1. Messenger RNA spike protein and host genome alteration the role of retrotransposons
The central dogma of molecular biology gives a key role to mRNAs considered critical mediators between inherited DNA encoded genetic information and proteomes/metabolomes that drive the multitude activities and functions at both cellular and organ levels [
1,
2,
3]. The free-floating spike proteins synthetized by cells targeted by the SARS-CoV-2 virus and destroyed by the immune response massively circulate in the blood and systematically approach ACE2 receptors expressed by a variety of cells, thereby promoting ACE2 internalization and degradation [
3,
4,
5,
6].
One possible explanation for the continued detection of SARS-CoV-2 viral spike protein mRNA in the absence of virus reproduction could be explained by the fact that in some cases, either DNA copies of viral subgenomic RNAs and mRNA of spike protein may eventually integrate into the host cells DNA by a reverse transcription (RT) mechanism [
7,
8,
9]. The formation of retrocopies in a human cell lineage is a quite well known process and predominantly includes LINE-1 or L1 (long interspersed element-1) retrotransposons-retrotransposition, albeit there there is also the possibility retrotransposition may take place through long terminal repeat (LTR) retrotransposons [
8,
9,
10].
Human L1 elements consist almost the 17% of the human genome, and are considered autonomous retrotransposons, which are able to retro-transpose themselves and other nonautonomous elements such as Alu, in fact L1 are a source of cellular endogenous RT [
11,
12,
13,
14]. Endogenous L1 elements have been shown to be expressed in aged human tissues and L1-mediated somatic retrotransposition is common in cancer patients and HIV patients. In addition, viral infection, including SARS-CoV-2 and Zika virus (ZIKV) infection are able to up-regulate the expression of endogenous L1 and other retrotransposons in host cells [
15,
16,
17,
18].
Based on the current knowledge, asserting that the only known mechanism by which RNA can integrate into the host genome should relies on the presence of a retrovirus particle containing RT may be deceiving as it does not consider the presence and significance of L1-driven retrotransposition in humans cells [
11,
12,
13]. Therefore, understanding the relationships between different mechanisms of mRNA spike protein cleavage and their accumulation would be consistent to presence of a unique pathophysiological mechanism which may eventually explain the risk of to severe forms of COVID-19 either Long-COVID or Spike protein syndrome [
19,
20,
21,
22].
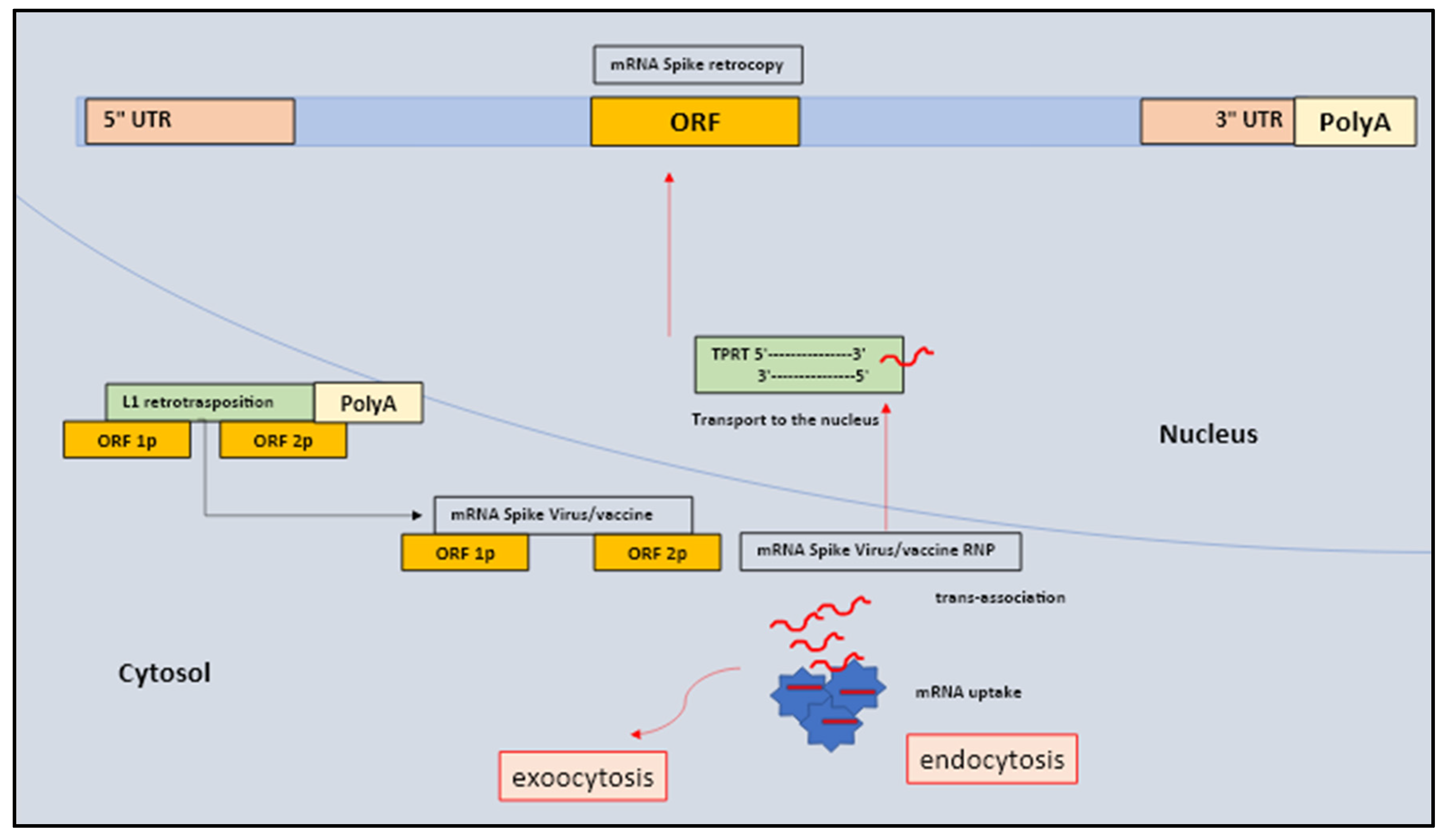
Early mammalian embryos undergo deep epigenetic reprogramming associated with chromatin re-organization. Retrotransposition is the molecular process in which transcribed and spliced mRNAs are accidentally reverse-transcribed and inserted into new genomic positions to form a retrogene at A/T-rich consensus target sites via the process termed target-primed reverse transcription (TPRT) [
19,
20,
21,
22]. The peculiar traits of these retrosequences are usually composed of a poly(A) tail tract, the loss of introns, and the presence of target site duplications that may differ either in size or sequence. In the antisense direction, L1 also codes for ORF0p-1p-2p small peptide with two subunits (1 and 2) located in the nucleus and capable of enhancing the efficiency of retrotransposition [
23]. Although, such retrogenes are often non-expressed pseudogenes due to the lack of regulatory sequences, some of them may accidentally recruit certain regulatory sequences and start acquiring new functions generating new functional retrogene, usually a chimera [
24,
25,
26,
27,
28,
29,
30,
31,
32,
33].
These chimeras tend to show either a mosaic structure with the retrogene coding regions combined with novel regulatory sequences that are not present in parental genes, or a hybrid coding region consisting of exons from unrelated genes, positioned near the insertion site in addition to a new regulatory sequences. Of note, the L1 protein machinery works on several levels preferentially targeting their encoding mRNA (cis-preference), mobilizing a variety of other RNAs present in the cell (trans-association) which then include non-autonomous mobile elements (Alu, SVA) and splicesomal RNAs and several protein-coding mRNAs [
28,
29,
30,
31,
32,
33,
34,
35].
As matter of fact, both transposomes and retrotransposomes showed to play big role in the genome, even though a great part have been truncated during genome evolution. Overall, the data have shown that LTR-mediated retrotransposition is highly conserved across a wide range of animal taxa and it represents one of the key mode of ongoing genetic evolution in eukaryotes. Therefore, many could be the factors and elements related to the deep damage of the mRNA spike protein, both by vaccines and viruses, seen as the possible cause that increases their L1-mediated retrotransposition and subsequent integration into the host genome [
28,
29,
30,
31,
32,
33,
34].
A matter of primary concern is the fact that all nucleated cells are competent for endocytosis and can become receptive to mRNA (macrophages, dendritic cells (DCs), professional antigen-presenting cells (APCs) as well as pluripotent and multipotent stem cells (SC). Several lines of evidence confirmed that the cellular uptake of lipid nanoparticles (LNP) mRNA relies on the binding of apolipoprotein E (ApoE) to LNPs and their subsequent endocytosis which is facilitated by lipoprotein a receptors. low density (LDL). The scenario suggests that virus mRNAs may eventually encounter the L1 machinery since the L1 elements are generally expressed in different human somatic tissues, including CNS competent cells (glial cells, neuronal progenitor cells, differentiating neurons and mature non-dividing neurons), liver, spleen, adrenal glands, lungs, heart as well as in lymphoblastoid cell lines, platelets, megakaryocyte, and T cells [
34,
35,
36,
37].
