Submitted:
18 August 2023
Posted:
21 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
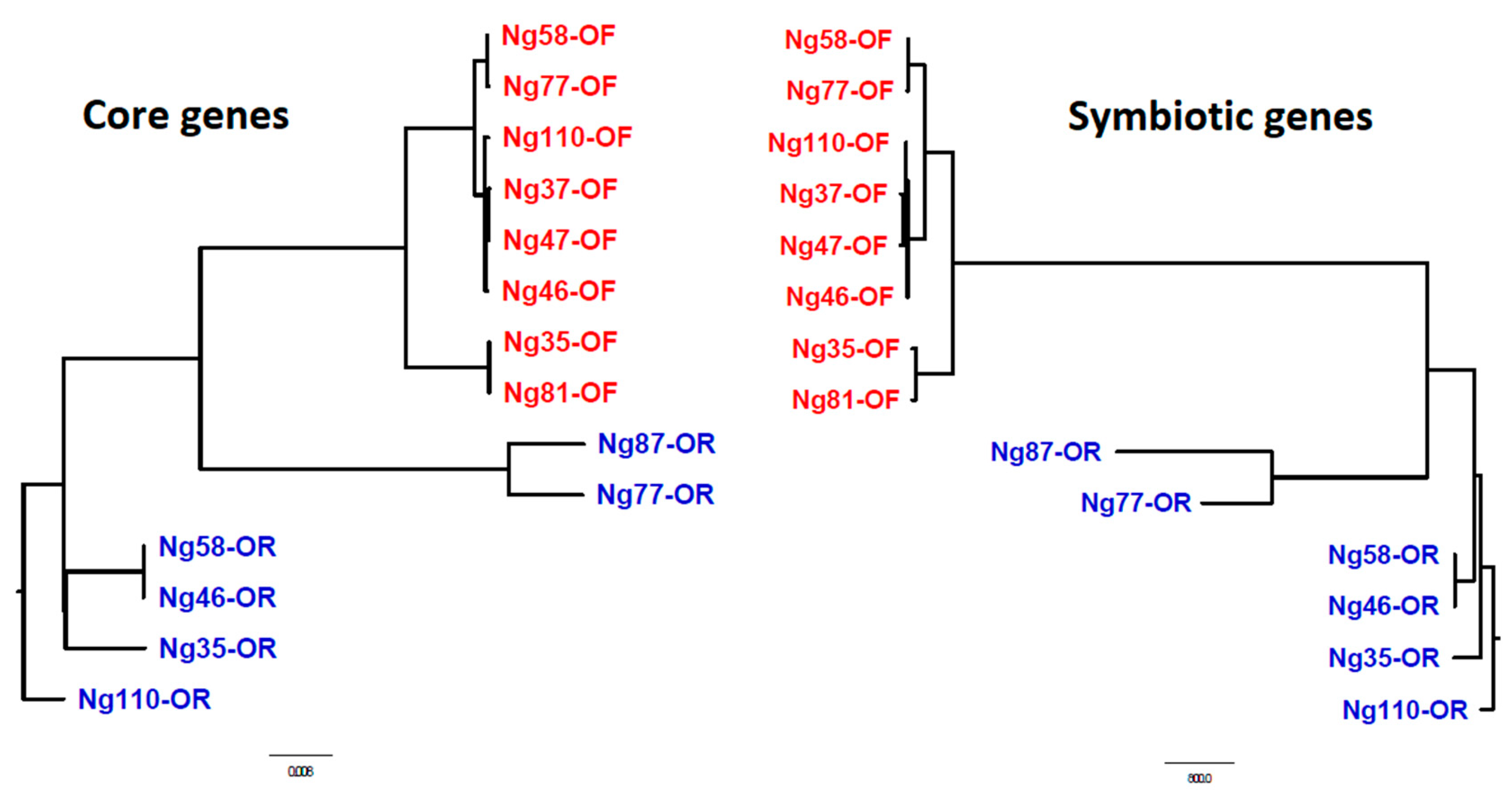
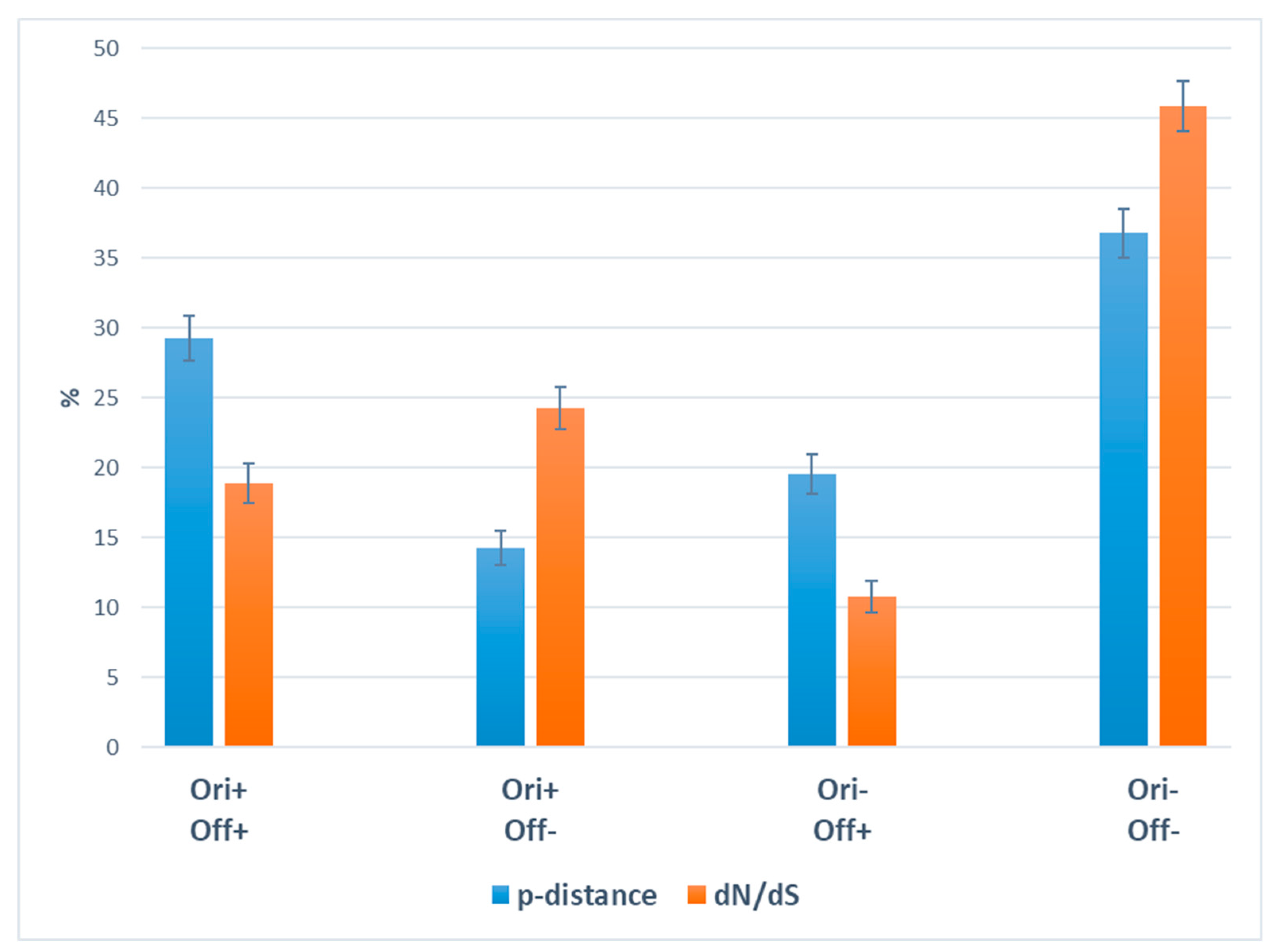
2.1. Gene polymorphism and natural selection
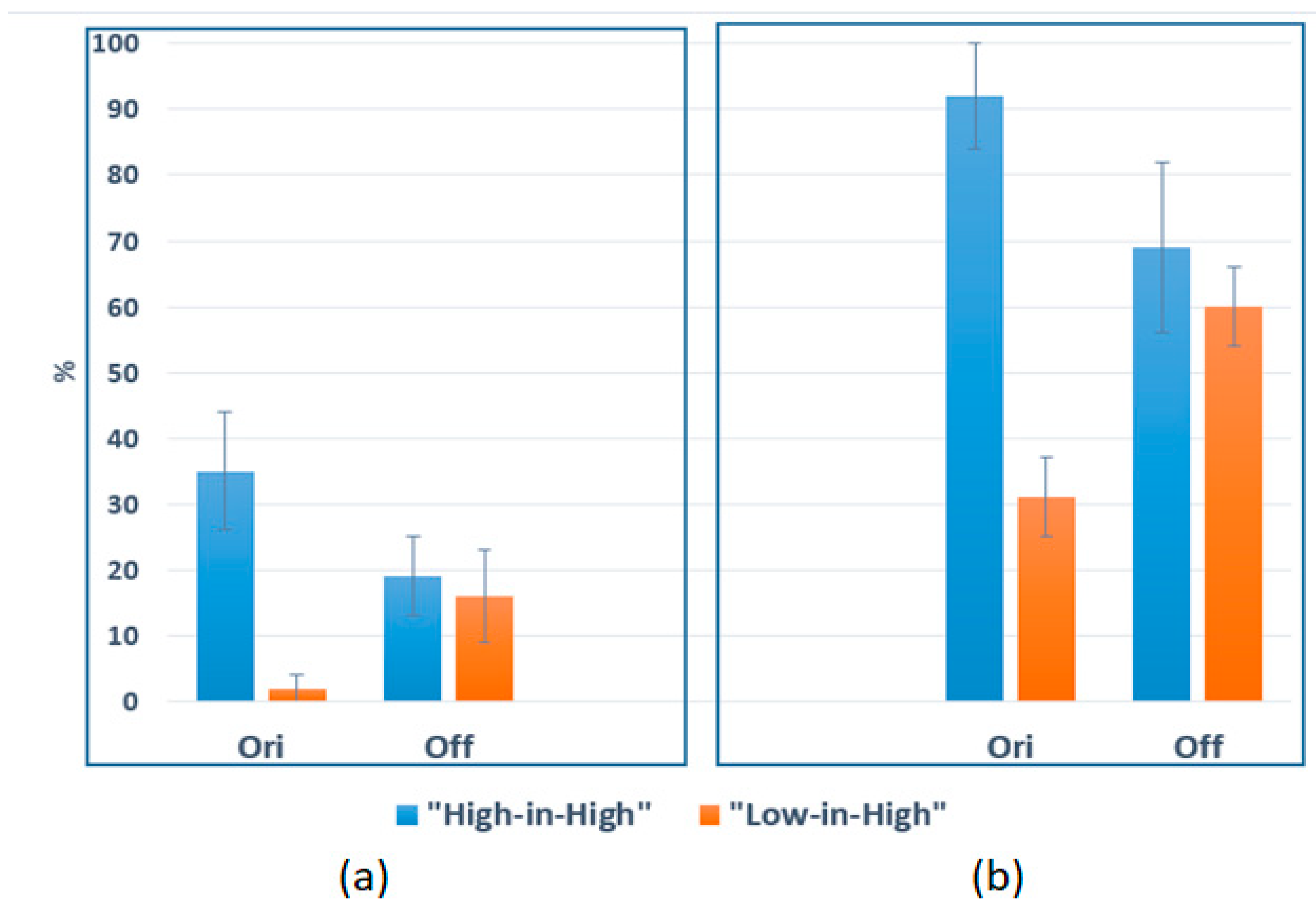
2.2. Gene ontology analysis
3. Discussion
4. Materials and Methods
4.1. Collection of strains and DNA sequencing
4.2. Finding core genes by global alignment
4.3. Symbiotic genes
4.4. Gene alignment using Muscle
4.5. Calculation of nucleotide polymorphism (p-distance)
4.6. Calculation of dN/dS index
4.7. Functional annotation of core genes, Gene Ontology (GO)
4.8. Determination of the predominance of functional groups of genes GO (gene Ontology) and statistical significance
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, P.W., Crossman, L.C., Johnston, A.WB., Thomson, N.R., Ghazoui, Z.F., Hull, K.H., Wexler, M., Curson, A., Todd, J.D., Poole, P.S., Mauchline, T.H., East, A.K., Quail, M.A., Churcher, C., Arrowsmith, C., Cherevach, I., Chillingworth, T., Clarke, K., Cronin, A., Davis, P., Fraser,A., Hance, Z., Hauser, H., Jagels, K., Moule, S., Mungall, K., Norbertczak, H., Rabbinowitsch, E., Sanders, M., Simmonds, M., Whitehead, S., Parkhill J. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol. 2006, 7, R34. [CrossRef]
