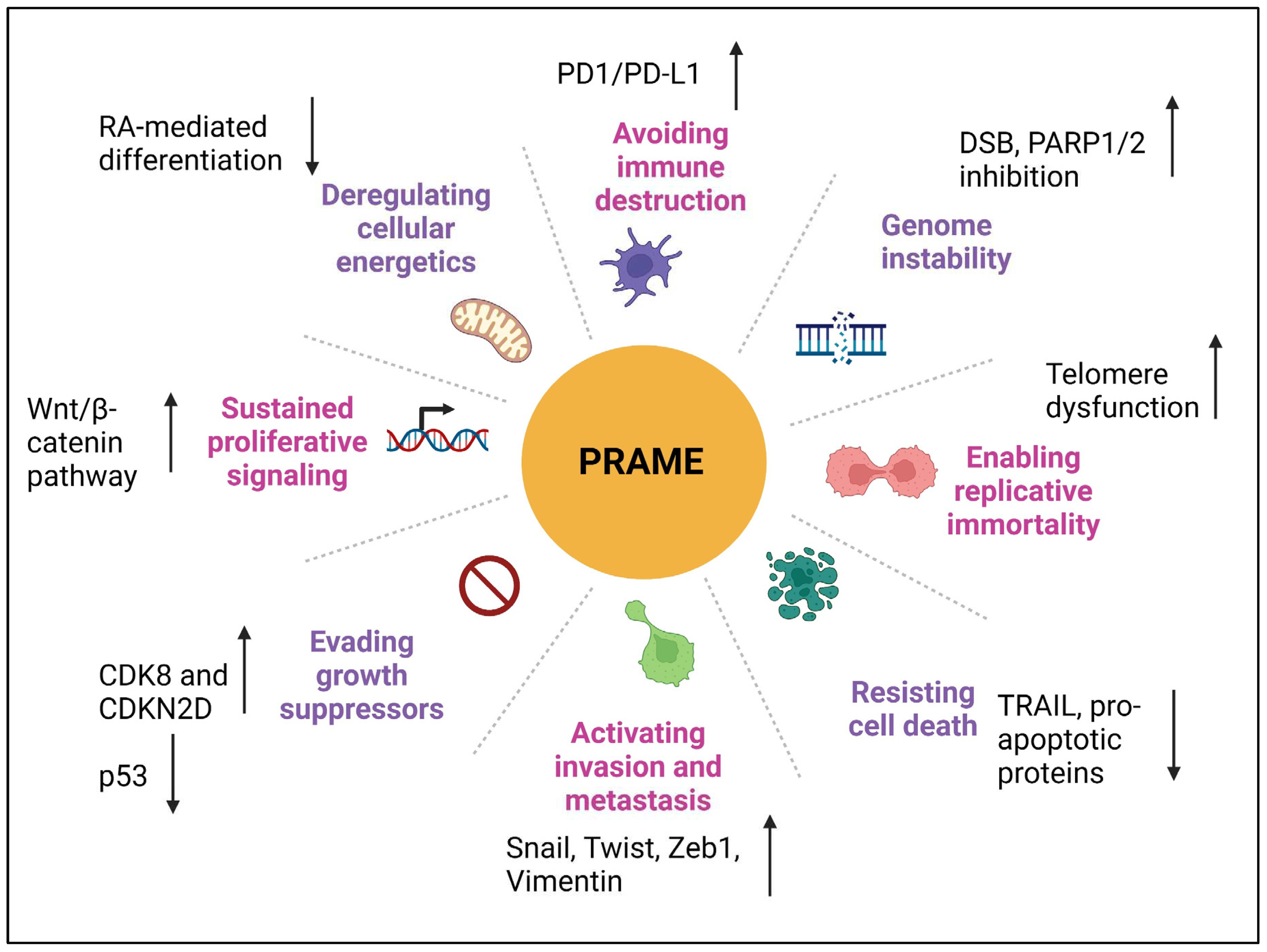
4. Role of PRAME in Different Cancer Hallmarks
4.1. Proliferation
4.2. Invasion and Metastasis
4.3. Epithelial-to-Mesenchymal Transition
4.4. Genomic Instability
4.5. Deregulating Cellular Energetics
4.6. Apoptosis and Chemoresistance
4.7. Immune Evasion
PRAME as a Biomarker
PRAME as a Target for Immunotherapy
Conclusions and Future Perspectives
- (1)
- its restricted expression in testis, ovaries and endometrium and overexpression in a number of cancer tissues, including 80-90% of primary and metastatic melanoma [9], 27-53% breast cancers [53], >90% neuroblastomas [53,80], 40-60% acute myeloid leukemia (AML) [82,89], 20-40% acute lymphoblastic leukemia (ALL) [9,82], 20-50% myeloma [53], and 30-40% chronic myeloid leukemia (CML) [53,134];
- (2)
- It has the ability to elicit T cell mediated immune response;
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ikeda, H.; et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997, 6, 199–208. [Google Scholar] [CrossRef]
- Williams, J.M.; et al. Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: Intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth. Mol. Microbiol. 1998, 27, 171–186. [Google Scholar] [CrossRef]
- Goodison, S.; Urquidi, V. The cancer testis antigen PRAME as a biomarker for solid tumor cancer management. Biomark. Med. 2012, 6, 629–632. [Google Scholar] [CrossRef]
- Hermes, N.; Kewitz, S.; Staege, M.S. Preferentially expressed antigen in melanoma (PRAME) and the PRAME family of leucine-rich repeat proteins. Curr. Cancer Drug Targets 2016, 16, 400–414. [Google Scholar] [CrossRef]
- Baren, V. PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br. J. Haematol. 1998, 102, 1376–1379. [Google Scholar] [CrossRef]
- Xu, Y.; et al. The role of the cancer testis antigen PRAME in tumorigenesis and immunotherapy in human cancer. Cell Prolif. 2020, 53, e12770. [Google Scholar] [CrossRef]
- Kaczorowski, M.; et al. PRAME expression in cancer. A systematic immunohistochemical study of> 5800 epithelial and nonepithelial tumors. Am. J. Surg. Pathol. 2022, 46, 1467. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wadelin, F.; et al. Leucine-rich repeat protein PRAME: Expression, potential functions and clinical implications for leukaemia. Mol. Cancer 2010, 9, 1–10. [Google Scholar] [CrossRef]
- Kawasaki, K.; et al. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997, 7, 250–261. [Google Scholar] [CrossRef]
- Schenk, T.; et al. Hypomethylation of PRAME is responsible for its aberrant overexpression in human malignancies. Genes Chromosomes Cancer 2007, 46, 796–804. [Google Scholar] [CrossRef]
- Varadi, M.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Birtle, Z.; Goodstadt, L.; Ponting, C. Duplication and positive selection among hominin-specific PRAME genes. BMC Genom. 2005, 6, 1–19. [Google Scholar] [CrossRef]
- Jumper, J.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kern, C.H.; Yang, M.; Liu, W.-S. The PRAME family of cancer testis antigens is essential for germline development and gametogenesis. Biol. Reprod. 2021, 105, 290–304. [Google Scholar] [CrossRef]
- Epping, M.T.; et al. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell 2005, 122, 835–847. [Google Scholar] [CrossRef]