2. Pluripotent, multipotent and progenitor adult stem cells the mRNA of spike protein the telomerasi, the retrotransposition integrated mechanism and the sequential cellular damages
Interestingly, both in SC and in adult neurogenesis, L1 retrotransposition plays a key role in the creation of an important neuronal mosaicism, which then leads to the onset of several neurological disorders [
36,
37,
38]. However, the main problem in this case may be attributable to progenitor cells, pluripotent and multipotent SCs because they possess a more open-state genome than adult somatic cells, which play a role in gene regulation by actively remodeling chromatin structure or providing more sites transcription factor binding [
36,
37,
38]. For example, the presence of an efficient L1 retrotransposition in mature non-dividing somatic and germ cells includes most of the cells in the respective organs, therefore restricted to a well-circumscribed area, while SCs can involve more cell phenotypes and therefore tissues and organs [
36,
37,
38].
There may also be a possibility that SARS-CoV-2 virus mRNA is epigenetically inherited via sperm RNA cargo. This can occur if testis cells of the male germinal lineage take up LNPs or EVs containing vaccine mRNA and if these mRNAs then end up in spermatozoa. Alternatively, during their functional maturation in the epididymis, spermatozoa could potentially actively internalize vaccine mRNAs supplied by epididymal EVs. The presence and integration of viral mRNAs in human sperm could be easily tested because the semen of vaccinated men, compared to other tissues relevant for integration, is a relatively accessible body fluid [
2,
39,
40,
41].
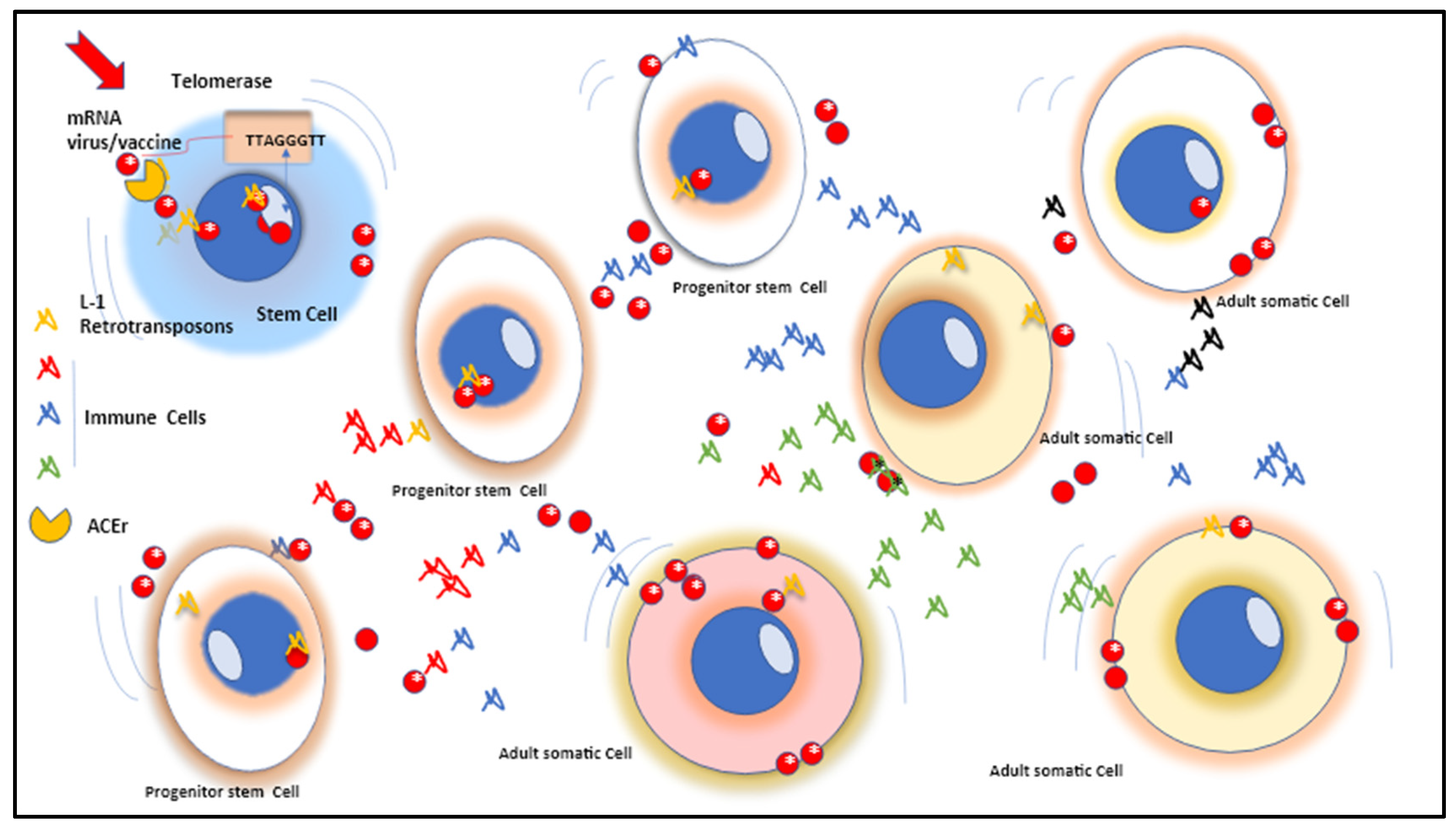
Regarding stem cells, one of the most important traits is the presence of telomeres and telomerase. Telomere stability and integrity are essential for genome stability, any dysregulation results in damage leading to early apoptosis, cell senescence or abnormal cell proliferation. It has been shown that telomerase and retrotransposon have a number of structural and mechanistic similarities, probably due to a common origin [
42,
43,
44,
45]. Retrotransposon telomerase itself ensures the telomere repeats in most eukaryotic organisms, which consists of a reverse transcriptase (RT) and RNA template key factors in the synthesis of the G-rich strand of telomere terminal repeats. Telomerase reverse transcriptase (TERT) is adopted with special N- and C-terminal extensions assisting a central RT-like domain. Telomerase includes two hallmarks of TERT which consist of a stable relationship with telomerase RNA and the ability to continuously reverse transcribe the RNA template segment [
46,
47,
48,
49] (
Figure 1).
Although telomerase activity in stem cells helps to stabilize stem cell genomes, it can also increase the risk of accumulating mutations that are transcribed and fixed in clonal cells and future phenotypes [
50,
51,
52,
53]. An interesting point is that the plasticity of the retroelements is that group II introns are expected to be of bacterial origin and are considered the archetype of the major eukaryotic spliceosomal introns. Thus, similar to TP-reverse transposition (TPRT) and telomere synthesis, after self-splicing, the intron can reverse splice into a DNA target and be reverse-transcribed by the TPRT [
50,
51,
52,
53].
Unlike telomeres which are typically confined to the ends of chromosomes, reverse transcriptions of viruses and transposons can target a plethora of sites. Furthermore, the RNA genomes of retrotransposons and LTR retroviruses are both reverse-transcribed within RNP complexes in the cytoplasm, thus the cDNA is integrated into the host genome. Host proteins transcribe retrotransposon or integrated provirus [
52,
53]. This generates another round of template RNA for protein synthesis. RNAs can also be used to generate additional genomes for VLPs (LTR retrotransposons) or virus particles (retroviruses) [
52,
53]. This highly complex mode of viral particle replication requires reverse transcription to be coordinated with other mandatory steps during cell replication. It is clear that the higher the doses, the greater the possibility of reverse transposition of the mRNA [
52,
53].
Reverse transposition of the virus mRNA molecule is basically a random event and can occur in any cell type that displays L1 elements [
2,
54,
55]. This event may lead to a unique scenario that should be considered the artifact of new idiopathic inflammatory conditions unresponsive to conventional treatments. Basically, the clonal expansion containing the new back-copy depends on its phenotypic effects and the proliferative ability of the mutated cells which will eventually lead to the persistent formation of affected tissues and clones which will become a constant target of the immune cells [
2,
54,
55].
In fact, a moderately affected or neutral backcopy emerged in a mature somatic cell is nothing but the derivation of highly proliferative progenitor cells such as precancerous cells or pluripotent and multipotent SCs that theoretically have infinite possibilities of propagating giving rise to an infinite number of abnormal descendant cells mostly present in somatic tissue [
56,
57,
58,
59,
60]. This phenomenon is closely related to the enzyme reverse transcriptase of stem cells which regulates the process of differentiation towards the mature phenotype. Through this mechanism, the stem cell can take up the microvesicles of mature cells containing mRNA which is subsequently released into the cytoplasm which can be detected both in the specific mRNA of the epithelial cell and in the protein translated from this mRNA [
2,
56,
57,
58,
59,
60].
