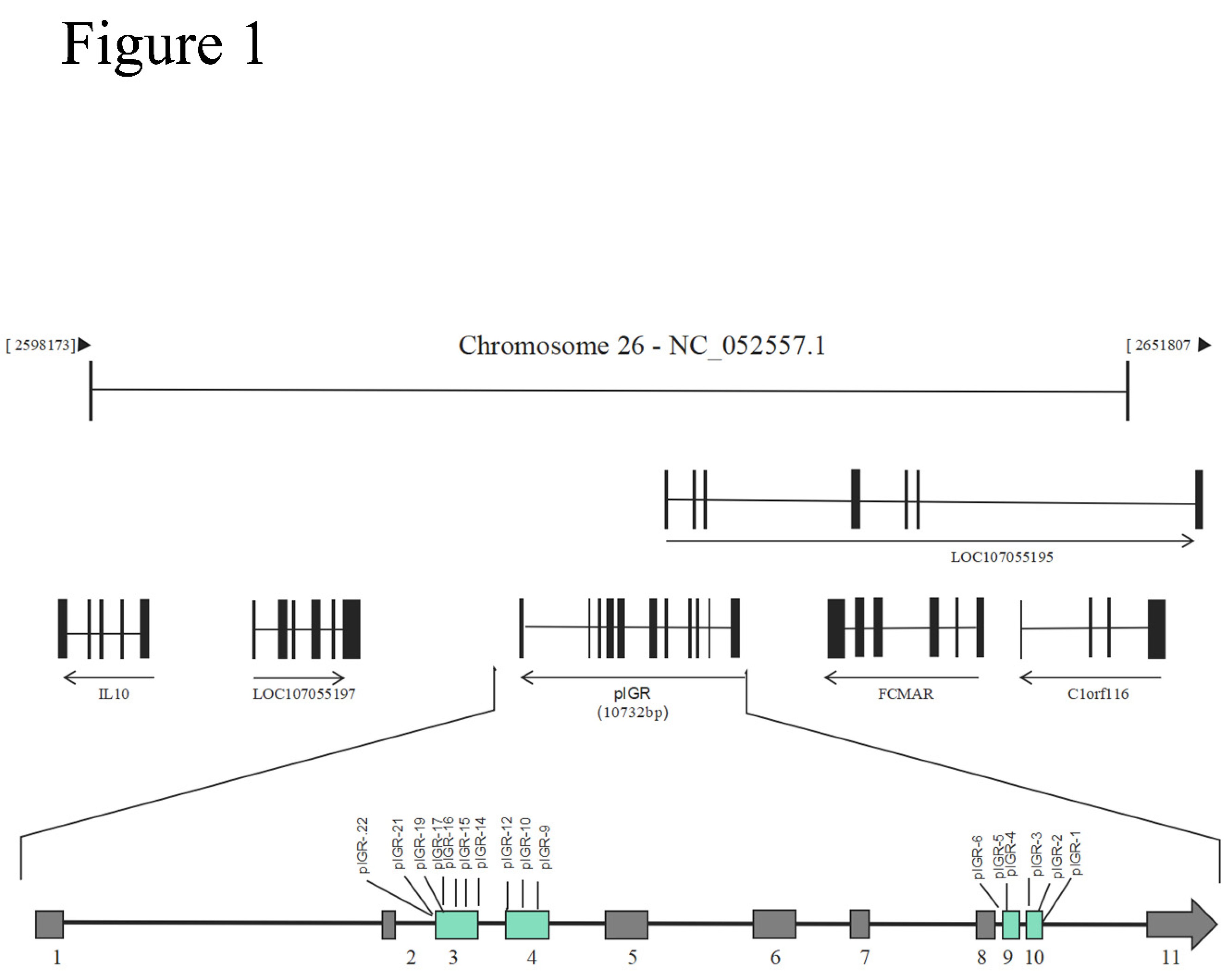
1. Introdution
Polymeric immunoglobulin receptor (
pIgR), a kind of type Ⅰ trans-membrane glycoprotein, also known as an integral membrane protein located at the basolateral surface of secretory epithelial cells. As a key component of the mucosal immune system,
pIgR is critical for the protective function of secretory immunoglobulins (SIgs) and mediates epithelial transcytosis of polymeric immunoglobulin A or polymeric immunoglobulin M (IgA or IgM) [
3,
35]. IgA class forms the first line of antigen-specific immune protection against inhaled, ingested, and sexually transmitted pathogens and antigens at mucosal surfaces [
12].
