1. Introduction
Aerobic methanotrophs are a unique subset of methylotrophic bacteria, which utilize methane (CH
4) as a sole source of carbon and energy [
1,
2,
3,
4]. A defining characteristic of these microorganisms is the use of methane monooxygenase (MMO) enzymes to catalyze the oxidation of methane to methanol [
5,
6]. A membrane-bound or particulate MMO (pMMO) is present in most currently described methanotroph species, while some methanotrophs may also contain a soluble form of this enzyme (sMMO).
Currently described aerobic methanotrophs comprise a number of genera within the
Gamma- and
Alphaproteobacteria (also known as type I and type II methanotrophs) as well as the
Verrucomicrobia [
7]. These bacteria inhabit a wide spectrum of habitats that differ with regard to temperature, pH, salinity, oxygen and methane availability, as well as other environmental variables [
1,
8,
9]. Known phenotypes of aerobic methanotrophs include psychro- and thermophiles, acido- and alkaliphiles, halophiles and salt-sensitive organisms.
Due to their ability to grow on methane and natural gas, methanotrophic bacteria have attracted considerable attention as potential producers of a single-cell protein (SCP) and various value-added products from C1 compounds [
10,
11,
12,
13,
14]. Methane SCP is one of the most advanced and accessible SCP production technologies, and is currently on the verge of large-scale commercialization [
13]. So far, the biotechnology of converting methane to microbial proteins implied using fast growing freshwater gammaproteobacterial methanotrophs of the genera
Methylococcus and
Methylomonas [
15,
16]. Many regions of the world, however, have limited freshwater resources, so that the search for methanotrophs capable of fast growth in seawater or saltwater is of high interest for further development of this biotechnology.
The first methanotroph isolated from seawater was ‘
Methylomonas pelagica’ [
17], which was further reclassified as ‘
Methylomicrobium pelagicum’, and recently placed in the genus
Methylotuvimicrobium [
18]. This methanotroph was obtained from Sargasso Sea and was characterized as requiring NaCl and growing well in seawater. Since that time, quite a number of halophilic and halotolerant methanotrophs have been isolated from various marine habitats as well as from hypersaline and soda lakes. These include
Methylosphaera hansonii from an Antarctic meromictic lake [
19],
Methylohalobius crimeensis from a hypersaline lake in Ukraine [
20],
Methylotuvimicrobium alcaliphilum from a soda lake [
21] and
Methylotuvimicrobium japanense from marine mud [
21],
Methylomarinum vadi and
Methylomarinovum caldicuralii from a shallow submarine system [
22,
23],
Methyloprofundus sedimenti from a deep marine sediment [
24], and some other methanotrophs (see
Table 1).
Highest tolerance to NaCl (up to 15%, w/v) was reported for
Methylohalobius crimeensis, although the strains representing this species demonstrated relatively low growth rates, 0.019-0.028 h
-1, in a medium with optimal NaCl concentration (6.5%, w/v). Good tolerance of NaCl in combination with high growth rates are characteristic for members of the genera
Methylomarinum and
Methylotuvimicrobium. Thus, the strains representing
Methylomarinum vadi grew in media with 1–8% (w/v) NaCl (optimum, 2–3% NaCl) and the highest specific growth rate was 0.33 h
-1 [
22]. However, the maximum cell yield reported for these methanotrophs was only 1×10
8 ml
-1 (OD
660 0.2). Representatives of the genus
Methylotuvimicrobium grow within a wide range of NaCl concentrations, from 0.03 to 1.5 M [
18,
21]. Among all above listed methanotrophs,
Methylotuvimicrobium species appear to be most suited for fast and robust growth in media with high salt content. Due to its high growth characteristics,
Methylotuvimicrobium alkaliphilum 20Z became one of the model organisms in methanotroph research [
31,
32]. Methane-consuming activity in these bacteria, however, is fastest at pH near 9 [
21], while SCP production technology relies on using near-neutral or slightly acidic media.
This study aimed at searching for novel methanotrophs capable of fast and robust growth in saltwater comparable in composition with seawater. The suitability of these bacteria for the purposes of SCP production from natural gas was verified in experiments on continuous cultivation in bioreactors.
2. Materials and Methods
2.1. Sampling site
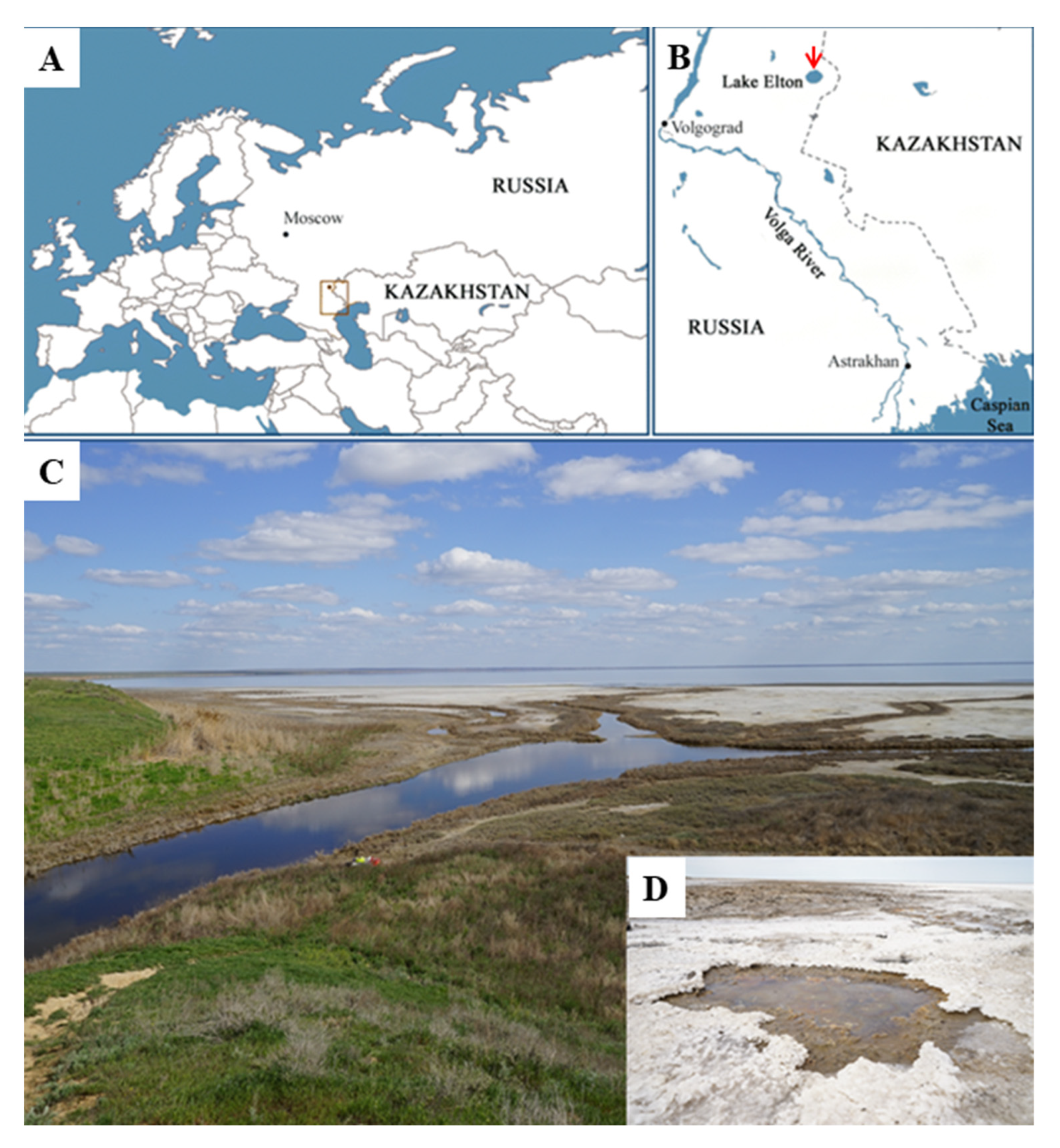
The sediment sample used in our study for isolation of halophilic methanotrophs was collected in August 2022 from beneath shallow water at the flowing of the river Chernavka into the hypersaline Lake Elton, Volgograd region, Russia (49.2085 N, 46.68024 E) (
Figure 1). At the time of sampling, the sediment temperature was 19 °C, pH 7.5, and the total salinity was 30 g L
-1. The sediment sample was transported to the laboratory and used for preparing enrichment cultures within two days after sampling.