Similarly to SARS-CoV-2 mRNA, synthetic mRNAs exhibit quite unique behavior related to their chemical structure, nucleotide sequence, and formulation. Interestingly, their concentration, stability and distribution on tissues, organs and cells also occur through genome integration via L1 elements [
61,
62,
63,
64]. This reflects either the amount of vaccine mRNA delivered in a single dose, the presence of SNPs that may impair the IFN-a (major somatic cell antiviral defense mechanism) expression, or the defection of the antiviral RNA interfering mechanism ( RNAi) typical of multipotent and pluripotent SCs [
65,
66,
67,
68]. An often overlooked point is that the efficacy of viral mRNA and vaccines is based on bypassing innate defense mechanisms, to reach the cytosol and then be translated by ribosomes. The adopted strategy relies on sequence optimizations and nucleoside modifications to achieve sufficient stability of owns mRNAs to access undetected by innate defense mechanisms such as IFN-y. In addition, the synthetic mRNA is protected by additional lipid nanoparticle (LNP) shielding facilitating its cellular uptake and entry into the cytosol via endosomal escape, which also favors the reverse transposition possibilities of vaccine mRNA [
65,
66,
67,
68].
3. Pluripotent, multipotent and progenitor stem cells contamination and further consequences
Retrotransposons identified in SCs along the differentiation process trigger serious considerations regarding the possibility of potentially harmful outcomes. Considering the ability of pluripotency and multipotency transcription factors to bind retrotransposons and regulate their activities in SCs, we hypothesized that retrotransposons may also be involved in embryogenesis and in the process of differentiation towards mature cells and tissues [
68,
69,
70,
71]. To support this hypothesis, we reported some advances from
in vivo and
in vitro retrotransposon studies conducted in mice and humans. The data highlighted the expression patterns of key transcriptional factors (such as transcription factors and epigenetic effectors) and Cis-regulatory elements (promoters, enhancers and super-enhancers) deeply involved in embryonic development. Among some pluripotency transcription factors that we can find are OCT4, SOX2, and NANOG, a group of genes that not only control SC activation by regulating pluripotency status and lineage differentiating expression, but also contribute to the architecture open chromatin stem cell [
71,
72,
73,
74,
75].
Indeed, some results demonstrated that L1 full-length mRNA and ORF1-encoded protein (L1TD1) are encoded in pluripotent and multipotent SCs (MSCs, ESCs, HSCs, and iPSCs). Of note, L1TD1 is a SC-specific RNA-binding protein and OCT4, SOX2, and NANOG are all capable of binding to the L1TD1 promoter. Further research confirmed that more than half of the new LINE-1 insertions are full-length and enriched in the specific protein-coding genes of SCs [
74,
75] (
Figure 2).
The mRNA genome integration was confirmed by an experiment performed using different mature somatic cell phenotypes from hESCs and hHSCs to test their ability to support L1 retrotransposition. Each differentiated cell type was infected with a hybrid adenovirus-retrotransposon virus containing a human L1 element with an EGFP reporter cassette [
2,
3,
31,
74,
75,
76]. This model was then confirmed by tests performed on the Zika virus (ZIKV), notorious for its ability to enter the host cell by integrating with the host DNA [
77,
78,
79,
80].
4. The Spike protein syndrome in searching for details
Although, still to be proven, concerns towards a possible association between SARS-CoV-2 and vaccine mRNA and the potential disruption of the SC differentiation process causing a severe debilitating condition are increasing day by day. Several results and discoveries have validated this hypothesis. We herein proposed a new pathogenesis model in agreement with the recent immunological advances in research findings on COVID-19-derived spike protein and mRNA vaccines. Central nervous system damage, peripheral neuropathies, venous thromboembolisms, and sudden myocardial infarction (MI) are classified according to the type of damage to brain, platelets, nerves, kidneys, and heart tissue following SARS-CoV infection -2 and mRNA vaccines and, what we then called Spike syndrome.
If we count the increased number of those debilitating severe idiopathic cases following the COVID-19 infection, Zika virus (ZIKV) would be of great help in understanding the mRNA tropism of SARS-CoV-2 and the role that SCs would eventually play in the etiopathogenesis of the current Spike syndrome. First of all, ZIKV and Sars-CoV-2 share many biological characteristics and infectious mechanisms. For example, in patients affected by ZIKV the analysis and measurement of CD34+ serum HSCs proved to be part of a very complex scenario made up of affected megakaryocytes-erythroid progenitors, erythrocyte lines and thrombocytes worsened also by the presence of single nucleotide polymorphism (SNP) on particular genes that control the immune responses (IL-6, IL-1a;b, TNF-α, IFN-γ and IL-10...) and the coagulation mechanism (MTHFR, Beta fibrinogen, ACE.. .). Immune analysis also confirmed the presence of overactive cytotoxic T cells, neutrophil NETs and M1 macrophages which ultimately explain the hyperinflammatory state and devastating thrombosis and sepsis as a consequence of virus mRNA tropism [
67,
81,
82,
83,
84].
Other studies have detected productively infected fetal cardiac MSCs (fcMSCs) via viral mRNA which was detected at 48 hours. Scientists were able to see the effects of ZIKV on the protein expression of several mesenchymal cell markers, indicating its ability to alter the differentiation process of fcMSCs and their derived cells and clones. ZIKV infections and cardiac complications in the form of myocarditis, pericarditis, atrial fibrillation, cardiac arrhythmias and heart failure are very close to the damages of SARS-CoV 2 and vaccine mRNA to cardiac tissues [
83,
84].
To evaluate whether ZIKV infection affects the differentiation potential of fcMSCs, cardiac and mesenchymal origin marker expression after ZIKV infection was also examined. It was observed that the expression of MSC markers CD90 and CD73 is substantially impaired in infected cells, the researchers were able to demonstrate that ZIKV infection increased CD90 expression and this phenomenon has previously been associated with increased differentiation ability of virus-infected MSCs, suggesting that the virus-induced increase in fcMSCs upon CD90 stimulus toward differentiation is a trick used by the virus itself to continue propagating.
There was also an increase in the CD73 receptor (ecto-5-prime-nucleotidase enzyme which, together with CD39, converts extracellular ATP to adenosine). The ability of CD73 to decrease local immune responses and to facilitate immune escape mechanisms is known. Thus, the ZIKV-mediated increase in CD73 has a role in facilitating viral replication by allowing the virus to escape host immune surveillance in cardiac tissue [
81,
82,
83,
84].
Intriguingly, one of the receptors most commonly implicated in ZIKV entry into neural progenitor cells is the AXL receptor tyrosine kinase (AXL). AXL also promotes the entry of SARS-CoV-2 into cells of the respiratory system, in fact this receptor has been shown to specifically interact with the N-terminal domain of SARS-CoV-2 S. But what is really interesting , both SARS-CoV and ZIKV first bind to Growth arrest-specific 6 (Gas6) via phosphatidylserine on the viral membrane and then bind to AXL. The GAS6/AXL axis is also expressed in many other cell types found in the nervous, cardiovascular, and immune systems, where they may play critical roles in influencing the normal development and function of specific mature cells and tissues [
85,
86,
87,
88].
Each phase of stem cell potency is characterized by its own distinct special morphology, which reflect the patterns of gene expression and epigenetic modification. Retrotransposons affected by spike protein mRNA show skewed stem cell specific expression and are considered a marker of the affected state. Therefore, it was hypothesized that the spike protein derived from Sars-CoV-2 mRNA, similarly to what happens with ZIKV, could compromise the cellular integrity of the host through the contamination of multipotent, pluripotent and progenitor SCs, a condition which was then aggravated by the possible presence of specific SNPs on genes responsible for inflammatory and immune regulation, such as interleukins and modulatory cytokines, such as IL6, TNFa, IFNy, IL10 etc... [
85,
86,
87,
88].
Therefore, taken together, these results could confirm that SARS-CoV-2 virus progresses similarly to other mRNA viruses, whereby spike protein integration via L1 back-deposition may occur via ACE2r: i) all types of differentiating stem cells; ii) progenitor cells; iii) differentiating adult cells such as neurons, myocytes, endothelial cells, chondrocytes and fibroblasts. Thus, it is logical to assume that L1 reactivation is necessary for pluripotency maintenance and self-renewal [
88].