- Provorov, N.A., Andronov, E.E., Kimeklis, A.K., Onishchuk, O.P., Igolkina, A.A., Karasev, E.S. Microevolution, speciation and macroevolution in rhizobia: genomic mechanisms and selective patterns. Front. Plant Sci. 2022, 13, 1026943. [CrossRef]
- Kimeklis, A., Chirak, E., Kuznetsova, I., Sazanova, A., Safronova, V., Belimov, A., Onishchuk, O., Kurchak, O., Aksenova, T., Pinaev, A., Andronov, E.E., Provorov N.A. Rhizobia Isolated from the Relict Legume Vavilovia formosa Represent a Genetically Specific Group within Rhizobium leguminosarum biovar viciae. Genes, 2019, 10, 991. [CrossRef]
- Onischuk O.P., Kurchak O.N., Kimeklis A.K., Aksenova T.S., Andronov E.E., Provorov N.A. Biodiversity of the symbiotic systems formed by nodule bacteria Rhizobium leguminosarum with the leguminous plants of galegoidcomplex. Sel’skokhozyaistvennaya Biologiya (Agricultural Biology) 2023, 58, 87–99. [CrossRef]
- Provorov, N.A., Andronov, E.E., Onishchuk, O.P. Forms of natural selection controlling the genomic evolution in nodule bacteria. Rus. J. Genet. 2017, 53, 411–419. [CrossRef]