- Oehler, V.G.; et al. The preferentially expressed antigen in melanoma (PRAME) inhibits myeloid differentiation in normal hematopoietic and leukemic progenitor cells. Blood J. Am. Soc. Hematol. 2009, 114, 3299–3308. [Google Scholar] [CrossRef]
- Costessi, A.; et al. The tumour antigen PRAME is a subunit of a Cul2 ubiquitin ligase and associates with active NFY promoters. EMBO J. 2011, 30, 3786–3798. [Google Scholar] [CrossRef]
- Lin, H.-C.; et al. CRL2 aids elimination of truncated selenoproteins produced by failed UGA/Sec decoding. Science 2015, 349, 91–95. [Google Scholar] [CrossRef]
- Tajeddine, N.; et al. Tumor-associated antigen preferentially expressed antigen of melanoma (PRAME) induces caspase-independent cell death in vitro and reduces tumorigenicity in vivo. Cancer Res. 2005, 65, 7348–7355. [Google Scholar] [CrossRef]
- Heery, D.M.; et al. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 1997, 387, 733–736. [Google Scholar] [CrossRef]
- Torchia, J.; et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 1997, 387, 677–684. [Google Scholar] [CrossRef]
- Rezvani, K.; et al. Ex vivo characterization of polyclonal memory CD8+ T-cell responses to PRAME-specific peptides in patients with acute lymphoblastic leukemia and acute and chronic myeloid leukemia. Blood J. Am. Soc. Hematol. 2009, 113, 2245–2255. [Google Scholar] [CrossRef]
- Takata, K.; et al. Tumor-associated antigen PRAME exhibits dualistic functions that are targetable in diffuse large B cell lymphoma. J. Clin. Investig. 2022, 132, e145343. [Google Scholar] [CrossRef]
- Mraz, M. Genetic mechanism for the loss of PRAME in B cell lymphomas. J. Clin. Investig. 2022, 132, e160983. [Google Scholar] [CrossRef]
- Zhang, W.; et al. Tumor-associated antigen Prame targets tumor suppressor p14/ARF for degradation as the receptor protein of CRL2Prame complex. Cell Death Differ. 2021, 28, 1926–1940. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Whiteside, T.L. Elevated PRAME expression: What does this mean for treatment of head and neck squamous cell carcinoma? Med. 2013, 7, 575–578. [Google Scholar] [CrossRef]
- Nin, D.S.; Deng, L.-W. Biology of Cancer-Testis Antigens and Their Therapeutic Implications in Cancer. Cells 2023, 12, 926. [Google Scholar] [CrossRef]
- Al-Khadairi, G.; Decock, J. Cancer testis antigens and immunotherapy: Where do we stand in the targeting of PRAME? Cancers 2019, 11, 984. [Google Scholar] [CrossRef]
- Roman-Gomez, J.; et al. Epigenetic regulation of PRAME gene in chronic myeloid leukemia. Leuk. Res. 2007, 31, 1521–1528. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Ortmann, C.A.; et al. Aberrant hypomethylation of the cancer–testis antigen PRAME correlates with PRAME expression in acute myeloid leukemia. Ann. Hematol. 2008, 87, 809–818. [Google Scholar] [CrossRef]
- Gutierrez-Cosio, S.; et al. Epigenetic regulation of PRAME in acute myeloid leukemia is different compared to CD34+ cells from healthy donors: Effect of 5-AZA treatment. Leuk. Res. 2012, 36, 895–899. [Google Scholar] [CrossRef]
- Watari, K.; et al. Identification of a melanoma antigen, PRAME, as a BCR/ABL-inducible gene. FEBS Lett. 2000, 466, 367–371. [Google Scholar] [CrossRef]