5. Proposed model of different Spike protein syndrome’s pathologies a possible physio pathogenesis related to affected stem cells and kidney/heart/brain axis
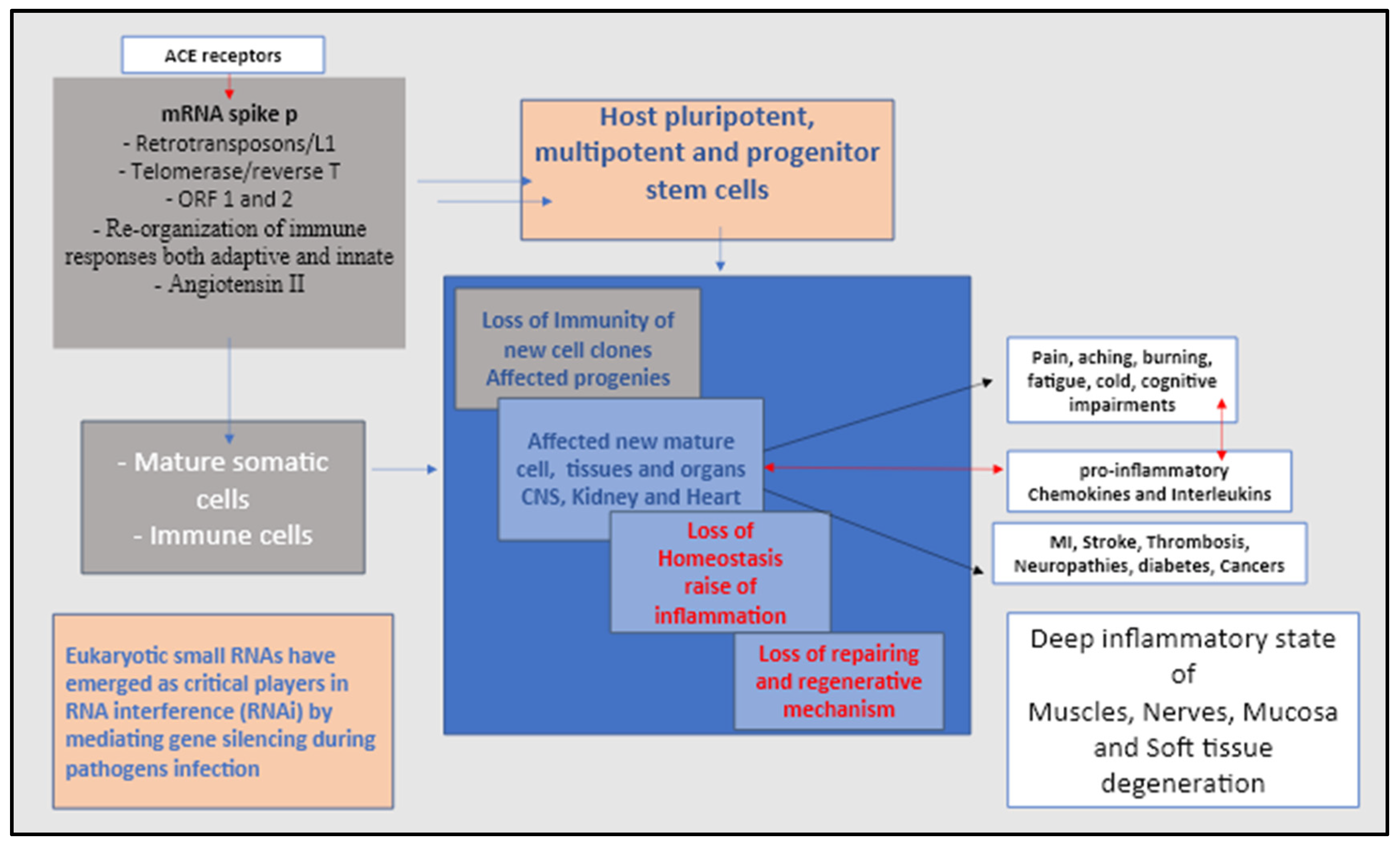
During initial COVID-19 infection, spike proteins typically break into smaller fragments once they have invaded human cells, however, spike proteins have been observed to remain intact in some cases. Intact spike protein that could be found in patient tissue and cell samples is indicative of continued attack by host immune system autoantibodies, the “
conditio sine qua non” of long COVID and spike protein syndrome [
88,
89] (
Figure 3).
Considering the group of axonal immune neuropathies which are a particular type of immune-mediated condition exhibited by Long-Covid and Spike protein syndrome, they have been shown to have a clinical presentation with a heterogeneous picture and relatively rapid progression of symptoms [
89,
90]. Clinically, Long-Covid and Spike Protein Syndrome are characterized by cold extremities, numbness, tingling, prickling sensations with paraesthesis events, general muscle weakness, perineal discomfort, and burning pains. The severe manifestation can eventually result in fulminant myocarditis, Guillaume Barret syndrome, fibromyalgia involving organs and gland dysfunction. Affected areas of the body often become extremally sensitive leading to unbearable intense or distorted experience of touch (allodynia). In such cases, pain is due to stimuli that does not normally provoke pain [
89,
90].
These signs have led us to carefully consider the importance of the brain-CNS/heart/kidney axis. The renin-angiotensin system (RAS), which plays a key role in regulating autonomic and neuroendocrine functions, maintaining cardiovascular and renal homeostasis, is probably the main affected mechanism after COVID-19 and mRNA vaccines. Angiotensin (Ang) receptors are scattered throughout the central nervous/cardiovascular/renal system and are involved in the synthesis of the peptides Ang-I, Ang-II, Ang-(1–7), Ang-(1–9) and the enzymes involved in their formation, angiotensin converting enzyme type 1 (ACE-1), type 2 (ACE-2) and chymases which are the preferred entry of SARS-CoV-2 mRNA [
91,
92,
93,
94] (
Figure 3).
In particular, the Ang-II is the main effector molecule of RAS exerting a central function in the regulation of blood pressure (BP), via the activation of AT1 receptors. Thus and presumably, any dysfunction occurring along the brain/CNS/heart/kidney axis may also affect the RAS by increasing the Ang-II synthesis, which in turn may drive to a sympathetic outflow and hypertension. Of note, Ang-II plays a key role for microglial activation by enhancing the production of pro-inflammatory cytokines that affect neuronal activity in the cerebral paraventricular nucleus (PVN) region causing perturbed sympathetic outflow, promoting vasoconstriction, tachycardia and hypertension [
91,
92,
93,
94].
The integrative responses of the kidneys to persistent inflammatory stress can negatively affect the thermoregulatory system, cardiovascular control, water and electrolyte levels. It is well known that persistent stress from the inflammatory spike protein may contribute to the onset of chronic kidney disease (CKD) by disrupting the RAS mechanism. It is known that in patients with chronic kidney disease anemia is a structural problem, which gives rise to a particular type of hypothermia that is often found in patients affected by Long Covid and Spike protein syndrome [
95].
Of note, multipotent cardiac SCs, progenitor cells, myocytes, and fibroblasts all express ACE2r which allows entry of the virus and its mRNA-derived spike protein. Similarly, pericytes and mural cells that support the maintenance and repair of microvasculature throughout the myocardium are also highly susceptible as they express the highest levels of ACE2 within cardiac tissues [
96].
It is worth mentioning that one of the most significant pathological events that clinicians have experienced thus far is consistent with sudden death related to myocardial infarction (MI), a condition that mostly affects young people and athletes. This event could be explained by a silent progressive decay of the coronary chemoreceptors and its neighboring nerves and ganglia [
96,
97,
98]. The inflammatory process can spread up to the fiber tracts located in the dorsal roots reaching the ganglion neurons that from there tend to influence neurons in the spinal cord, reaching the sympathetic efferent neurons. The direct involvement of stem cells is therefore assumed, derived infected mature and progenitor neuro cells may also contribute to the disorders of sympathetic afferent neurons that are associated with multiple districts such as heart, esophagus and bronchi through: i), the affection of vagal afferent axons that are associated with myocardial receptors, which ascend in the vagal nerves up to the node ganglion reaching the nucleus of the tractus solitarius and interneurons; and, ii) these interneurons will in turn affect the vagus and sympathetic preganglionic efferent nerves in the bulbospinal tracts [
97,
98,
99,
100].
Interestingly, an experiment conducted by Marchiano and colleagues, on the contractile properties of human pluripotent SCs-derived cardiomyocytes on 3D engineered cardiac tissues, showed the progression of infection by virus mRNA. The results showed that the viral mRNA to affect the contractile ability of cells has reached the dedifferentiated state. Maximal contraction force in infected tissue has decreased as early as 72 hpi (hypotension predictor), and contractions decrease to less than 25% of the force measured at baseline at 144 hpi. This event, the authors referred, could contribute to a progressive loss of force production [
94].
It should be added that the reduction in pericyte vascular coverage observed in the heart or lungs of patients with COVID-19 indicated that SARS-CoV-2 may affect the microvasculature by targeting pericytes. It has been noted that an increase in adverse cardiovascular events, which often affect young and athletic people as we mentioned earlier, could be an event due to either the greater accumulation of Ang II or to the higher level of active circulating SCs much more present in younger individuals and those who practice sport [
96,
97,
98,
99,
100].
The anomalous and misleading data is confirmed by the fact that the procedures for detecting immune axonal neuropathies in most cases revealed absent or reduced nerve amplitudes with normal latencies and conduction velocities. We assume that probably the final stage of stem cell differentiation does not express particular modified morphologies, or particular patterns of gene expression and epigenetic modifications that often go undetected. However, the role of the L1 retrotransposon mechanism in the life cycle of affected SCs carrying the viral mRNA may be accounted for those abnormal features that are not shown using conventional detection tests [
100,
101].