- Österman, J., Marsh, J., Laine, P.K., Zeng, Z., Alatalo, E., Sullivan, J.T., Young, P.W., Thomas-Oates, J., Paulin, L., Lindström K. Genome sequencing of two Neorhizobium galegae strains reveals a noeT gene responsible for the unusual acetylation of the nodulation factors. BMC Genomics. 2014, 15, 500. [CrossRef]
- Karasev, E.S., Andronov, E.E., Aksenova, T.S., Tupikin, A.E., Provorov, N.A. Evolution of goat’s rue rhizobia (Neorhizobium galegae): an analysis of the polymorphism of the nitrogen fixation genes and the genes of nodule formation. Russ. J. Genetics. 2019, 55, 234–238. [CrossRef]
- Provorov, N.A., Andronov, E.E., Kimeklis, A.K., Onishchuk, O.P., Igolkina, A.A., Karasev, E.S. Microevolution, speciation and macroevolution in rhizobia: genomic mechanisms and selective patterns. Front. Plant Sci. 2022, 13, 1026943. [CrossRef]
- Xin, Z., Cai, Y., Dang, L.T. Burke H.M.S., Revote J., Charitakis N., Bienroth D., Nim H.T., Li H.Y., Ramialison M. MonaGO: a novel gene ontology enrichment analysis visualisation system. BMC Bioinformatics, 2022, 23, 69. [CrossRef]
- Andrews, C.A. Natural Selection, Genetic Drift, and Gene Flow Do Not Act in Isolation in Natural Populations. Nature Education Knowledge 2010, 3, 5. [Google Scholar]