2.2. Enrichment procedure
An aliquot of the sediment sample was used as inoculum to obtain enrichment culture of methanotrophic bacteria using the mineral medium MS containing (in grams per liter of distilled water) KNO3, 0.25; NH4Cl, 0.25; MgSO4 × 7H2O, 0.4; CaCl2 × 2H2O, 0.1; NaCl, 20.0; KCl. 1.5; 100 mM phosphate buffer, pH 7.5, 1% (vol/vol); and trace element solution 0.1% (vol/vol), containing the following (g/L): EDTA, (in grams per liter) EDTA, 5; FeSO4 × 7H2O, 2; ZnSO4 × 7H2O, 0.1; MnCl2 × 4H2O, 0.03; CoCl2 × 6H2O, 0.2; CuSO4 × 5H2O, 0.1; NiCl2 × 6H2O, 0.02; and Na2MoO4, 0.03. One gram of the sediment was added to 500 ml bottle containing 100 ml of MS medium. The bottle was sealed with silicone rubber septa, and methane was added aseptically using a syringe equipped with a disposable filter (0.22 µm) to achieve a 10–20% mixing ratio in the headspace. The incubation was performed on a rotary shaker (120 r.p.m.) at 30 °C. Every 10 days of incubation, an aliquot of the developed cell suspension was transferred to a bottle with a fresh medium MS (in ratio 1:10) and incubated with methane in the gas phase under the same conditions. After 4 passages, the enriched methane-oxidizing microbial consortium, designated Ch1, was subjected to 16S rRNA gene-based molecular analysis to identify its composition.
2.3. Molecular analysis of methanotroph community composition
Total DNA was isolated from 1 ml of the examined cell suspension using the DNeasy PowerMax Soil Kit (Qiagen, Carlsbad, CA, United States). PCR fragments of the 16S rRNA gene were obtained with the universal primers 341F (5'-CCTAYGG-GDBGCWSCAG-3') and 806R (5'-GGACTACNVG- GGTHTCTAAT-3') [
33]. The PCR fragments were barcoded with Nextera XT Index Kit v.2 (Illumina, United States) and purified using Agencourt AMPure beads (Beckman Coulter, Brea, CA, United States). The concentrations of the obtained PCR products were calculated using the Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA, United States). All PCR fragments were then mixed in equal amounts and sequenced on an Illumina MiSeq (2 × 300 nt from both ends). Pairwise reads were combined using FLASH v.1.2.11 [
34]. The sequences were clustered into operational taxonomic units (OTUs) at 97% identity using Usearch [
35]; low-quality reads, chimeras, and singletons were eliminated during clusterization using the Usearch algorithm. Taxonomic identification was carried out using the SILVA v.132 database and the VSEARCH algorithm [
36].
2.4. Cultivation in a bioreactor
The enriched methane-oxidizing microbial consortium Ch1 was used for inoculating a bioreactor. The cultivation on natural gas (CH
4 content 97.3%) was performed in a 1.5 l bioreactor (GPC BIO, France) with a working volume of 1 l. The following process parameters were used for all experiments: temperature, 30 °C; agitation, 1000 rpm; gas flow rate, 6000 ccm; air flow rate, 18000 ccm; pH 7.0. pH was controlled via titration with 1% NH
4OH which also served as an additional nitrogen source. Three media compositions that differed to each other with regard to the total salt content were tested in growth experiments with the consortium Ch1 (
Table 2). Each of these media contained the same trace elements of the following composition (mg/L): FeSO
4 × 7H
2O, 18.9; ZnSO
4 × 7H
2O, 3.3; MnSO
4 × 5H
2O, 21.3; CoSO
4×6H
2O, 0.48; CuSO
4 × 5H
2O, 16.0; NiCl
2 × 6H
2O, 1.0; Na
2MoO
4, 0.4, and H
3BO
3, 7.5.
Bioreactor was inoculated at OD600 = 0.5 with a CH4-grown seed culture of consortium Ch1 cultivated in MS medium. Growth was monitored by measuring the OD600 using a Spectroquant Prove 300 spectrophotometer (Merck, Germany) every two hours. Dissolved oxygen was maintained below 20% of the full (100%) air saturation during the entire run by adjusting the agitation speed. Gas fermentations were run in continuous modes with the average dilution rate of 0.20 h-1.
To determine the cell dry weight (CDW), cells were collected on filters (0.22 µm) using vacuum filtration, washed with distilled water and dried at 105 ºC to a constant weight. To determine the content of protein, lipids, carbohydrate and solid base ash in the biomass, cells were collected after the dilution rate of 0.2 h
-1 was achieved. Cell suspensions were centrifuged at 10000 ×g for 5 min. Collected cells were washed with distilled water and freeze-dried at -70 °C. The Kjeldahl technique was used to determine the protein concentration [
37] in lyophilized biomass with the help of Kjeldahl analyzer (FOSS, Sweden). Lipid extraction and content determination was performed by Bligh-Dyer method [
38]. Total carbohydrates and solid base ash were determined according to the standard procedures [
39].