6. The virus non-codingRNA the mechanism to avoid immune surveillance and deactivating stem cells
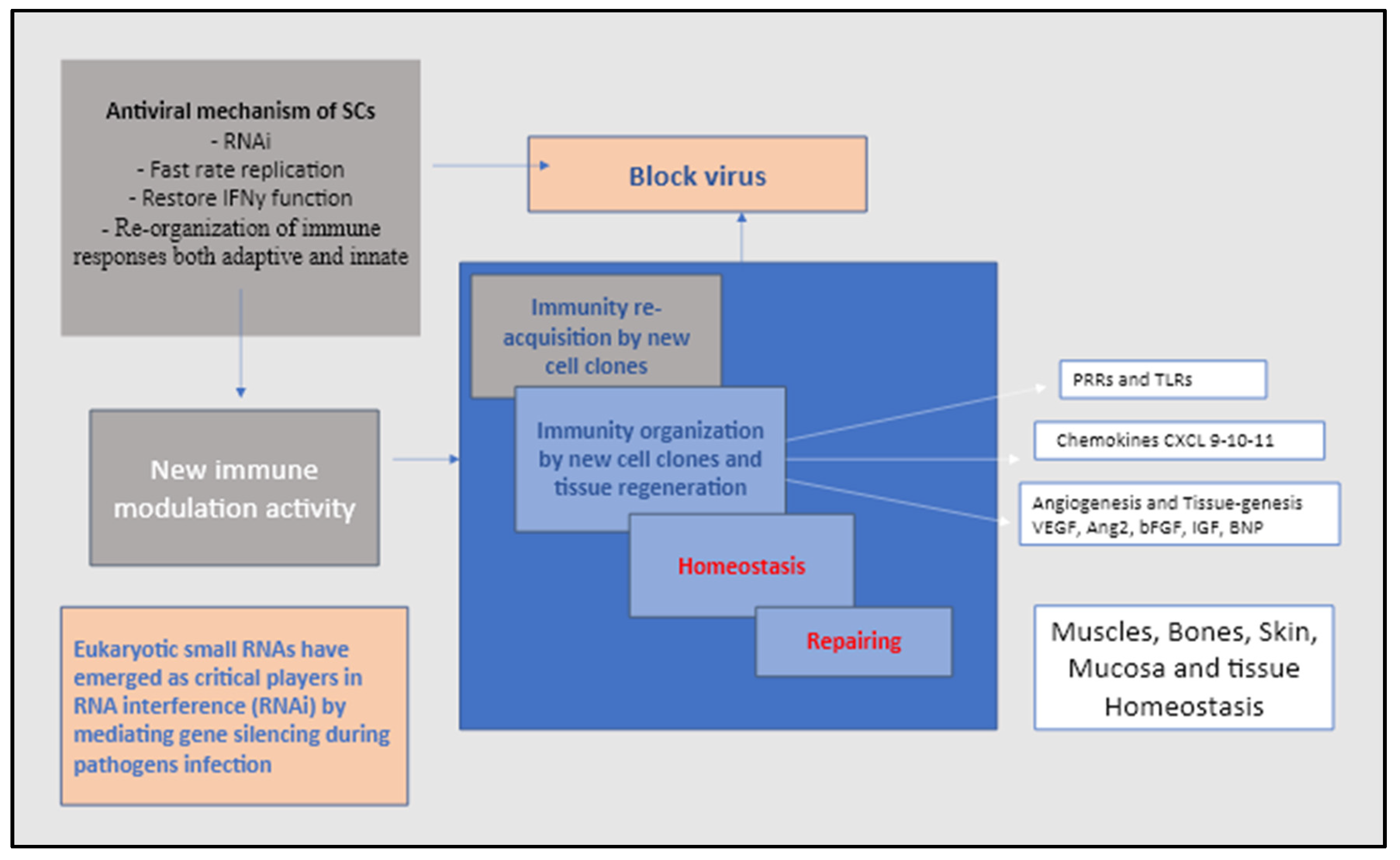
However, it should be mentioned that SCs play an equally important role in immunity against viruses [
102]. Until very recently, the lack of biogenetic and physiological evidence for the existence of siRNA (small interfering RNA) suggested that the RNAi (interfering RNA) machinery has not to be considered a key device in antiviral immunity of pluripotent and multipotent human SCs [
102,
103,
104] (
Figure 4). However, the results of pluripotent embryonic SCs (ESCs) studies in mice showed something quite unique demonstrating that the siRNA of viral origin was detected in ESCs infected with encephalomyocarditis virus or Nodamura virus, suggesting that the RNAi pathway might be functional in these. cells [
104,
105,
106,
107,
108].
Thus, it appears that during the millennia in which humans and viruses have co-evolved in a sort of competitive race, somehow the virus has adopted a strategy to knock down SC RNAi by targeting and using SC RNAi. Although the mechanism is still unclear, in line with other authors, we have proposed that RNA viruses such as SARS-CoV-2 and vaccine analogues may use or interfere with RNAi-related pathways in the control or inhibition of expression, epigenetic modification and regulation of heterochromatin. By blocking dsRNA from possible contact with the RNAi pathway, these viruses are known to block or affect the RNAi once the viRNAs have been produced and inserted into host cells [
107,
108,
109,
110,
111,
112].
We assumed that the key pathway adopted by RNA viruses such as the SARS-CoV-2 to subvert the stem cells RNAi and making them as own multiplication carriers takes place by using the non-coding RNA (ncRNA) [
113]. The internal structure of ncRNAs composed of 200bp include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR. The long ncRNAs indicate two main groups one circular and one linear. Since the ncRNAs are not presented via major histocompatibility complex (MHC) the virus would not be recognized as danger by the adaptive immune system which facilitate them the invasion allowing them to evade host innate immune as well [
109]. Of note, also the SARS-CoV-2 virus is equipped with ncRNAs that is usually upregulated in COVID-19 active infection that would eventually be used as a biomarker for disease progression and severity. Thus, the inhibition of IFN and RNAi damages remain the most important tools adopted by viruses to weaken antiviral response by mature progenitor and somatic cells on one side, and pluripotent, multipotent SCs on the other [
110].
7. Final considerations
Whether SARS-CoV-2 or vaccine mRNAs could possibly be integrated into the host genome, we are aware that this issue remains an open question for further screening and analysis. However, at present this hypothesis is the only one that could explain the relatively new and varied clinical condition of both Long-Covid and the Spike Protein Syndrome. That sais, it remains unclear why it was neglected the retroposition biology of L1 retroelements, the important presence of circulating stem cells and the role of their telomerase (reverse transcriptase’RT) in locating the mRNA retroposition into the DNA transcription mechanism [
103,
104,
105]. The field of stem cells began its development more than 30 years ago and telomerase and retroelements in humans have been studied for more than 40 years [
103,
104,
105] but obviously without any attempt to look for a link between the two elements. Unfortunately, all of this tends to give the impression that stem cells and L1-driven retroposition are something of a forgotten topic in mRNA science.
A significant part of most evolutionary eukaryotic genomes is the result of regeneration processes in which the stem cell activities, the RNA transcription mechanism and the formation of "retrogenes" necessarily foresee the role of reverse transcript synthesis allowing to integrate new coding transcripts of DNA into the host genome. The mRNA of the SARS-CoV-2 virus and vaccines is closely associated with retrotransposons and the telomerase machinery and all have played a major role in this genomic sculpting [
1,
2,
103,
104,
105,
106,
107,
108,
109,
110,
111,
112,
113].
Furthermore, the current hypotheses would also be able to explain the mechanism of the inflammatory disease secondary to Long-COVID syndrome and mRNA spike syndrome, trying to give a correct analysis of why and how some subjects are more prone to experience devastating complication. The data presented in this paper, suggested that the higher numbers of circulating affected stem cells that continuously working by replacing old tissues and cells with new generation of affected mature somatic cells may prevalently occur in young and healthy individuals due to their cells’ longer telomeres and faster telomerase activity. As a result, these affected offspring tend to generate large chunks of almost irreversibly contaminated tissues and organs. The higher the regeneration mode, the higher the risk of receiving inflammatory patterns imprinted by the mRNA Spike protein. As earlier mentioned, viral mRNA can either alter the expression of host RNAi or IFN using cellular machinery to form viral miRNAs, new patterns that are eventually integrated via L1 retroposition mechanism into the host genome [
102,
103,
104,
105,
106,
107,
108,
109,
110,
111,
112,
113].
8. Conclusion
We conclude by writing that the retrotransposition machinery of mature somatic and stem cells can use their RNA templates to pick up and include new mRNA into host DNA at the ends of chromosome breaks, unfortunately, whether this process is compatible for the future adaptation, we don’t know yet. We should consider the phylogenetic and functional activities of these retroelements as the functional mechanism that our body uses in an attempt to respond and adapt to specific new stimuli and new insults. This implies that current assessments of the mRNA spike protein as a direct consequence of both the SARS-CoV-2 virus and the vaccine, may foster a new line of investigation that can specifically address the mechanism of genome integration and the role of stem cells.
Author Contributions
Conceptualization, C.G.I. and N.C.D.K.; investigation, C.G.I. and M.G.B., P.D., R.L.; figures, C.G.I. and N.C.D.K,; data curation, L.S., A.M., V.H.P., T.C.T. and S.K.A.; writing, review and editing, C.G.I., L.S., S.K.A., A.M.K., R.D.P. and M.G.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted did not involving humans or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xu, S.; Yang, K.; Li, R.; Zhang, L. mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef] [PubMed]
- Domazet-Lošo, T. mRNA Vaccines: Why Is the Biology of Retroposition Ignored? Genes 2022, 13, 719. [Google Scholar] [CrossRef]
- Zhang, L.; Bisht, P.; Flamier, A.; Barrasa, M.I.; Friesen, M.; Richards, A.; Hughes, S.H.; Jaenisch, R. LINE1-Mediated Reverse Transcription and Genomic Integration of SARS-CoV-2 mRNA Detected in Virus-Infected but Not in Viral mRNA-Transfected Cells. Viruses 2023, 15, 629. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Milicic Stanic B, Maddox S, de Souza AMA, Wu X et al. Male bias in ACE2 basic science research: Missed opportunity for discovery in the time of COVID-19. Am. J. Physiol. Regul. 2021, 320, R925–R937.