- Passeron, T.; et al. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J. Clin. Investig. 2009, 119, 954–963. [Google Scholar] [CrossRef]
- Lee, Y.-K.; et al. Tumor antigen PRAME is up-regulated by MZF1 in cooperation with DNA hypomethylation in melanoma cells. Cancer Lett. 2017, 403, 144–151. [Google Scholar] [CrossRef]
- Sakurai, E.; et al. Downregulation of microRNA-211 is involved in expression of preferentially expressed antigen of melanoma in melanoma cells. Int. J. Oncol. 2011, 39, 665–672. [Google Scholar]
- Nettersheim, D.; et al. The cancer/testis-antigen PRAME supports the pluripotency network and represses somatic and germ cell differentiation programs in seminomas. Br. J. Cancer 2016, 115, 454–464. [Google Scholar] [CrossRef]
- Meng, D.; et al. A transcriptional target of androgen receptor, miR-421 regulates proliferation and metabolism of prostate cancer cells. Int. J. Biochem. Cell Biol. 2016, 73, 30–40. [Google Scholar] [CrossRef]
- Yu, L.; et al. HDAC5-mediated PRAME regulates the proliferation, migration, invasion, and EMT of laryngeal squamous cell carcinoma via the PI3K/AKT/mTOR signaling pathway. Open Med. 2023, 18, 20230665. [Google Scholar] [CrossRef]
- Zhu, H.; et al. Downregulation of PRAME suppresses proliferation and promotes apoptosis in hepatocellular carcinoma through the activation of P53 mediated pathway. Cell. Physiol. Biochem. 2018, 45, 1121–1135. [Google Scholar] [CrossRef]
- Tan, P.; et al. Expression and prognostic relevance of PRAME in primary osteosarcoma. Biochem. Biophys. Res. Commun. 2012, 419, 801–808. [Google Scholar] [CrossRef]
- Chen, X.; et al. PRAME Promotes Cervical Cancer Proliferation and Migration via Wnt/β-Catenin Pathway Regulation. Cancers 2023, 15, 1801. [Google Scholar] [CrossRef]
- Sun, Z.; et al. PRAME is critical for breast cancer growth and metastasis. Gene 2016, 594, 160–164. [Google Scholar] [CrossRef]
- Field, M.G.; et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget 2016, 7, 59209. [Google Scholar] [CrossRef]
- Al-Sulaiti, B.; et al. PRAME, cell migration and invasion of triple negative breast cancer cells. Ann. Oncol. 2017, 28, xi26. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; et al. PRAME expression in head and neck cancer correlates with markers of poor prognosis and might help in selecting candidates for retinoid chemoprevention in pre-malignant lesions. Oral Oncol. 2013, 49, 144–151. [Google Scholar] [CrossRef]
- Huang, H.; et al. Extracellular domain shedding of the ALK receptor mediates neuroblastoma cell migration. Cell Rep. 2021, 36, 109363. [Google Scholar] [CrossRef]
- Huang, Q.; et al. Preferentially expressed antigen of melanoma prevents lung cancer metastasis. PLoS ONE 2016, 11, e0149640. [Google Scholar] [CrossRef]
- Al-Khadairi, G.; et al. PRAME promotes epithelial-to-mesenchymal transition in triple negative breast cancer. J. Transl. Med. 2019, 17, 1–14. [Google Scholar] [CrossRef]
- Hedrich, V.; et al. PRAME Is a Novel Target of Tumor-Intrinsic Gas6/Axl Activation and Promotes Cancer Cell Invasion in Hepatocellular Carcinoma. Cancers 2023, 15, 2415. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; et al. PRAME induces genomic instability in uveal melanoma. 2023.