2.5. Isolation studies
To isolate methane-oxidizing bacteria, cell suspensions from both the original enrichment culture and the bioreactor culture were subjected to multiple serial dilutions in 120-ml flasks containing 10 ml of MS medium. The flasks were incubated on a rotary shaker (120 r.p.m.) at 30 °C. Culture purity was verified by examination using phase-contrast microscopy and by plating on 10-fold diluted Luria–Bertani agar (1.0% tryptone, 0.5% yeast extract, 2.0% NaCl).
2.6. Morphological characterization and microscopy
Morphological observations and cell-size measurements were made with a Zeiss Axioplan 2 microscope and Axiovision 4.2 software (Zeiss, Germany). For electron microscopy, cells of exponentially growing cultures were collected by centrifugation and pre-fixed with 2.5% (w/v) glutaraldehyde in 0.05 M cacodylate buffer (pH 7.2) for 1 h at 4°C and then fixed in OsO
4 (1% (w/v) OsO
4 + 0.7% (w/v) ruthenium red solution) in the same buffer for 4 h at 20 °C. After fixation, the samples were sequentially kept in a 3% uranyl acetate solution in 30% ethanol for 4 h, then in 70% ethanol for 12 h at 4 °C. The material was dehydrated in 96% ethanol (2 treatments for 15 min), then in absolute acetone (3 treatments for 10 min). Then the samples were embedded in Epon 812 epoxy resin (Eрoху Embedding Medium Epon® 812, Sigma-Aldrich, USA). Ultrathin sections were cut on an LKB-III microtome (LKB, Sweden), stained with an aqueous solution of 3% uranyl acetate (at 37 °C for 20 min) and then post-stained with lead citrate [
40] at 37 °C for 20 min. Specimen samples were examined with a JEM 100CXII transmission electron microscope (Jeol, Japan) at an 80 kV accelerating voltage. To test the presence of flagella, cell suspensions from the cultures were dried onto grids and treated with 1% (w/v) phosphotungstic acid solution. Negatively stained cells were examined with a JEM 100CXII electron microscope (Jeol, Japan) at an 80 kV accelerating voltage. Photo documentation of the materials was carried out using a Morada G2 (Olympus, Japan) digital optical image output system.
2.7. DNA Extraction
Strain Ch1-1 was grown in the liquid medium MS as described above. The cells were harvested after incubation at 30°C on a rotary shaker at 120 rpm for 2 days. Genomic DNA extraction was done using the standard CTAB and phenol-chloroform protocol [
41].
2.8. Genome Sequencing and Annotation
Nanopore sequencing library was prepared using the 1D ligation sequencing kit (SQK-LSK108, Oxford Nanopore, UK). Sequencing was performed on an R9.4 flow cell (FLO-MIN106) using MinION device. Adapters were trimmed using Porechop v0.2.4 (
https://github.com/rrwick/Porechop). Assembly of long reads was performed using Flye v.2.8.1 [
42] and evaluated with Quast 5.0 [
43] and Busco 5.1.2 [
44]. Annotation was performed using PROKKA [
45] and GhostKOALA [
46].
2.9. Phylogenomic analysis
The genome-based tree of strain Ch1-1 and phylogenetically related members of the family
Methylococcaceae was reconstructed using the Genome Taxonomy Database and GTDB-Tk package (
https://github.com/Ecogenomics/GtdbTk), release 04-RS89. The maximum likelihood genome-based phylogenetic tree was constructed using MegaX software [
47].
2.10. Sequence accession numbers
The 16S rRNA gene sequence and the assembled genome sequence of strain Ch1-1 have been deposited in the GenBank under the accession numbers OR427371 and JAUZWD000000000, respectively.
3. Results and Discussion
3.1. Enriched methane-oxidizing consortium and its composition
The methane-oxidizing microbial community enriched from the studied sediment sample was represented by morphologically different cells that showed a tendency to clamp together. Two major cell morphotypes were thick rods and spiral-shaped cells. Express analysis of the microbial community composition by Illumina-based 16S rRNA gene sequencing revealed the presence of two methanotrophs affiliated with the family Methylococcaceae, namely Methylomarinum- and Methylotuvimicrobium-like bacteria. The relative abundance of 16S rRNA gene reads from Methylomarinum- and Methylotuvimicrobium-like methanotrophs was 20.80% and 0.12%, respectively. Major groups of satellite bacteria were represented by methylotrophs of the genus Methylophaga (27.5% of all 16S rRNA gene reads) and halophilic heterotrophs of the genus Thalassospira (35.7%). Minor groups of satellite bacteria included Terasakiella (3.5%), Oceanobacter (2.9%), Pseudoalteromonas (1.8%), and Vibrio (1.5%). This methane-oxidizing consortium, designated Ch1, was tested for the ability to grow in a bioreactor on natural gas in media with high salt content.