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger, N et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [CrossRef]
- Devaux, C.A.; Camoin-Jau, L. Molecular Mimicry of the Viral Spike in the SARS-CoV-2 Vaccine Possibly Triggers Transient Dysregulation of ACE2, Leading to Vascular and Coagulation Dysfunction Similar to SARS-CoV-2 Infection. Viruses 2023, 15, 1045. [Google Scholar] [CrossRef]
- Zhang, L.; Bisht, P.; Flamier, A.; Barrasa, M.I.; Friesen, M.; Richards, A.; Hughes, S.H.; Jaenisch, R. LINE1-Mediated Reverse Transcription and Genomic Integration of SARS-CoV-2 mRNA Detected in Virus-Infected but Not in Viral mRNA-Transfected Cells. Viruses 2023, 15, 629. [Google Scholar] [CrossRef]
- Wang S, Yao X, But S, Ping Y et al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat. Cell Biol. 2021, 23, 1314–1328. [CrossRef]
- Wei, W.; Gilbert, N.; Ooi, S.L.; Lawler, J.F.; Ostertag, E.M.; Kazazian, H.H.; Boeke, J.D.; Moran, J.V. Human L1 Retrotransposition: cisPreference versus trans Complementation. Mol. Cell. Biol. 2001, 21, 1429–1439. [Google Scholar] [CrossRef]
- Dewannieux, M.; Esnault, C.; Heidmann, T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003, 35, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, J.L.; Doucet, A.J.; Bucheton, A.; Moran, J.V.; Gilbert, N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007, 17, 602–611. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H.; Vinckenbosch, N.; Long, M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009, 10, 19–31. [Google Scholar] [CrossRef]
- Salgado-Albarran M, Navarro-Delgado E. I, Del Moral-Morales A, Alcaraz N et al. Comparative transcriptome analysis reveals key epigenetic targets in SARS-CoV-2 infection. NPJ Syst. Biol. Appl. 2021, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Song, H.; Xu, Y.; Garrison, K.E.; Buzdin, A.A.; Anwar, N.; Hunter, D.V.; Mujib, S.; Mihajlovic, V.; Martin, E.; et al. LINE-1 Retrotransposable Element DNA Accumulates in HIV-1-Infected Cells. J. Virol. 2013, 87, 13307–13320. [Google Scholar] [CrossRef] [PubMed]
- Sudhindar, P.D.; Wainwright, D.; Saha, S.; Howarth, R.; McCain, M.; Bury, Y.; Saha, S.S.; McPherson, S.; Reeves, H.; Patel, A.H.; et al. HCV Activates Somatic L1 Retrotransposition—A Potential Hepatocarcinogenesis Pathway. Cancers 2021, 13, 5079. [Google Scholar] [CrossRef]
- Schöbel, A.; Nguyen-Dinh, V.; Schumann, G.G.; Herker, E. Hepatitis C virus infection restricts human LINE-1 retrotransposition in hepatoma cells. PLOS Pathog. 2021, 17, e1009496. [Google Scholar] [CrossRef]
- Bonenfant, G.; Meng, R.; Shotwell, C.; Badu, P.; Payne, A.F.; Ciota, A.T.; Sammons, M.A.; Berglund, J.A.; Pager, C.T. Asian Zika Virus Isolate Significantly Changes the Transcriptional Profile and Alternative RNA Splicing Events in a Neuroblastoma Cell Line. Viruses 2020, 12, 510. [Google Scholar] [CrossRef]
- Bellavite, P.; Ferraresi, A.; Isidoro, C. Immune Response and Molecular Mechanisms of Cardiovascular Adverse Effects of Spike Proteins from SARS-CoV-2 and mRNA Vaccines. Biomedicines 2023, 11, 451. [Google Scholar] [CrossRef]
- Carelli, F.N.; Hayakawa, T.; Go, Y.; Imai, H.; Warnefors, M.; Kaessmann, H. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016, 26, 301–314. [Google Scholar] [CrossRef]
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.M.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Parrish, N.F.; Fujino, K.; Shiromoto, Y.; Iwasaki, Y.W.; Ha, H.; Xing, J.; Makino, A.; Kuramochi-Miyagawa, S.; Nakano, T.; Siomi, H.; et al. piRNAs derived from ancient viral processed pseudogenes as transgenerational sequence-specific immune memory in mammals. RNA 2015, 21, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Son, J.-H.; Do, H.; Han, J. Intragenic L1 Insertion: One Possibility of Brain Disorder. Life 2022, 12, 1425. [Google Scholar] [CrossRef]
- Gundry, SF.; mRNA COVID Vaccines Dramatically Increase Endothelial Inflammatory Markers and ACS Risk as Measured by PULS Cardiac Test: A Warning. Circulation. 2021. Available online: www.ahajournals.org›abs›circ.144.suppl_1.10712 (accessed on 6 April 2023).
- Angels F, Spanevello A, Reboldi G, Visca D, Verdecchia, P. SARS-CoV-2 vaccines: Lights and shadows. Eur. J. Intern. Med. 2021, 88, 1–8. [CrossRef]
- Symer, D.E.; Connelly, C.; Szak, S.T.; Caputo, E.M.; Cost, G.J.; Parmigiani, G.; Boeke, J.D. Human L1 Retrotransposition Is Associated with Genetic Instability In Vivo. Cell 2002, 110, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 Elements in Structural Variation and Disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef]
- Chen, J.-M.; Férec, C.; Cooper, D.N. LINE-1 Endonuclease-Dependent Retrotranspositional Events Causing Human Genetic Disease: Mutation Detection Bias and Multiple Mechanisms of Target Gene Disruption. J. Biomed. Biotechnol. 2006, 2006, 1–9. [Google Scholar] [CrossRef]
- Burns, K.H. Our Conflict with Transposable Elements and Its Implications for Human Disease. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 51–70. [Google Scholar] [CrossRef]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef]
- Chesnokova, E.; Beletskiy, A.; Kolosov, P. The Role of Transposable Elements of the Human Genome in Neuronal Function and Pathology. Int. J. Mol. Sci. 2022, 23, 5847. [Google Scholar] [CrossRef]
- Denli, A.M.; Narvaiza, I.; Kerman, B.E.; Pena, M.; Benner, C.; Marchetto, M.C.; Diedrich, J.K.; Aslanian, A.; Ma, J.; Moresco, J.J.; et al. Primate-Specific ORF0 Contributes to Retrotransposon-Mediated Diversity. Cell 2015, 163, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Maestre, J.; Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000, 24, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Sultana, T.; van Essen, D.; Siol, O.; Bailly-Bechet, M.; Philippe, C.; El Aabidine, A.Z.; Pioger, L.; Nigumann, P.; Saccani, S.; Andrau, J.-C.; et al. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection. Mol. Cell 2019, 74, 555–570. [Google Scholar] [CrossRef] [PubMed]
- A Erwin, J.; Paquola, A.C.M.; Singer, T.; Gallina, I.; Novotny, M.; Quayle, C.; A Bedrosian, T.; I A Alves, F.; Butcher, C.R.; Herdy, J.R.; et al. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 2016, 19, 1583–1591. [Google Scholar] [CrossRef] [PubMed]
- Zhang L, Richards A, Barrasa MI, Stephen HH et al. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118. [CrossRef] [PubMed]
- Hessien, M.; Donia, T.; Tabll, A.A.; Adly, E.; Abdelhafez, T.H.; Attia, A.; Alkafaas, S.S.; Kuna, L.; Glasnovic, M.; Cosic, V.; et al. Mechanistic-Based Classification of Endocytosis-Related Inhibitors: Does It Aid in Assigning Drugs against SARS-CoV-2? Viruses 2023, 15, 1040. [Google Scholar] [CrossRef]
- Wang, J.; Huang, J.; Shi, G. Retrotransposons in pluripotent stem cells. Cell Regen. 2020, 9, 1–10. [Google Scholar] [CrossRef]
- Patlar, B. On the Role of Seminal Fluid Protein and Nucleic Acid Content in Paternal Epigenetic Inheritance. Int. J. Mol. Sci. 2022, 23, 14533. [Google Scholar] [CrossRef]
- Poiani, A. Complexity of seminal fluid: a review. Behav. Ecol. Sociobiol. 2006, 60, 289–310. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, H.; Kvist, U.; Ernerudh, J.; Sanz, L.; Calvete, J.J. Seminal Plasma Proteins: What Role Do They Play? Am. J. Reprod. Immunol. 2011, 66 (Suppl. 1), 11–22. [Google Scholar] [CrossRef]
- Lupatov, A.Y.; Yarygin, K.N. Telomeres and Telomerase in the Control of Stem Cells. Biomedicines 2022, 10, 2335. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Passos, J.F.; Birket, M.J.; Beckmann, T.; Brings, S.; Peters, H.; Birch-Machin, M.A.; von Zglinicki, T.; Saretzki, G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell Sci. 2008, 121, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Kaul, Z.; Cesare, A.J.; I Huschtscha, L.; A Neumann, A.; Reddel, R.R. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 2011, 13, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Leem, S.-H.; Londoño-Vallejo, J.A.; Kim, J.-H.; Bui, H.; Tubacher, E.; Solomon, G.; Park, J.-E.; Horikawa, I.; Kouprina, N.; Barrett, J.C.; et al. The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene 2002, 21, 769–777. [Google Scholar] [CrossRef]