- Epping, M.T.; Bernards, R. A causal role for the human tumor antigen preferentially expressed antigen of melanoma in cancer. Cancer Res. 2006, 66, 10639–10642. [Google Scholar] [CrossRef]
- Costantini, L.; et al. Retinoic acids in the treatment of most lethal solid cancers. J. Clin. Med. 2020, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Siddikuzzaman, C.G.; Grace, V.B. All trans retinoic acid and cancer. Immunopharmacol. Immunotoxicol. 2010, 33, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Kewitz, S.; Staege, M.S. Knock-down of PRAME increases retinoic acid signaling and cytotoxic drug sensitivity of Hodgkin lymphoma cells. PLoS ONE 2013, 8, e55897. [Google Scholar] [CrossRef]
- De Carvalho, D.; et al. PRAME/EZH2-mediated regulation of TRAIL: A new target for cancer therapy. Curr. Mol. Med. 2013, 13, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Mello, B.P.; et al. Regulation of TRAIL expression by PRAME and EZH2 as potential therapeutic target against solid tumors. BMC Proceedings. 2013, 7, 10. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; et al. Expression of MAGE-A3, NY-ESO-1, LAGE-1 and PRAME in urothelial carcinoma. Br. J. Cancer 2012, 107, 116–122. [Google Scholar] [CrossRef]
- Xu, Y.; et al. PRAME promotes in vitro leukemia cells death by regulating S100A4/p53 signaling. Eur Rev Med Pharmacol Sci 2016, 20, 1057–1063. [Google Scholar]
- Wadelin, F.R.; et al. PRAME is a golgi-targeted protein that associates with the Elongin BC complex and is upregulated by interferon-gamma and bacterial PAMPs. PLoS ONE 2013, 8, e58052. [Google Scholar] [CrossRef]
- Babiak, A.; et al. Frequent T cell responses against immunogenic targets in lung cancer patients for targeted immunotherapy. Oncol. Rep. 2014, 31, 384–390. [Google Scholar] [CrossRef]
- Matko, S.; et al. PRAME peptide-specific CD8+ T cells represent the predominant response against leukemia-associated antigens in healthy individuals. Eur. J. Immunol. 2018, 48, 1400–1411. [Google Scholar] [CrossRef]
- Setiadi, A.F.; et al. Identification of mechanisms underlying transporter associated with antigen processing deficiency in metastatic murine carcinomas. Cancer Res. 2005, 65, 7485–7492. [Google Scholar] [CrossRef]
- Vyas, J.M.; Van der Veen, A.G.; Ploegh, H.L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 2008, 8, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Setiadi, A.F.; et al. Epigenetic control of the immune escape mechanisms in malignant carcinomas. Mol. Cell. Biol. 2007, 27, 7886–7894. [Google Scholar] [CrossRef] [PubMed]
- Roszik, J.; et al. Overexpressed PRAME is a potential immunotherapy target in sarcoma subtypes. Clin. Sarcoma Res. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Iura, K.; et al. Cancer-testis antigens PRAME and NY-ESO-1 correlate with tumour grade and poor prognosis in myxoid liposarcoma. J. Pathol. Clin. Res. 2015, 1, 144–159. [Google Scholar] [CrossRef]
- Wang, W.-L.; et al. RNA expression profiling reveals PRAME, a potential immunotherapy target, is frequently expressed in solitary fibrous tumors. Mod. Pathol. 2021, 34, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Naik, A.; et al. Cancer testis antigen PRAME: An anti-cancer target with immunomodulatory potential. J. Cell. Mol. Med. 2021, 25, 10376–10388. [Google Scholar] [CrossRef]
- Gassenmaier, M.; et al. Diffuse PRAME expression is highly specific for thin melanomas in the distinction from severely dysplastic nevi but does not distinguish metastasizing from non-metastasizing thin melanomas. Cancers 2021, 13, 3864. [Google Scholar] [CrossRef]
- Bahmad, H.F.; Oh, K.S.; Alexis, J. Potential diagnostic utility of PRAME and p16 immunohistochemistry in melanocytic nevi and malignant melanoma. J. Cutan. Pathol. 2023, 50, 763–772. [Google Scholar] [CrossRef]
- Chen, Y.P.; et al. PRAME is a useful marker for the differential diagnosis of melanocytic tumours and histological mimics. Histopathology 2023, 82, 285–295. [Google Scholar] [CrossRef]
- Kunc, M.; et al. Diagnostic test accuracy meta-analysis of PRAME in distinguishing primary cutaneous melanomas from benign melanocytic lesions. Histopathology 2023, 83, 3–14. [Google Scholar] [CrossRef]
- Parra, O.; et al. PRAME expression in cutaneous melanoma does not correlate with disease-specific survival. J. Cutan. Pathol. 2023, 50, 903–912. [Google Scholar] [CrossRef]
- Doolan, P.; et al. Prevalence and prognostic and predictive relevance of PRAME in breast cancer. Breast Cancer Res. Treat. 2008, 109, 359–365. [Google Scholar] [CrossRef]
- Epping, M.; et al. PRAME expression and clinical outcome of breast cancer. Br. J. Cancer 2008, 99, 398–403. [Google Scholar] [CrossRef]
- Conte, F.; et al. In silico recognition of a prognostic signature in basal-like breast cancer patients. PLoS ONE 2022, 17, e0264024. [Google Scholar] [CrossRef]
- Orlando, D.; et al. Adoptive Immunotherapy Using PRAME-Specific T Cells in MedulloblastomaTCR-Based Immunotherapy for Medulloblastoma. Cancer Res. 2018, 78, 3337–3349. [Google Scholar] [CrossRef]
- Oberthuer, A.; et al. The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin. Cancer Res. 2004, 10, 4307–4313. [Google Scholar] [CrossRef]
- Steinbach, D.; et al. Clinical implications of PRAME gene expression in childhood acute myeloid leukemia. Cancer Genet. Cytogenet. 2002, 133, 118–123. [Google Scholar] [CrossRef]
- Ding, K.; et al. PRAME gene expression in acute leukemia and its clinical significance. Cancer Biol. Med. 2012, 9, 73. [Google Scholar]
- Fu, R.; Ding, K.; Shao, Z. The Expression and Clinical Significance of PRAME Gene in Acute Leukemia; American Society of Hematology: Washington, DC, USA, 2007. [Google Scholar]
- Steger, B.; et al. WT1, PRAME, and PR3 mRNA expression in acute myeloid leukemia (AML). J. Immunother. 2020, 43, 204–215. [Google Scholar] [CrossRef]
- Kulkarni, N.V.; et al. Correlation of preferentially expressed antigen of melanoma (PRAME) gene expression with clinical characteristics in acute leukemia patients. J. Genet. Eng. Biotechnol. 2022, 20, 97. [Google Scholar] [CrossRef]
- Santamaría, C.; et al. The relevance of preferentially expressed antigen of melanoma (PRAME) as a marker of disease activity and prognosis in acute promyelocytic leukemia. 2008.