3.2. Growth of the methanotrophic consortium Ch1 in a bioreactor
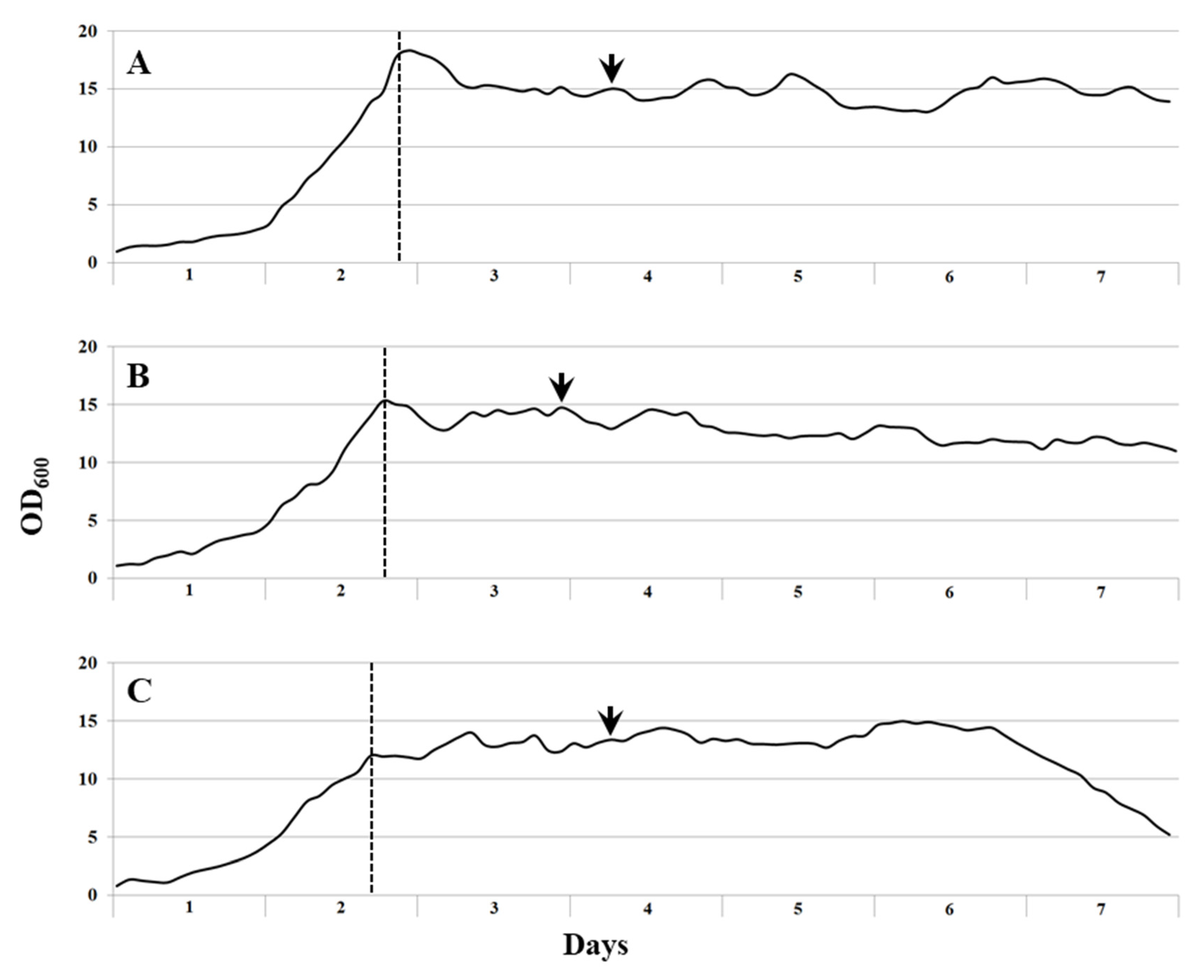
Three media compositions that differed with regard to the total salt content were tested in growth experiments on continuous cultivation of the consortium Ch1 on natural gas (
Table 2). Stable growth with the specific growth rate 0.21±0.01 h
-1 was recorded for the media with total salt contents of 23 and 29 g L
-1, while the consortium growing in a medium with 35.9 g salts L
-1 demonstrated a bit lower growth rate of 0.19±0.01 h
-1 (
Figure 2,
Table 3). The highest biomass yield of 5.77±1.16 g CDW L
-1 was obtained during continuous cultivation of the consortium Ch1 in a medium with a total salt content of 29 g L
-1 (
Table 3). To determine protein content and other nutritional values of the cell biomass, cell suspensions were collected during stable fermentation process with the highest productivity and high growth rates. The content of protein in dry cell biomass showed a tendency to decrease with increase in a total salt content (
Table 3). The highest protein content in dry cell biomass (65.4%) was recorded in the medium containing 23 g salts L
-1. The same tendencies were observed with regard to the contents of total lipids and carbohydrates. The content of solid base ash varied between 0.15% (23 g salts L
-1) and 0.19% (in media with 35.9 and 29 g salts L
-1).
Molecular analysis of the consortium Ch1 composition was performed with the cell suspensions collected during the stable fermentation process (sampling points are indicated by arrows in
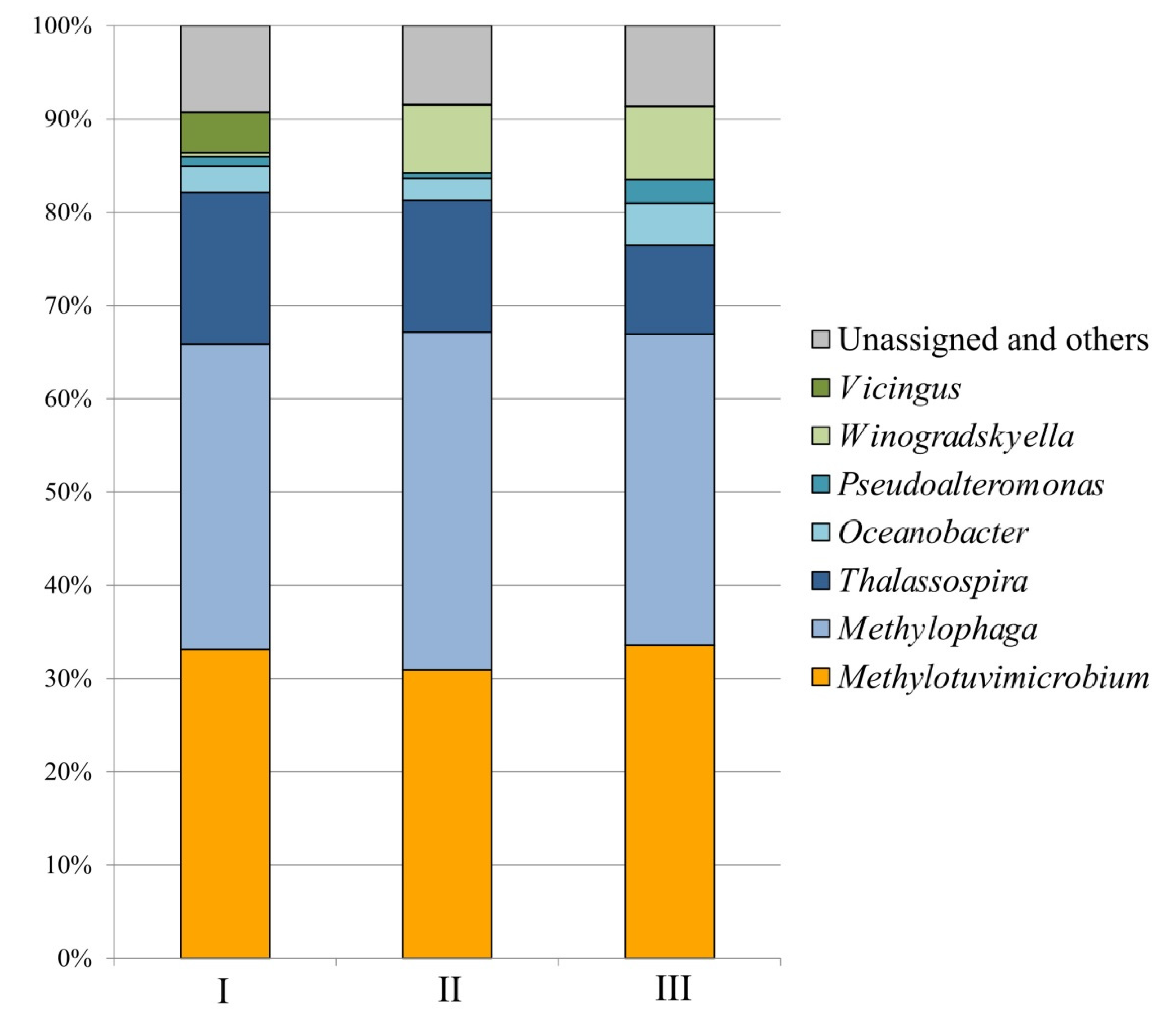
Figure 2). Overall, the microbial composition in fermenters running with media of different salinities was quite similar (
Figure 3). Major methanotrophic component of the studied consortium was represented by
Methylotuvimicrobium-like bacterium. The 16S rRNA gene fragments from this methanotroph comprised 30.9-33.6% of total 16S rRNA gene reads retrieved from the examined samples. The reads affiliated with
Methylomarinum-like methanotrophs were also detected but these were present in a low relative abundance only (0.04-0.25%). Notably, highest relative abundance of
Methylomarinum-like bacteria was detected in the bioreactor operating with the medium of a highest salt content, 35.9 g salts L
-1. Methylotrophs of the genus
Methylophaga (32.7-36.2% of all 16S rRNA gene reads) and halophilic heterotrophs of the genus
Thalassospira (9.5-16.3%) were the two groups of numerically dominant satellite bacteria. Representatives of the genus
Winogradskyella developed mostly in the bioreactors with high salt contents, 29 and 35.9 g salts L
-1, with the relative abundances of 7.3 and 7.8%, respectively. The opposite trend was observed for bacteria of the genus
Vicingus, whose growth was pronounced in the bioreactor with lowest salinity only (4.4% of all 16S rRNA gene reads). Other numerically significant community members included satellite bacteria of the genera
Oceanobacter (2.3-4.6%) and
Pseudoalteromonas (0.6-2.5%), which developed in all bioreactor cultures.