- Casacuberta, E. Drosophila: Retrotransposons Making up Telomeres. Viruses 2017, 9, 192. [Google Scholar] [CrossRef]
- Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: The promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell 1997, 88, 647–655. [CrossRef]
- Jenner, L.P.; Peska, V.; Fulnečková, J.; Sýkorová, E. Telomeres and Their Neighbors. Genes 2022, 13, 1663. [Google Scholar] [CrossRef]
- Vaquero-Sedas, M.I.; Vega-Palas, M.A. Assessing the Epigenetic Status of Human Telomeres. Cells 2019, 8, 1050. [Google Scholar] [CrossRef]
- Cubiles, M.D.; Barroso, S.; I Vaquero-Sedas, M.; Enguix, A.; Aguilera, A.; A Vega-Palas, M. Epigenetic features of human telomeres. Nucleic Acids Res. 2018, 46, 2347–2355. [Google Scholar] [CrossRef]
- Gámez-Arjona, F.M.; López-López, C.; Vaquero-Sedas, M.I.; Vega-Palas, M.A. On the organization of the nucleosomes associated with telomeric sequences. Biochim. et Biophys. Acta (BBA) - Mol. Cell Res. 2010, 1803, 1058–1061. [Google Scholar] [CrossRef]
- Vaquero-Sedas, M.I.; Vega-Palas, M.A. On the chromatin structure of eukaryotic telomeres. Epigenetics 2011, 6, 1055–1058. [Google Scholar] [CrossRef] [PubMed]
- Hughes, S.H.; Chandler, M.; Gellert, M.; Lambowitz, A.M.; Rice, P.A.; Sandmeyer, S.B. Reverse Transcription of Retroviruses and LTR Retrotransposons. Microbiol Spectr. 2015, MDNA3-0027-2014. [Google Scholar] [CrossRef]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 Elements in Structural Variation and Disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef]
- Kaer, K.; Speek, M. Retroelements in human disease. Gene 2013, 518, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Żyżyńska-Galeńska, K.; Bernat, A.; Piliszek, A.; Karasiewicz, J.; Szablisty, E.; Sacharczuk, M.; Brewińska-Olchowik, M.; Bochenek, M.; Grabarek, J.; Modliński, J.A. Embryonic Environmental Niche Reprograms Somatic Cells to Express Pluripotency Markers and Participate in Adult Chimaeras. Cells 2021, 10, 490. [Google Scholar] [CrossRef] [PubMed]
- Petkov, S.; Dressel, R.; Rodriguez-Polo, I.; Behr, R. Controlling the Switch from Neurogenesis to Pluripotency during Marmoset Monkey Somatic Cell Reprogramming with Self-Replicating mRNAs and Small Molecules. Cells 2020, 9, 2422. [Google Scholar] [CrossRef] [PubMed]
- Ciomborowska-Basheer, J.; Staszak, K.; Kubiak, M.R.; Makałowska, I. Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts. Cells 2021, 10, 912. [Google Scholar] [CrossRef]
- Staszak, K.; Makałowska, I. Cancer, Retrogenes, and Evolution. Life 2021, 11, 72. [Google Scholar] [CrossRef]
- Aoyama, K.; Itokawa, N.; Oshima, M.; Iwama, A. Epigenetic Memories in Hematopoietic Stem and Progenitor Cells. Cells 2022, 11, 2187. [Google Scholar] [CrossRef]
- Demongeot, J.; Fougère, C. mRNA COVID-19 Vaccines—Facts and Hypotheses on Fragmentation and Encapsulation. Vaccines 2022, 11, 40. [Google Scholar] [CrossRef]
- Aldén, M.; Olofsson Falla, F.; Yang, D.; Barghouth, M.; Luan, C.; Rasmussen, M.; De Marinis, Y. Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line. Curr. Issues Mol. Biol. 2022, 44, 1115–1126. [Google Scholar] [CrossRef] [PubMed]
- Giannotta, G.; Murrone, A.; Giannotta, N. COVID-19 mRNA Vaccines: The Molecular Basis of Some Adverse Events. Vaccines 2023, 11, 747. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Lavine, K.J.; Lin, C.-Y. Myocarditis after Covid-19 mRNA Vaccination. New Engl. J. Med. 2021, 385, 1332–1334. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Kim, M.C.; Kim, K.H.; Jeong, I.-S.; Cho, Y.S.; Choi, Y.D.; Lee, J.E. Case Report: Acute Fulminant Myocarditis and Cardiogenic Shock After Messenger RNA Coronavirus Disease 2019 Vaccination Requiring Extracorporeal Cardiopulmonary Resuscitation. Front. Cardiovasc. Med. 2021, 8, 758996. [Google Scholar] [CrossRef]
- Wilkins, C.; Dishongh, R.; Moore, S.C.; Whitt, M.A.; Chow, M.; Machaca, K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 2005, 436, 1044–1047. [Google Scholar] [CrossRef]
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Pham, V.H.; Tran, T.C.; Dipalma, G.; Bianco, A.; Serlenga, E.M.; Aityan, S.K.; Pierangeli, V.; et al. Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection. Diagnostics 2022, 12, 2824. [Google Scholar] [CrossRef]
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; D’ettorre, E.; Nico, A.; Inchingolo, F.; Dipalma, G.; Tomassone, D.; Serlenga, E.M.; Dalagni, G.; et al. New Translational Trends in Personalized Medicine: Autologous Peripheral Blood Stem Cells and Plasma for COVID-19 Patient. J. Pers. Med. 2022, 12, 85. [Google Scholar] [CrossRef]
- Sinibaldi-Vallebona, P.; Matteucci, C.; Spadafora, C. Retrotransposon-Encoded Reverse Transcriptase in the Genesis, Progression and Cellular Plasticity of Human Cancer. Cancers 2011, 3, 1141–1157. [Google Scholar] [CrossRef]
- Lavia, P.; Sciamanna, I.; Spadafora, C. An Epigenetic LINE-1-Based Mechanism in Cancer. Int. J. Mol. Sci. 2022, 23, 14610. [Google Scholar] [CrossRef]
- Maupetit-Mehouas, S.; Vaury, C. Transposon Reactivation in the Germline May Be Useful for Both Transposons and Their Host Genomes. Cells 2020, 9, 1172. [Google Scholar] [CrossRef]
- Kravchuk, E.V.; Ashniev, G.A.; Gladkova, M.G.; Orlov, A.V.; Vasileva, A.V.; Boldyreva, A.V.; Burenin, A.G.; Skirda, A.M.; Nikitin, P.I.; Orlova, N.N. Experimental Validation and Prediction of Super-Enhancers: Advances and Challenges. Cells 2023, 12, 1191. [Google Scholar] [CrossRef] [PubMed]
- Seymour, T.; Twigger, A.-J.; Kakulas, F. Pluripotency Genes and Their Functions in the Normal and Aberrant Breast and Brain. Int. J. Mol. Sci. 2015, 16, 27288–27301. [Google Scholar] [CrossRef] [PubMed]
- Balzanelli, M.G.; Distratis, P.; Catucci, O.; Cefalo, A.; Lazzaro, R.; Inchingolo, F.; Tomassone, D.; Aityan, S.K.; Ballini, A.; Nguyen, K.C.D.; et al. Mesenchymal Stem Cells: The Secret Children’s Weapons against the SARS-CoV-2 Lethal Infection. Appl. Sci. 2021, 11, 1696. [Google Scholar] [CrossRef]
- Närvä, E.; Rahkonen, N.; Emani, M.R.; Lund, R.; Pursiheimo, J.; Nästi, J.; Autio, R.; Rasool, O.; Denessiouk, K.; Lähdesmäki, H.; et al. RNA-Binding Protein L1TD1 Interacts with LIN28 via RNA and is Required for Human Embryonic Stem Cell Self-Renewal and Cancer Cell Proliferation. STEM CELLS 2011, 30, 452–460. [Google Scholar] [CrossRef]
- Zhang L, Richards A, Barrasa MI, Stephen HH et al. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118. [CrossRef]
- Dorneles, M.L.d.A.; Cardoso-Lima, R.; Souza, P.F.N.; Rosa, D.S.; Magne, T.M.; Santos-Oliveira, R.; Alencar, L.M.R. Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties. Viruses 2022, 14, 1727. [Google Scholar] [CrossRef]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.-C.; Medits, I.; Sharma, A.; Simon-Lorière, E.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Haouz, A.; et al. Structural basis of potent Zika–dengue virus antibody cross-neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Figueiredo, C.P.; Barros-Aragão, F.G.Q.; Neris, R.L.S.; Frost, P.S.; Soares, C.; Souza, I.N.O.; Zeidler, J.D.; Zamberlan, D.C.; de Sousa, V.L.; Souza, A.S.; et al. Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Azevedo RS, de Sousa JR, Araujo MTF, Filho AJM et al. In Situ Immune Response and Mechanisms of Cell Damage in Central Nervous System of Fatal Cases Microcephaly by Zika Virus. Sci. Rep. 2017, 8, 5.