- Abdelmalak, C.; et al. PRAME gene expression in childhood acute lymphoblastic leukemia: Impact on prognosis. Clin. Lab. 2014, 60, 55–61. [Google Scholar] [CrossRef]
- Zhang, W.; et al. PRAME expression and promoter hypomethylation in epithelial ovarian cancer. Oncotarget 2016, 7, 45352. [Google Scholar] [CrossRef] [PubMed]
- Greiner, J.; et al. Expression of tumor-associated antigens in acute myeloid leukemia: Implications for specific immunotherapeutic approaches. Blood 2006, 108, 4109–4117. [Google Scholar] [CrossRef] [PubMed]
- Abramenko, I.В.; et al. Expression of PRAME gene in multiple myeloma. Ter. Arkhiv 2004, 79, 77–81. [Google Scholar]
- Misurin, A.; et al. Expression of PRAME and WT1 in Multiple Myeloma Patients during High Dose Chemotherapy and Auto-SCT; American Society of Hematology: Washington, DC, USA, 2008. [Google Scholar]
- Yang, L.; et al. PRAME gene copy number variation is related to its expression in multiple myeloma. DNA Cell Biol. 2017, 36, 1099–1107. [Google Scholar] [CrossRef]
- Yang, L.; et al. Both Methylation and Copy Number Variation Participated in the Varied Expression of PRAME in Multiple Myeloma. OncoTargets Ther. 2020, 13, 7545. [Google Scholar] [CrossRef]
- Takata, K.; et al. PRAME Expression Is Correlated with Treatment Outcome and Specific Features of the Tumor Microenvironment in Classical Hodgkin Lymphoma. Blood 2019, 134, 1509. [Google Scholar] [CrossRef]
- Mitsuhashi, K.; et al. Prognostic significance of PRAME expression based on immunohistochemistry for diffuse large B-cell lymphoma patients treated with R-CHOP therapy. Int. J. Hematol. 2014, 100, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Baba, H.; et al. PRAME expression as a potential biomarker for hematogenous recurrence of esophageal squamous cell carcinoma. Anticancer Res. 2019, 39, 5943–5951. [Google Scholar] [CrossRef] [PubMed]
- Oyama, K.; et al. Impact of preferentially expressed antigen of melanoma on the prognosis of hepatocellular carcinoma. Gastrointest. Tumors 2016, 3, 128–135. [Google Scholar] [CrossRef]
- Pan, S.H.; et al. Gene expression of MAGE-A3 and PRAME tumor antigens and EGFR mutational status in Taiwanese non–small cell lung cancer patients. Asia-Pac. J. Clin. Oncol. 2017, 13, e212–e223. [Google Scholar] [CrossRef] [PubMed]
- Thongprasert, S.; et al. The prevalence of expression of MAGE-A3 and PRAME tumor antigens in East and South East Asian non-small cell lung cancer patients. Lung Cancer 2016, 101, 137–144. [Google Scholar] [CrossRef]
- Gezgin, G.; et al. PRAME as a potential target for immunotherapy in metastatic uveal melanoma. JAMA Ophthalmol. 2017, 135, 541–549. [Google Scholar] [CrossRef]
- Ludzik, J.; et al. Dermoscopic features associated with 3-GEP PLA: LINC00518, PRAME, and TERT expression in suspicious pigmented lesions. Ski. Res. Technol. 2023, 29, e13323. [Google Scholar] [CrossRef]
- Kuruwitage Ishikawa, A.S.; et al. Quantitative expression evaluation of PRAME gene in osteosarcoma. Mol. Biol. Rep. 2023, 50, 4301–4307. [Google Scholar] [CrossRef]
- Cammareri, C.; et al. PRAME immunohistochemistry in soft tissue tumors and mimics: A study of 350 cases highlighting its imperfect specificity but potentially useful diagnostic applications. Virchows Archiv 2023, 483, 145–156. [Google Scholar] [CrossRef]
- Partheen, K.; et al. Four potential biomarkers as prognostic factors in stage III serous ovarian adenocarcinomas. Int. J. Cancer 2008, 123, 2130–2137. [Google Scholar] [CrossRef]
- Partheen, K.; et al. Expression analysis of stage III serous ovarian adenocarcinoma distinguishes a sub-group of survivors. Eur. J. Cancer 2006, 42, 2846–2854. [Google Scholar] [CrossRef]
- Ricci, C.; et al. PRAME Expression in Mucosal Melanoma of the Head and Neck Region. Am. J. Surg. Pathol. 2023, 47, 599. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; et al. Clinicopathological and prognostic significance of PRAME overexpression in human cancer: A meta-analysis. BioMed Res. Int. 2020, 2020, 8828579. [Google Scholar] [CrossRef] [PubMed]
- Misyurin, V.; et al. Prognostic value of PRAME activity in tumor cells of follicular lymphoma. Clin. Oncohematol 2019, 12, 173–178. [Google Scholar] [CrossRef]