3.3. Isolation of methanotrophic bacteria
Our major isolation efforts were focused on obtaining two target methanotrophs,
Methylomarinum- and
Methylotuvimicrobium-like bacteria in pure cultures. Cell suspensions from both the original enrichment culture and bioreactor-grown cultures were used as isolation sources. All attempts to isolate target methanotrophs by plating cell suspensions onto the agar medium MS were unsuccessful. Apparently, methanotrophs did not form colonies on this agar medium. The use of multiple serial dilutions in the liquid medium MS was more helpful. This approach allowed obtaining one methanotrophic isolate, strain Ch1-1, from the original methane-oxidizing enrichment culture. The 16S rRNA gene sequence of strain Ch1-1 displayed 97.09-97.24% similarity to the corresponding gene fragments of two characterized representatives of
Methylomarinum vadi, strains IT-4
T and T2-1, methanotrophs isolated from two distinct marine habitats [
22]. Among taxonomically uncharacterized bacterial isolates, the highest 16S rRNA gene similarity of 99.45% was observed with
Methylomarinum strain SSMP-1, which was obtained from a terrestrial saline mud pot at the northern terminus of the Eastern Pacific Rise [
48].
Attempts to isolate methanotrophs from bioreactor-grown cultures were unsuccessful, most likely due to a high cell aggregation and tight metabolic interactions of methanotrophs and heterotrophs under conditions of continuous cultivation.
3.4. Characterization of methanotrophic isolate
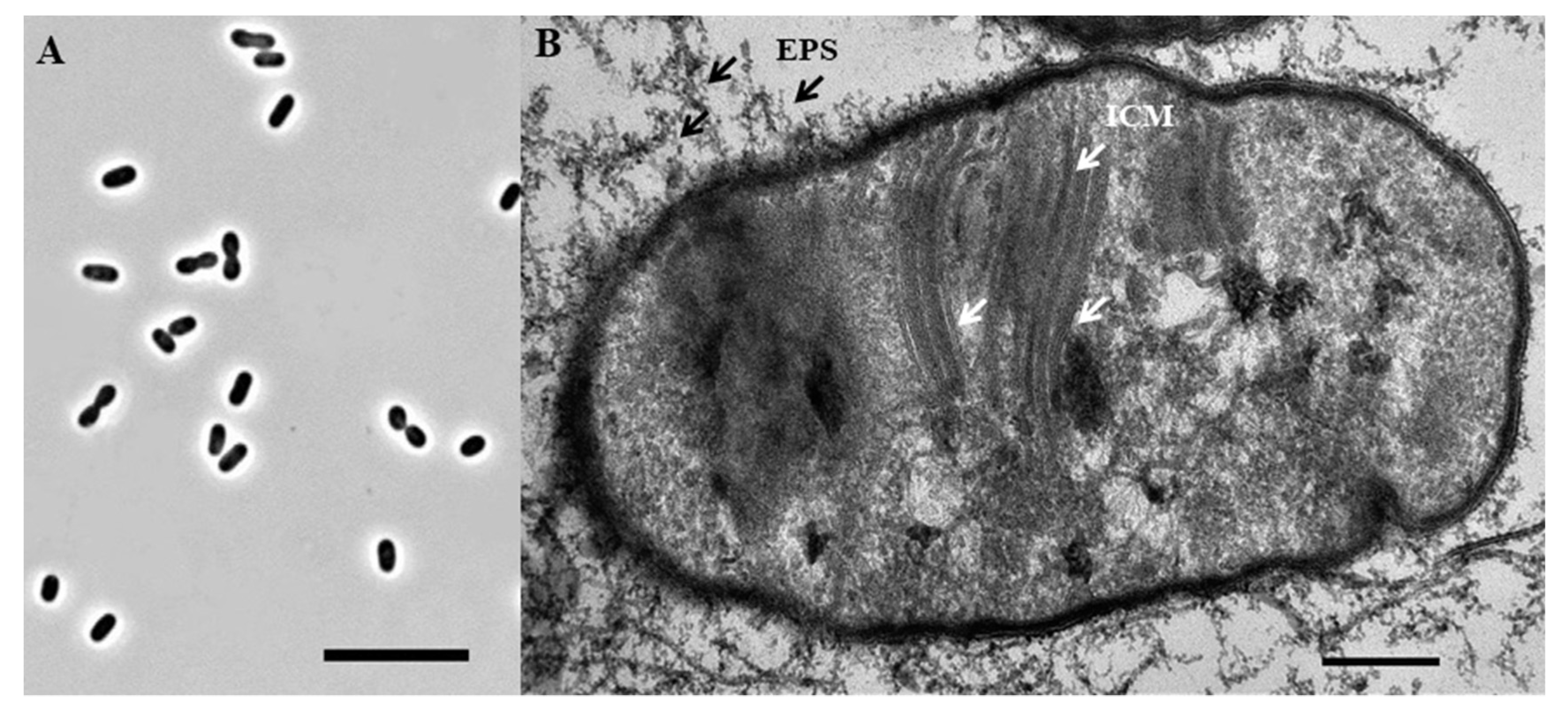
Strain Ch1-1 was represented by short motile rods or ovoids, which were 0.85 ± 0.05 μм wide by 1.50 ± 0.10 μм long (
Figure 4A). Cells multiplied by binary fission. Formation of short cell chains (up to 4 cells) was occasionally observed. Strain Ch1-1 did not grow on agar media. Liquid cultures were pink in color. Analysis of ultrathin cell sections showed a typical Gram-negative structure of the cell wall and the presence of intracytoplasmic membranes (ICM), arranged as stacks of vesicular disks (
Figure 4B). This ICM arrangement is characteristic of type I methanotrophs. Cells produced large amounts of exopolysaccharide, which appeared as dense long threads (
Figure 4B).