- Roth, H.; Schneider, L.; Eberle, R.; Lausen, J.; Modlich, U.; Blümel, J.; Baylis, S.A. Zika virus infection studies with CD34 + hematopoietic and megakaryocyte-erythroid progenitors, red blood cells and platelets. Transfusion 2020, 60, 561–574. [Google Scholar] [CrossRef]
- Balzanelli, M.G.; Distratis, P.; Dipalma, G.; Vimercati, L.; Inchingolo, A.D.; Lazzaro, R.; Aityan, S.K.; Maggiore, M.E.; Mancini, A.; Laforgia, R.; et al. Sars-CoV-2 Virus Infection May Interfere CD34+ Hematopoietic Stem Cells and Megakaryocyte–Erythroid Progenitors Differentiation Contributing to Platelet Defection towards Insurgence of Thrombocytopenia and Thrombophilia. Microorganisms 2021, 9, 1632. [Google Scholar] [CrossRef]
- Heazlewood, S.Y.; Ahmad, T.; Cao, B.; Cao, H.; Domingues, M.; Sun, X.; Heazlewood, C.K.; Li, S.; Williams, B.; Fulton, M.; et al. High ploidy large cytoplasmic megakaryocytes are hematopoietic stem cells regulators and essential for platelet production. Nat. Commun. 2023, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Balzanelli, M.G.; Distratis, P.; Lazzaro, R.; Pham, V.H.; Tran, T.C.; Dipalma, G.; Bianco, A.; Serlenga, E.M.; Aityan, S.K.; Pierangeli, V.; et al. Analysis of Gene Single Nucleotide Polymorphisms in COVID-19 Disease Highlighting the Susceptibility and the Severity towards the Infection. Diagnostics 2022, 12, 2824. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Josey, B.; Sayitoglu, E.C.; Potens, R.; Sultu, T.; Duru, A.D.; Beljanski, V. Characterization of zika virus infection of human fetal cardiac mesenchymal stromal cells. PLOS ONE 2020, 15, e0239238. [Google Scholar] [CrossRef] [PubMed]
- Strange, D.P.; Jiyarom, B.; Pourhabibi Zarandi, N.; Xie, X.; Baker, C.; Sadri-Ardekani, H.; Shi, P.-Y.; Verma, S. Axl Promotes Zika Virus Entry and Modulates the Antiviral State of Human Sertoli Cells. mBio 2019, 10. [Google Scholar] [CrossRef]
- Hastings, A.K.; Hastings, K.; Uraki, R.; Hwang, J.; Gaitsch, H.; Dhaliwal, K.; Williamson, E.; Fikrig, E. Loss of the TAM Receptor Axl Ameliorates Severe Zika Virus Pathogenesis and Reduces Apoptosis in Microglia. iScience 2019, 13, 339–350. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Shastri, A.; Al Aiyan, A.; Kishore, U.; Farrugia, M.E. Immune-Mediated Neuropathies: Pathophysiology and Management. Int. J. Mol. Sci. 2023, 24, 7288. [Google Scholar] [CrossRef]
- Tulbă, D.; Popescu, B.O.; Manole, E.; Băicuș, C. Immune Axonal Neuropathies Associated With Systemic Autoimmune Rheumatic Diseases. Front. Pharmacol. 2021, 12. [Google Scholar] [CrossRef]
- Martyniak, A.; Tomasik, P.J. A New Perspective on the Renin-Angiotensin System. Diagnostics 2022, 13, 16. [Google Scholar] [CrossRef]
- Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS-CoV-2. Cardiovascular 2020, 116, 1097–1100. [CrossRef] [PubMed]
- Dolhnikoff M, Ferreira Ferranti J, de Almeida Monteiro RA, Duarte-Neto A.N et al. SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet Child Adolescent. Health 2020, 4, 790–794. [CrossRef] [PubMed]
- Marchiano, S.; Hsiang, T.-Y.; Khanna, A.; Higashi, T.; Whitmore, L.S.; Bargehr, J.; Davaapil, H.; Chang, J.; Smith, E.; Ong, L.P.; et al. SARS-CoV-2 Infects Human Pluripotent Stem Cell-Derived Cardiomyocytes, Impairing Electrical and Mechanical Function. Stem Cell Rep. 2021, 16, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Jura, M.; Garus, M.; Krakowska, K.; Urban, S.; Błaziak, M.; Iwanek, G.; Zymliński, R.; Biegus, J.; Paleczny, B. A Methodological Perspective on the Function and Assessment of Peripheral Chemoreceptors in Heart Failure: A Review of Data from Clinical Trials. Biomolecules 2022, 12, 1758. [Google Scholar] [CrossRef]
- Grassi, G.; Seravalle, G.; Cattaneo, B.M.; Lanfranchi, A.; Vailati, S.; Giannattasio, C.; Del Bo, A.; Sala, C.; Bolla, G.B.; Pozzi, M.; et al. Sympathetic Activation and Loss of Reflex Sympathetic Control in Mild Congestive Heart Failure. Circulation 1995, 92, 3206–3211. [Google Scholar] [CrossRef]
- DeBurgh Daly M, Scott MJ. An Analysis of the Primary Cardiovascular Reflex Effects of Stimulation of the Carotid Body Chemoreceptors in the Dog. J. Physiol. 1962, 162, 555–573. [CrossRef]
- Daly MD, Scott MJ. The Cardiovascular Responses to Stimulation of the Carotid Body Chemoreceptors in the Dog. J. Physiol. 1963, 165, 179–197. [CrossRef]
- Lenka, N.; Krishnan, S.; Board, P.; Rangasamy, D. Exploiting the power of LINE-1 retrotransposon mutagenesis for identification of genes involved in embryonic stem cell differentiation. Stem Cell Rev. Rep. 2014, 10, 408–416. [Google Scholar] [CrossRef]
- Esposito, M.; Gualandi, N.; Spirito, G.; Ansaloni, F.; Gustincich, S.; Sanges, R. Transposons Acting as Competitive Endogenous RNAs: In-Silico Evidence from Datasets Characterised by L1 Overexpression. Biomedicines 2022, 10, 3279. [Google Scholar] [CrossRef]
- Blackburn, E.H. TELOMERASES. Annu. Rev. Biochem. 1992, 61, 113–129. [Google Scholar] [CrossRef]
- Balzanelli M, Distratis P, Lazzaro Rita, Pham VH et al. The Anti-Viral Activity of Stem Cells: A Rational Explanation for their Use in Clinical Application. Bentham Science Publishers 2023, 23, 738–747.
- Blackburn, E.H.; Collins, K. Telomerase: An RNP Enzyme Synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2010, 3, a003558–a003558. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo Isacco C, Pham Hung V, Nguyen Cao DK, Vo Lh, T et al. Autologous Peripheral Blood Stem Cells Increase the Telomere Length in Patients: A Case Report of 13 Patients. Journal of Stem Cell Research & Therapy 2016, 6, 1–6.
- AbdelMassih, A.; Agha, H.; El-Saiedi, S.; El-Sisi, A.; El Shershaby, M.; Gaber, H.; Ismail, H.-A.; El-Husseiny, N.; Amin, A.R.; ElBoraie, A.; et al. The role of miRNAs in viral myocarditis, and its possible implication in induction of mRNA-based COVID-19 vaccines-induced myocarditis. Bull. Natl. Res. Cent. 2022, 46, 1–8. [Google Scholar] [CrossRef]
- Siu, R.W.C.; Fragkoudis, R.; Simmonds, P.; Donald, C.L.; Chase-Topping, M.E.; Barry, G.; Attarzadeh-Yazdi, G.; Rodriguez-Andres, J.; Nash, A.A.; Merits, A.; et al. Antiviral RNA Interference Responses Induced by Semliki Forest Virus Infection of Mosquito Cells: Characterization, Origin, and Frequency-Dependent Functions of Virus-Derived Small Interfering RNAs. J. Virol. 2011, 85, 2907–2917. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived miRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.-Q.; Li, X.-F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human Virus-Derived Small RNAs Can Confer Antiviral Immunity in Mammals. Immunity 2017, 46, 992–1004. [Google Scholar] [CrossRef]
- Henzinger, H.; Barth, D.A.; Klec, C.; Pichler, M. Non-Coding RNAs and SARS-Related Coronaviruses. Viruses 2020, 12, 1374. [Google Scholar] [CrossRef]
- Zambon, R.A.; Vakharia, V.N.; Wu, L.P. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell. Microbiol. 2006, 8, 880–889. [Google Scholar] [CrossRef]
- Ding, S.-W.; Voinnet, O. Antiviral Immunity Directed by Small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Shiu, P.K.T.; Ilieva, M.; Holm, A.; Uchida, S.; DiStefano, J.K.; Bronisz, A.; Yang, L.; Asahi, Y.; Goel, A.; Yang, L.; et al. The Non-Coding RNA Journal Club: Highlights on Recent Papers—12. Non-Coding RNA 2023, 9, 28. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).