- Ricci, C.; et al. Immunohistochemical Expression of Preferentially Expressed Antigen in Melanoma (PRAME) in the Uninvolved Background Testis, Germ Cell Neoplasia in Situ, and Germ Cell Tumors of the Testis. Am. J. Clin. Pathol. 2022, 157, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Ercolak, V.; et al. PRAME expression and its clinical relevance in Hodgkin's lymphoma. Acta Haematol. 2015, 134, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Vulcani-Freitas, T.M.; et al. Perfil de expressão do gene PRAME em meduloblastoma. Arquivos Neuro-Psiquiatria 2011, 69, 9–12. [Google Scholar] [CrossRef]
- Brenne, K.; et al. PRAME (preferentially expressed antigen of melanoma) is a novel marker for differentiating serous carcinoma from malignant mesothelioma. Am. J. Clin. Pathol. 2012, 137, 240–247. [Google Scholar] [CrossRef]
- Neumann, E.; et al. Heterogeneous expression of the tumor-associated antigens RAGE-1, PRAME, and glycoprotein 75 in human renal cell carcinoma: Candidates for T-cell-based immunotherapies? Cancer Res. 1998, 58, 4090–4095. [Google Scholar]
- Pankov, D.; et al. In vivo immuno-targeting of an extracellular epitope of membrane bound preferentially expressed antigen in melanoma (PRAME). Oncotarget 2017, 8, 65917. [Google Scholar] [CrossRef]
- Pujol, J.-L.; et al. Safety and immunogenicity of the PRAME cancer immunotherapeutic in patients with resected non–small cell lung cancer: A phase I dose escalation study. J. Thorac. Oncol. 2016, 11, 2208–2217. [Google Scholar] [CrossRef] [PubMed]
- Kirkey, D.C.; et al. Therapeutic targeting of PRAME with mTCRCAR T cells in acute myeloid leukemia. Blood Adv. 2023, 7, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Loeb, A.M.; et al. Targeting PRAME with TCR-Mimic CAR T Cells in AML. Blood 2021, 138, 733. [Google Scholar] [CrossRef]
- Fløisand, Y.; et al. WT1 and PRAME RNA-loaded dendritic cell vaccine as maintenance therapy in de novo AML after intensive induction chemotherapy. Leukemia 2023, 37, 1842–1849. [Google Scholar] [CrossRef]
- Sailer, N.; et al. T-cells expressing a highly potent PRAME-specific T-cell receptor in combination with a chimeric PD1-41BB co-stimulatory receptor show a favorable preclinical safety profile and strong anti-tumor reactivity. Cancers 2022, 14, 1998. [Google Scholar] [CrossRef]
- Dwivedi, R.; et al. A systematic review of potential immunotherapies targeting PRAME in retinoid resistant oral potentially malignant disorders and oral cancer. Curr. Mol. Med. 2022, 22, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, M.K.; Gjerstorff, M.F. CAR T-cell cancer therapy targeting surface cancer/testis antigens. Front. Immunol. 2020, 11, 1568. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, R.E.; et al. Beyond Checkpoint Inhibitors: Enhancing Antitumor Immune Response in Lung Cancer. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Bunk, S.; et al. Effective Targeting of PRAME-Positive Tumors with Bispecific T Cell-Engaging Receptor (TCER®) Molecules. Blood 2019, 134, 3368. [Google Scholar] [CrossRef]
- Vasileiou, S.; et al. T-cell therapy for lymphoma using nonengineered multiantigen-targeted T cells is safe and produces durable clinical effects. J. Clin. Oncol. 2021, 39, 1415. [Google Scholar] [CrossRef]
- Kinoshita, H.; et al. Outcome of donor-derived TAA-T cell therapy in patients with high-risk or relapsed acute leukemia post allogeneic BMT. Blood Adv. 2022, 6, 2520–2534. [Google Scholar] [CrossRef]
- Lulla, P.D.; et al. Clinical effects of administering leukemia-specific donor T cells to patients with AML/MDS after allogeneic transplant. Blood 2021, 137, 2585–2597. [Google Scholar] [CrossRef]
- Hont, A.B.; et al. Immunotherapy of relapsed and refractory solid tumors with ex vivo expanded multi-tumor associated antigen specific cytotoxic T lymphocytes: A phase I study. J. Clin. Oncol. 2019, 37, 2349. [Google Scholar] [CrossRef]
- Hoyos, V.; et al. Multi-antigen-targeted T-cell therapy to treat patients with relapsed/refractory breast cancer. Ther. Adv. Med. Oncol. 2022, 14, 17588359221107113. [Google Scholar] [CrossRef] [PubMed]
- Smaglo, B.G.; et al. A phase I trial targeting advanced or metastatic pancreatic cancer using a combination of standard chemotherapy and adoptively transferred nonengineered, multiantigen specific T cells in the first-line setting (TACTOPS); American Society of Clinical Oncology: Alexandria, VA, USA, 2020. [Google Scholar]
- Cheever, M.A.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef] [PubMed]