Strain Ch1-1 was capable of growth within the temperature range of 5-38 ºC. NaCl was required for growth, which was observed within NaCl concentration range of 0.1-10% (w/v). Best growth was recorded at NaCl concentration of 1.5-2.0% (w/v).
3.5. General genome features of strain Ch1-1 and genome-based phylogeny
Sequencing of strain Ch1-1 using Oxford Nanopore technology yielded 123,765 reads with a total length of 1.34 Gb. The genome assembly comprises 4 contigs totaling 4.8 Mbp, with an average G+C content of 50.7 mol %. Genome contains three identical rrn operon copies (16S-23S-5S rRNA), 46 tRNA genes, 4469 predicted protein-coding sequences and 7 Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) loci.
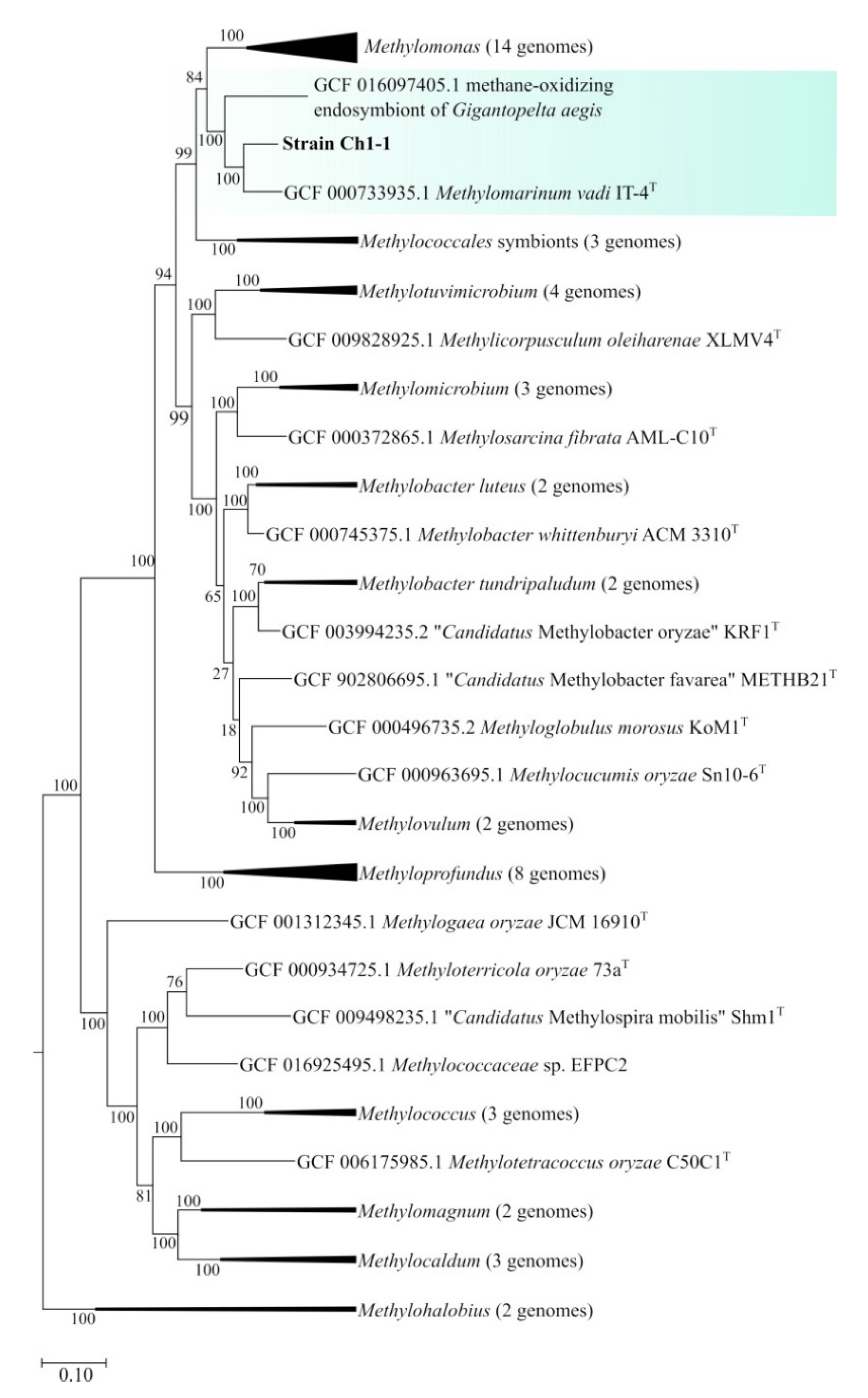
The genome-based phylogeny of strain Ch1-1 was determined based on the comparative analysis of 120 ubiquitous single-copy proteins (
Figure 5). Strain Ch1-1 belonged to the phylogenetic lineage defined by
Methylomarinum vadi IT
T [
22]. One additional member of this lineage was represented by methane-oxidizing endosymbiont of the deep-sea hydrothermal vent snail
Gigantopelta aegis [
49].
The genome contains a single pmoCAB gene cluster encoding conventional particulate MMO. Gene cluster encoding soluble MMO was not identified. Methanol oxidation capability of strain Ch1-1 is explained by the presence of gene clusters encoding MxaFI- and XoxF-methanol dehydrogenases. Genes involved in tetrahydromethanopterin and tetrahydrofolate-linked pathways as well as formate oxidation were identified. Complete set of genes for the function of the ribulose monophosphate pathway is present. Genome harbors genes encoding serine-glyoxylate aminotransferase, 3-glycerate kinase and putative hydroxypyruvate reductase, which are the key enzymes of the serine pathway of formaldehyde assimilation. Genome also contains genes for malyl-CoA lyase and malate thiokinase as well as PEP carboxylase for glyoxylate biosynthesis. Genes encoding large and the small subunits of RuBisCo (cbbL and cbbS) were not detected. Genome harbors genes required for conducting both dissimilatory and assimilatory nitrate reduction. Strain Ch1-1 also possesses all genes necessary for ectoine biosynthesis.