In past decades, studies on
pIgR increased significantly in various vertebrate species, including human, mouse, and fish. Single nucleotide polymorphism studies have found that the human
pIgR gene was significantly associated with IgA nephropathy [
23,
25], and 4 SNPs in the
pIgR gene were susceptible cause for nosopharyngeal cancer [
10]. Moreover, the
pIgR gene had a role in the protection of mycobacterial disease which was demonstrated by mice gene-targeting knockout [
32]. The possible role of
pIgR’s associations with body weight and disease efficiency in fish has been widely investigated by researchers [
20]. Therefore, the analysis of
pIgR subunit polymorphisms is beneficial for understanding the potential variants which affect disease-resistant. However, there are poor studies of
pIgR’s polymorphisms and their related functions in poultry.
The objective of this study is to investigate the pIgR gene’s polymorphisms and elucidate the association between single nucleotide polymorphisms and growth in chickens. Furthermore, in order to clearly reveal the function of the pIgR gene, we also analyzed the associations of its polymorphisms with disease-resistant traits and their significance levels. These polymorphic sites (SNP 3, 9, 12, 15, 16, 19 and 21) could be used to improve breeding method on body weights, shank lengths, and disease-resistant in broiler breeders.
4. Discussion
Polymeric immunoglobulin receptors play an important role in cellular immunity in chickens. It has a critical role in the maintenance of barrier and intestinal homeostasis by transporting polymeric IgA antibodies across intestinal epithelial cells (IECs) into gut secretions [
13,
24]. In this study, the association analysis was evaluated between the
pIgR SNPs and disease resistance traits (NDV, ALV, and IBD) as well as growth traits in chickens.
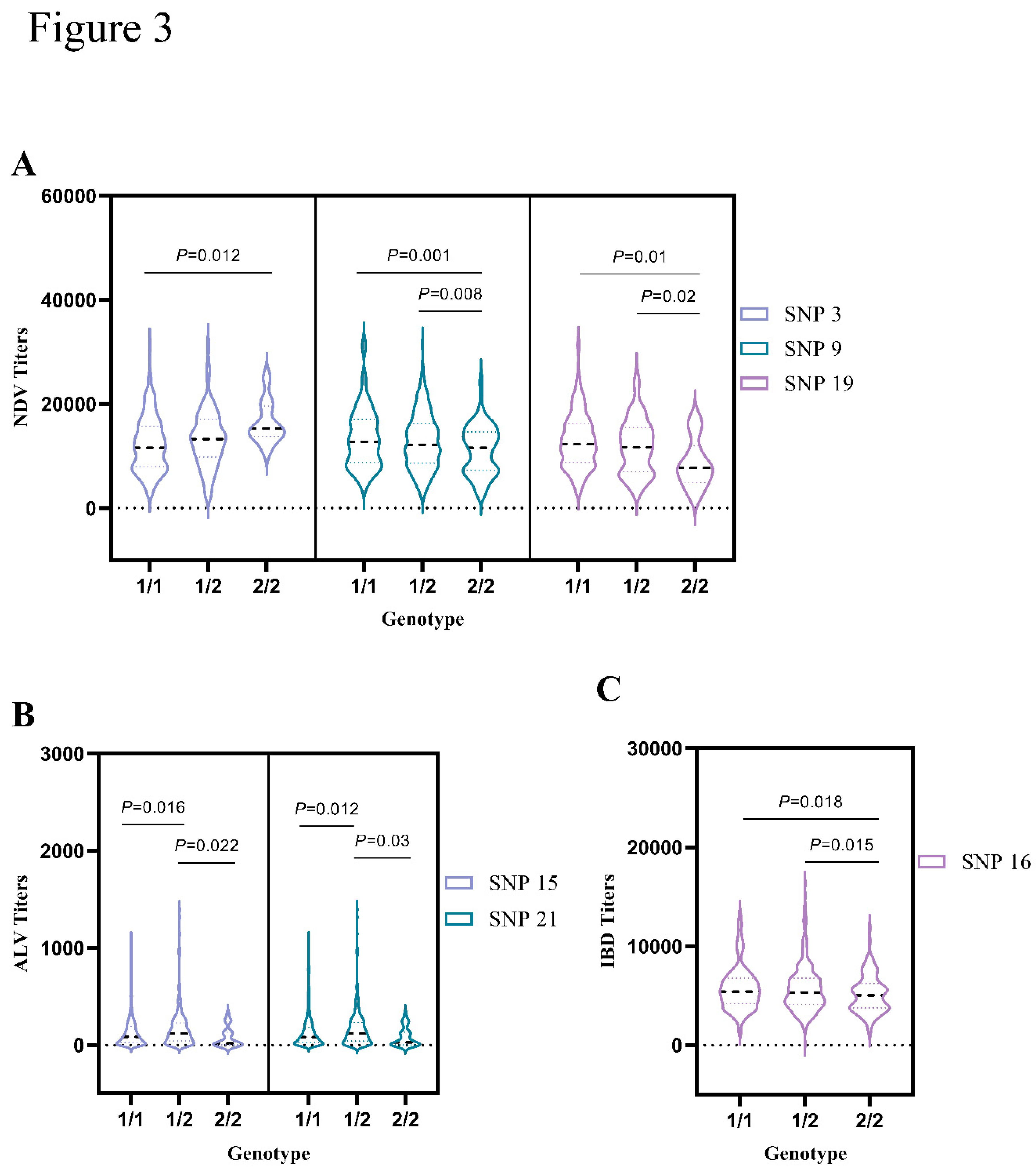
Newcastle disease is caused by a single-stranded RNA virus. In this disease, the viral protein is translated through the transcription of the negative-sense RNA genome. The viral protein of the single-stranded RNA virus known as Newcastle disease. It’s translated by reading the genome of the virus's negative-sense RNA [
19]. It is a highly contagious and infectious disease which occurs frequently in poultry flocks. Historically, the outcome of polymorphisms for chicken has been demonstrated in a number of studies. Major histocompatibility complex (
MHC) class II genetic polymorphisms were found to have substantial effects on NDV titer and body weight for the SNPs 209 and SNP 254 [
21]. Seven genes (TLR7, MX, IFI27L2, SLC5A1, HSPA2, IFRD1, IL1R1) were significantly affected after being exposed to a lentogenic strain of NDV [