- Sahraei, M.; et al. Repression of MUC1 Promotes Expansion and Suppressive Function of Myeloid-Derived Suppressor Cells in Pancreatic and Breast Cancer Murine Models. Int. J. Mol. Sci. 2021, 22, 5587. [Google Scholar] [CrossRef]
- Bose, M.; et al. Overexpression of MUC1 Induces Non-Canonical TGF-β Signaling in Pancreatic Ductal Adenocarcinoma. Front. Cell Dev. Biol. 2022, 39, 821875. [Google Scholar] [CrossRef]
- Bose, M.; Mukherjee, P. Microbe–MUC1 crosstalk in cancer-associated infections. Trends Mol. Med. 2020, 26, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Radich, J.P.; et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 2794–2799. [Google Scholar] [CrossRef]
- Gerber, J.M.; et al. Characterization of chronic myeloid leukemia stem cells. Am. J. Hematol. 2011, 86, 31–37. [Google Scholar] [CrossRef]
- Bose, M.; Mukherjee, P. Potential of anti-MUC1 antibodies as a targeted therapy for gastrointestinal cancers. Vaccines 2020, 8, 659. [Google Scholar] [CrossRef] [PubMed]
- Zahavi, D.; Weiner, L. Monoclonal antibodies in cancer therapy. Antibodies 2020, 9, 34. [Google Scholar] [CrossRef]
- Chang, A.Y.; et al. A therapeutic T cell receptor mimic antibody targets tumor-associated PRAME peptide/HLA-I antigens. J. Clin. Investig. 2017, 127, 2705–2718. [Google Scholar] [CrossRef]
- Weber, G.; et al. Generation of tumor antigen-specific T cell lines from pediatric patients with acute lymphoblastic leukemia—Implications for immunotherapy. Clin. Cancer Res. 2013, 19, 5079–5091. [Google Scholar] [CrossRef] [PubMed]
- Quintarelli, C.; et al. Cytotoxic T lymphocytes directed to the preferentially expressed antigen of melanoma (PRAME) target chronic myeloid leukemia. Blood J. Am. Soc. Hematol. 2008, 112, 1876–1885. [Google Scholar] [CrossRef]
- Griffioen, M.; et al. Detection and functional analysis of CD8+ T cells specific for PRAME: A target for T-cell therapy. Clin. Cancer Res. 2006, 12, 3130–3136. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.H.; et al. Efficient identification of novel Hla-A* 0201–Presented Cytotoxic T Lymphocyte epitopes in the widely expressed tumor antigen prame by Proteasome-Mediated digestion analysis. J. Exp. Med. 2001, 193, 73–88. [Google Scholar] [CrossRef]
- Weber, J.S.; et al. A phase 1 study of a vaccine targeting preferentially expressed antigen in melanoma and prostate-specific membrane antigen in patients with advanced solid tumors. Journal of immunotherapy Hagerstown 2011, 34, 556. [Google Scholar] [CrossRef]
- Gutzmer, R.; et al. Safety and immunogenicity of the PRAME cancer immunotherapeutic in metastatic melanoma: Results of a phase I dose escalation study. ESMO Open 2016, 1, e000068. [Google Scholar] [CrossRef]
- Sakamoto, S.; et al. Prospect and progress of personalized peptide vaccinations for advanced cancers. Expert Opin. Biol. Ther. 2016, 16, 689–698. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar] [PubMed]
- Huber, E.M.; et al. Immuno-and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, B.; et al. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J. Immunol. 2012, 189, 3538–3547. [Google Scholar] [CrossRef]
- Hinrichs, C.S.; Rosenberg, S.A. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol. Rev. 2014, 257, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; et al. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene 2020, 39, 4909–4924. [Google Scholar] [CrossRef] [PubMed]
| Cohort | PRAME detection frequency | Disease | Clinico-pathological parameters | References |
|---|---|---|---|---|
| Egyptian | 20-40% | Acute Lymphoblastic Leukemia | Correlated with increased overall and disease-free survival and lower relapse | [87] |
| German | 40-60% | Acute Myeloid Leukemia | Positively correlated with increased overall and disease-free survival and negatively correlated to the white blood cell count at diagnosis | [81,89] |
| Dutch, Irish | 27%-53% | Breast cancer | Independent marker for poor disease-free and overall survival and distant metastases, correlates with negative estrogen receptor status | [76,77] |
| Chinese | 80-90% | Cervical cancer | Associated with increased proliferation and migration in CC cells. | [43] |
| 30-40% | Chronic Myeloid Leukemia | |||