3.6. Growth of the methanotrophic isolate Ch1-1 in a bioreactor
Since the above reported growth characteristics of salt-tolerant methanotrophs in continuous bioreactor cultures refer to the consortium dominated with
Methylotuvimicrobium-like methanotroph (
Table 3), we tested strain Ch1-1 for the ability to grow under the same conditions in a medium with a total salt content of 29 g L
-1. Stable growth with the specific growth rate 0.15±0.03 h
-1 and the maximum OD
600 5.93 were recorded for this bacterium. The highest biomass yield was 2.11±0.13 g CDW L
-1 and the productivity was 0.32 g CDW L
-1 h
-1. These values, however, were obtained during our first experiment on a continuous cultivation of strain Ch1-1 in a bioreactor. Long-term cultivation of this bacterium in a bioreactor may result in better growth rates and productivity.
In summary, our study demonstrated a possibility of modifying the earlier developed technology of single-cell protein production for the use in regions with limited freshwater resources. The methanotrophic consortium obtained from the sediments of the hypersaline Lake Elton was capable of fast and highly productive growth in saltwater comparable in composition with seawater. The mineral medium with a total salt content of 35.9 g L
-1 (
Table 2) is identical in composition with seawater from the Bay of Biscay [
50]. Although the total salt content/composition may vary in water from different locations, the methanotrophic consortium described in our study demonstrated a stable growth in a wide range of salinities (
Table 3). The same was true for the isolate of
Methylomarinum-like methanotroph, strain Ch1-1, which was capable of growth within NaCl concentration range of 0.1-10% (w/v). With the only exception of
Methylohalobius crimeensis, the upper limit of NaCl tolerance recorded for strain Ch1-1 (10%, w/v) is higher than those reported for other halophilic and halotolerant methanotrophs (
Table 1). Interestingly, the closest phylogenetic relative of strain Ch1-1,
Methylomarinum strain SSMP-1, was also isolated not from a marine habitat, but from a terrestrial saline mud pot [
48]. These two methanotrophs display a number of phenotypic differences from described strains of
Methylomarinum vadi. In particular, cells of strains Ch1-1 and SSMP-1 are pink-pigmented in a contrast to unpigmented representatives of
Methylomarinum vadi. Temperature and salinity growth ranges of these bacteria also displayed some differences. Apparently, strains Ch1-1 and SSMP-1 represent a potentially new species of the genus
Methylomarinum.
Unfortunately, the
Methylotuvimicrobium-like methanotroph could not be obtained in a pure culture in this study. The 16S rRNA gene reads from this bacterium displayed 98.7% similarity to the corresponding gene fragment from
Methylotuvimicrobium japanense NI
T, isolated from marine mud in Japan [
21]. So far, no data were reported on growth characteristics of
Methylotuvimicrobium japanense in continuous cultures in a bioreactor. Further work is needed to investigate the biotechnological potential of both
Methylotuvimicrobium- and
Methylomarinum-like methanotrophs as well as the functional role of satellite bacteria that develop in bioreactor cultures of these potential single-cell protein producers in saltwater.
Author Contributions
Conceptualization, S.N.D.; resources, N.V.P.; investigation, E.N.T., R.Z.S., I.Y.O., A.A.K. and D.V.F.; data curation, E.N.T. and I.Y.O.; writing-original draft preparation, E.N.T., I.Y.O. and S.N.D.; writing-review and editing, S.N.D. and N.V.P.; funding acquisition, S.N.D. and N.V.P. All authors have read and agreed to the published version of the manuscript.
Funding
The article was made with support of the Ministry of Science and Higher Education of the Russian Federation in accordance with agreement № 075-15-2022-318 date April 20, 2022 on providing a grant in the form of subsidies from the Federal budget of Russian Federation. The grant was provided for state support for the creation and development of a World-class Scientific Center "Agrotechnologies for the Future".
Data Availability Statement
The 16S rRNA gene sequence and the assembled genome sequence of strain Ch1-1 have been deposited in the GenBank under the accession numbers OR427371 and JAUZWD000000000, respectively.
Acknowledgments
The authors thank Timur Kanapatskiy for the help with sampling.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic Aspects of Aerobic Obligate Methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry. 2013, 267–285.
- Khmelenina, V.N.; Colin Murrell, J.; Smith, T.J.; Trotsenko, Y.A. Physiology and Biochemistry of the Aerobic Methanotrophs. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids; Springer: Cham, Switzerland, 2019; pp. 1–25. [Google Scholar]
- Murrell, J.C.; Gilbert, B.; McDonald, I.R. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Sazinsky, M.H.; Lippard, S.J. Methane Monooxygenase: Functionalizing Methane at Iron and Copper. Met. Ions Life Sci. 2015, 15, 205–256. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Knief, C. Diversity and Phylogeny of Described Aerobic Methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability. 2018; 17–42. [Google Scholar] [CrossRef]
- Trotsenko, Y.A.; Khmelenina, V.N. Biology of extremophilic and extremotolerant methanotrophs. Arch. Microbiol. 2002, 177, 123–131. [Google Scholar] [CrossRef]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bac-teria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Strong, P.; Xie, S.; Clarke, W.P. Methane as a resource: can the methanotrophs add value? Environ. Sci. Technol. 2015, 49, 4001–4018. [Google Scholar] [CrossRef]
- Mühlemeier, I.M.; Speight, R.; Strong, P.J. Biogas, bioreactors and bacterial methane oxidation. In Methane Biocatalysis: Paving the Way to Sustainability. 2018, 213–235.
- Risso, C.; Choudhary, S.; Johannessen, A.; Silverman, J. Methanotrophy goes commertial: Challenges, opportunities, and brief history. In Methane Biocatalysis: Paving the Way to Sustainability. 2018, 293–298.
- Martínez, J.B.G.; Pearce, J.M.; Throup, J.; Cates, J.; Lackner, M.; Denkenberger, D.C. Methane Single Cell Protein: Potential to Secure a Global Protein Supply Against Catastrophic Food Shocks. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef]
- Le, H.T.Q.; Lee, E.Y. Methanotrophs: Metabolic versatility from utilization of methane to multi-carbon sources and perspectives on current and future applications. Bioresour. Technol. 2023, 384, 129296. [Google Scholar] [CrossRef]
- Skrede, A.; Berge, G.; Storebakken, T.; Herstad, O.; Aarstad, K.; Sundstøl, F. Digestibility of bacterial protein grown on natural gas in mink, pigs, chicken and Atlantic salmon. Anim. Feed. Sci. Technol. 1998, 76, 103–116. [Google Scholar] [CrossRef]
- Koffas, M.; Odom, J.M.; Square, K. High growth methanotrophic bacterial strain. US Patent 6 689 601. 2003.