28]. The chicken Myxovirus (
Mx) gene promoter’s polymorphisms were demonstrated, which illustrated the SNP4 G > A mutation was associated with chicken embryos’ susceptibility to the virulent NDV challenge [
22]. In this study, we detected a significant association between NDV and
pIgR SNPs (
P < 0.05). Three SNPs, such as SNP 3, SNP 9, and SNP 19, were first demonstrated. The combination of those SNPs may increase the resistant effect on NDV infected chicken. The
pIgR polymorphisms associated with NDV resistance may predispose the host of the group (
TT/AA/CC) to resist preferentially. Thus, NDV resistance related gene has a great potential to affect the immune system and conduct seed selection in chickens.
Avian Leukosis Virus, a single-stranded RNA retrovirus, is composed of six closely related envelope subgroups, which are identified as A, B, C, D, E, and J [
1,
37]. It mostly affects primarily chickens and can not only cause tumors but also immunosuppression, decreased productivity, and other production issues in infected flocks [
8,
9]. Previous studies demonstrated significant associations between three highly pathogenic receptor genes (
NHE1,
tva, and
tvb) and conferring resistance to ALV [
4,
16]. Here, our results also indicated that the resistance of ALV was significantly associated with two SNPs (SNP 15 and 21) in chickens
pIGR gene. The heterozygous (
AG/CT) SNPs mentioned in the previous text have exhibited higher antibody titers in response to the ALV challenge. Thus, provided complementary tools of this gene may reveal the potential of two genotypes with hybridization dominance resistance to ALV in chickens.
Infectious bursal disease (IBD) is an immunosuppressive virus of double-stranded RNA, which primarily not only infects B-cells in the bursa follicles, but also activates T cells and macrophages in peripheral lymphoid organs [
29]. After IBD virus exposure, the levels of total serum and spleen IgA, IgG, and IgM were not affected by the reduction of B cell in the bursa [
26]. From an immunological standpoint, previous studies demonstrated that the
BF2121 alleles of
MHC gene’s exhibition were highly associated with the resistance to IBD infection [
7]. Knockdown of the ribosomal protein L18 (
RPL18) gene by RNA interference was significantly associated with affecting viral replication [
33]. However, for the polymeric immunoglobulin receptor gene, our results also indicated its
C allele had a resistant effect on chicken infected by germs IBD. Its implication will have great potential for progressing animal breeding, and can be used as a diagnostic tool for specificity and sensitivity, and provides effective protection
.