| Canadian,Japanese | Deleted in >13% of tumors in the Canadian cohort>30% overexpressed in the Japanese cohort | Diffuse Large B-cell Lymphoma | Deletion of PRAME was associated with decreased overall and disease-free survival in the Canadian cohort.PRAME overexpression was correlated with shorter progression-free and overall survival, and decreased response to chemotherapy in the Japanese cohort. | [24,95] |
| Japanese | 87% tumor tissues | Esophageal cancer | Shorter disease-specific survival and hematogenous recurrence | [96] |
| Russian | 42-86% in lymph node, bone marrow and blood | Follicular lymphoma | Higher Ki-67 activity and larger tumor mass. Survival parameters were worse with high PRAME expression level. Combination of both high FLIPI-1/FLIPI-2 risk, and high PRAME expression level determines extremely unfavorable prognosis. | [108] |
| Polish | >75% in HNSCC tissues and lymph nodes | Head and neck squamous carcinoma | Correlates with the tumor grade, size, nodal involvement, and the clinical status of HNSCC patients. | [47] |
| Chinese, Japanese | 27-60% | Hepatocellular carcinoma | Correlated with high Ki-67 activity, AJCC stage, tumor size, metastasis, invasion, poor overall survival | [41,97] |
| Turkish | 10-69% | Hodgkin’s lymphoma | Correlated with shorter disease-free survival and overall survival | [110] |
| Taiwanese,East and South-East Asian | >30%>50% | Non-small Cell Lung carcinoma | Increased in squamous cell carcinomas compared to adenocarcinomas in all the cohorts, and in smokers compared to non-smokers in the East and South-eat Asian cohort.No correlation with survival in the Taiwanese cohort | [98,99] |
| Roman, Brazilian | >80% | Medulloblastoma | No significant association | [79,111] |
| American,German | 80-90% | Melanoma | No prognostic significance in thin melanoma.PRAME+/p16- melanocytic lesion is unlikely to be a nevus but most nevi were PRAME-/p16+ | [71,72] |
| Chinese, Russian | 20-68% | Multiple myeloma | Correlated with lower progression-free survival, unfavorable prognosis | [90,91,92,93] |
| German | >90% | Neuroblastoma | Correlated with higher tumor stage and the age of patients at diagnosis | [80] |
| Chinese | >68% | Osteosarcoma | Associated with poor prognosis and lung metastasis | [42,102] |
| Swedish, Norwegian, Chinese | 60-90% | Ovarian cancer | Associated with overall survival, disease-free survival, grade, stage, metastasis, | [88,104,105,112] |
| German | >40% | Renal cell carcinoma | Associated with unfavorable prognosis | [113] |
| Italian | >70% | Testicular cancer | Associated with seminomas | [109] |
| Danish | >20% | Urothelial cancer | Correlated with high grade and stage and poor response to chemotherapy | [59] |
| NCT number and name | Disease | Treatment | Start Year | Publication |
|---|---|---|---|---|
| NCT01333046, ACTAL | Hodgkin lymphoma, non-Hodgkin lymphoma, Hodgkin disease |
Multi TAA T cells (NY-ESO-1, MAGEA4, PRAME, Survivin and SSX), and Azacitidine |
2012 | [124] |
| NCT02203903, RESOLVE | Relapsed/refractory hematopoietic malignancies (ALL, AML, CML, MDS) |
Multi TAA T cells (WT1, PRAME, and Survivin) |
2015 | [125] |
| NCT02239861, TACTASOM | Rhabdomyosarcoma | Multi TAA T cells (NY-ESO-1, MAGEA4, PRAME, Survivin, and SSX) |
2015 | NR |
| NCT02291848, TACTAM | Multiple myeloma | Multi TAA T cells (NY-ESO-1, MAGEA4, PRAME, Survivin, and SSX) |
2015 | NR |
| NCT02475707, STELLA | Leukemia, ALL | Multi TAA T cells (WT1, PRAME, and Survivin) |
2016 | NR |
| NCT02494167, ADSPAM | AML, MDS | Multi TAA T cells (WT1, NY-ESO-1, PRAME, and Survivin) |
2016 | [126] |
| NCT02743611, BP-011 | AML, MDS, uveal melanoma | BPX-701 and Rimiducid | 2017 | NR |
| NCT02789228, REST | Solid tumors (Wilms tumor, neuroblastoma, rhabdomyosarcoma, adenocarcinoma, and esophageal cancer) | Multi TAA T cells (WT1, PRAME, and/or Survivin) |
2016 | [127] |
| NCT03093350, TACTIC | Breast cancer | Multi TAA T cells (NY-ESO-1, MAGEA4, PRAME, Survivin, and SSX2) |
2017 | [128] |
| NCT03192462, TACTOPS | Pancreatic cancer | Multi TAA T cells (NY-ESO-1, MAGEA4, PRAME, Survivin, |
2018 | [129] |
| NCT03503968, CD-TCR-001 | High risk myeloid andlymphoid neoplasms | MDG1011 | 2018 | NR |
| NCT03652545, REMIND | Brain tumorTAA, | Multi TAA T cells (WT1, PRAME, and/or Survivin) |
2018 | NR |
| NCT03686124 | Solid tumor | Autologous PRAME-targeting TCR-engineered T-cells ACTengine® IMA203/IMA203CD8 as Monotherapy or in Combination with Nivolumab |
2019 | NR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