- Sieburth, J.M.; Johnson, P.W.; Eberhardt, M.A.; Sieracki, M.E.; Lidstrom, M.; Laux, D. The first me-thane-oxidizing bacterium from the upper mixing layer of the deep ocean: Methylomonas pelagica sp. nov. Curr. Microbiol. 1987, 14, 285–293. [Google Scholar] [CrossRef]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic Analysis of the Gammapro-teobacterial Methanotrophs (Order Methylococcales) Calls for the Reclassification of Members at the Genus and Species Levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.P.; McCammon, S.A.; Skerrat, J.H. Methylosphaera hansonii gen. nov., sp. nov., a psychrophilic, group I methanotroph from Antarctic marine-salinity, meromictic lakes. Microbiology 1997, 143, 1451–1459. [Google Scholar] [CrossRef]
- Heyer, J.; Berger, U.; Hardt, M.; Dunfield, P.F. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 2005, 55, 1817–1826. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Khmelenina, V.; Eshinimaev, B.; Sorokin, D.; Fuse, H.; Lidstrom, M.; Trotsenko, Y. Classification of halo(alkali)philic and halo(alkali)tolerant methanotrophs provisionally assigned to the genera Methylomicrobium and Methylobacter and emended description of the genus Methylomicrobium. Int. J. Syst. Evol. Microbiol. 2008, 58, 591–596. [Google Scholar] [CrossRef]
- Hirayama, H.; Fuse, H.; Abe, M.; Miyazaki, M.; Nakamura, T.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinum vadi gen. nov., sp. nov., a methanotroph isolated from two distinct marine en-vironments. Int. J. Syst. Evol. Microbiol. 2013, 63, 1073–1082. [Google Scholar] [CrossRef]
- Hirayama, H.; Abe, M.; Miyazaki, M.; Nunoura, T.; Furushima, Y.; Yamamoto, H.; Takai, K. Methylomarinovum caldicuralii gen. nov., sp. nov., a moderately thermophilic methanotroph isolated from a shallow submarine hydrothermal system, and proposal of the family Methylothermaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 989–999. [Google Scholar] [CrossRef]
- Tavormina, P.L.; Hatzenpichler, R.; McGlynn, S.; Chadwick, G.; Dawson, K.S.; Connon, S.A.; Orphan, V.J. Methyloprofundus sedimenti gen. nov., sp. nov., an obligate methanotroph from ocean sediment belonging to the 'deep sea-1' clade of marine methanotrophs. Int. J. Syst. Evol. Microbiol. 2015, 65, 251–259. [Google Scholar] [CrossRef]
- Bowman, J.P.; Sly, L.I.; Nichols, P.D.; Hayward, A.C. Revised Taxonomy of the Methanotrophs: Description of Methylobacter gen. nov., Emendation of Methylococcus, Validation of Methylosinus and Methylocystis Species, and a Proposal that the Family Methylococcaceae Includes Only the Group I Methanotrophs. Int. J. Syst. Evol. Microbiol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Lees, V.; Owens, N.J.P.; Murrell, J.C. Nitrogen metabolism in marine methanotrophs. Arch. Microbiol. 1991, 157. [Google Scholar] [CrossRef]
- Khmelenina, V.N.; Starostina, N.G.; Tsvetkova, M.G.; Sokolov, A.P.; Suzina, N.E.; Trotsenko, Y.A. Methanotrophic bacteria in saline reservoirs of Ukraine and Tuva. Microbiol. (English translation of Mikrobiologiia). 1996, 65, 609–615. [Google Scholar]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Suzina, N.E.; Lysenko, A.M.; Trotsenko, Y.A. New methanotrophic isolates of the southern Transbaikal region. Microbiol. (English translation of Mikrobiologiia). 1999, 68, 592–600. [Google Scholar]
- Kalyuzhnaya, M.G.; Khmelenina, V.N.; Eshinimaev, B. Suzina, N.; Nikitin, D.; Solonin, A.; Lin, J-L.; McDonald, I.; Murrell, C.; Trotsenko, Y. Taxonomic characterization of new alkaliphilic and alkalitolerant methanotrophs from soda lakes of the southeastern Transbaikal region and description of Methylomicro-bium buryatense sp. nov. Syst. Appl. Microbiol. 2001, 24, 166–176. [Google Scholar]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W.; et al. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.A.N.; Raftery, D.; Fu, Y.; Bringel, F.; Vuilleumier, S.; Beck, D.A.C.; Trotsenko, Y.A.; Khmelenina, V.N.; Lidstrom, M.E. Highly efficient methane biocatalysis re vealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef]
- Akberdin, I.R.; Thompson, M.; Hamilton, R.; Desai, N.; Alexander, D.; Henard, C.A.; Guarnieri, M.T.; Kalyuzhnaya, M.G. Methane utilization in Methylomicrobium alcaliphilum 20ZR: a systems approach. Sci. Rep. 2018, 8, 2512. [Google Scholar] [CrossRef]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: a versatile open source tool for met-agenomics. Peer.J. Preprints. 2016, 4, e2409v1. [Google Scholar]
- Kjeldahl, J. Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern. Anal. Bioanal. Chem. 1883, 22, 366–382. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Tsao, R.; Li, Y.; Miao, M. Food Safety: Food Analysis Technologies/Techniques. Encyclopedia of Agriculture and Food Systems. 2014, 273–288. [Google Scholar] [CrossRef]
- Reynolds, E.S. THE USE OF LEAD CITRATE AT HIGH pH AS AN ELECTRON-OPAQUE STAIN IN ELECTRON MICROSCOPY. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K. Preparation of Genomic DNA from Bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGAX: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Fradet, D.T.; Tavormina, P.L.; Orphan, V.J. Members of the methanotrophic genus Methylomarinum inhabit inland mud pots. PeerJ. 2016, 4, e2116. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Sun, J.; Chen, C.; Sun, Y.; Zhou, Y.; Yang, Y.; Zhang, W.; Li, R.; Zhou, K.; Wong, W.C.; et al. Hologenome analysis reveals dual symbiosis in the deep-sea hydrothermal vent snail Gigantopelta aegis. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Lazure, P.; Jegou, A-M. ; Kerdreux, M. Analysis of salinity measurments near islands on the French con-tinental shelf of the Bay of Biscay. Sci. Mar. 2006, 70S1, 7–14. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).