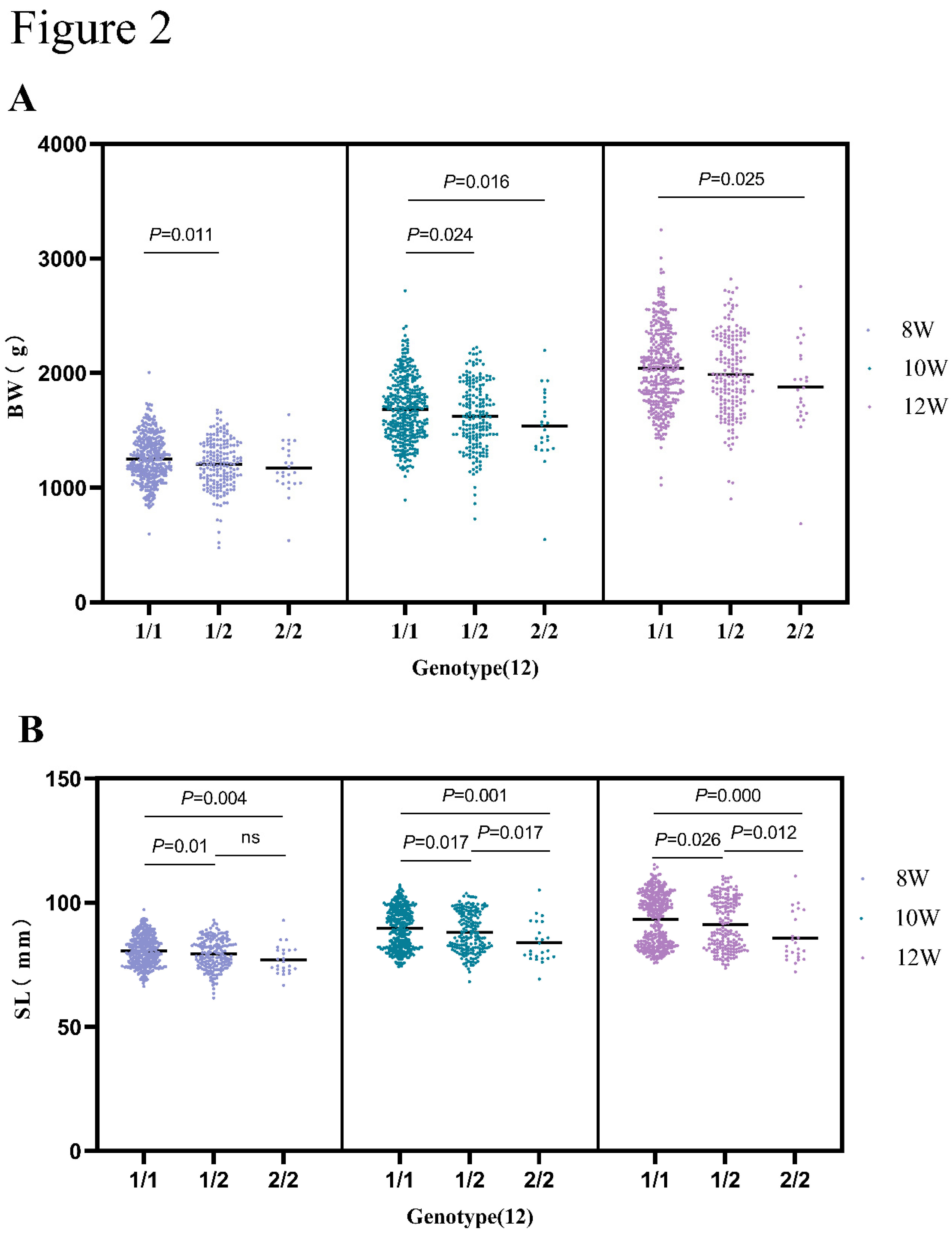
Growth traits are the most essential and economic trait in poultry breeding, especially body weight and shank lengths. Historically, DNA polymorphisms have been widely concerned to affect growth traits [
6,
14,
15]. In Amakusa Daioh cross chickens, significant associations were found between a
CCKAR SNP and growth traits [
31]. Recently, the SNPs of growth-related genes, including
GH,
IGF2, and
TSHB, were associated with traits that influence body size and were potentially involved in bone growth [
36]. It is well known that growth traits are microeffect polygene regulated. By Genome-wide association studies, 113 quantitative trait nucleotides were discovered with significant effects on chicken growth traits, including nucleotides on chromosome 26 [
5]. Interestingly, another gene that is chosen for this study, SNP 12 in the
pIgR gene, was found on chromosome 26 too. Although the effect of
pIgR on growth in chickens remains unclear, our findings discovered new associated SNPs and enriched the information on existing SNPs. Moreover, our results indicated
pIgR could be used as a genetic marker. By doing that, new polymorphisms could integrate into the breeding program to increase growth performance and the disease-resistance of populations.
This variant may alter the efficiency of pIgR to release IgA complex and consequently increase the disease resistance for the chicken population. This might supplement antibody-mediated defense because pIgR polymorphisms may affect disease occurrence by modifying IgA immune selection. There was a new and valuable discovery about associations of the pIgR gene polymorphisms with disease resistance traits in chickens. However, we assessed that there was a statistical correlation between inhere five viruses' genetic resistance and other economic growth traits. In further studies, the relevant genetic functions of these SNPs are required to confirm by gene editing technology of poultry. In addition, the majority need is to verify the mechanism of chicken pIgR genes related to disease-resistant and economic traits in large populations with different chicken lines. The trade-off issue between disease resistance and growth performance could be resolved with a fuller understanding of this inheritance mechanism. Achieving this genetic equilibrium gets us breeds with satisfactory commercial traits in both disease resistance and growth.