- Cheng, C., Kirkpatrick, M. Molecular evolution and the decline of purifying selection with age. Nat Commun 2021, 12, 2657. [CrossRef] [PubMed]
- Lee C.-R., Mitchell-Olds T. Environmental Adaptation Contributes to Gene Polymorphism across the Arabidopsis thaliana. Genome, Molecular Biology and Evolution 2012, 29, 3721–3728. [CrossRef] [PubMed]
- Marchinko, K.B., Matthews, B., Arnegard, M.E., Rogers, S.M., Schluter, D. Maintenance of a Genetic Polymorphism with Disruptive Natural Selection in Stickleback. Current Biology 2014, 2014 24, 1289–1292. [CrossRef]
- Rahman, S., Kosakovsky, P.S.L., Webb, A., He,y J. Weak selection on synonymous codons substantially inflates dN/dS estimates in bacteria. PNAS 2021, 118, e2023575118. [CrossRef] [PubMed]
- Taub, D.R., Page, J. Molecular Signatures of Natural Selection for Polymorphic Genes of the Human Dopaminergic and Serotonergic Systems: A Review. Front Psychol 2016, 8, 857. [CrossRef]
- Moon, S.U., Na, B.K., Kang, J.M., Kim, J.Y., Cho, S..H, Park, Y.K., Sohn, W.M., Lin, K., Kim, T.S. Genetic polymorphism and effect of natural selection at domain I of apical membrane antigen-1 (AMA-1) in Plasmodium vivax isolates from Myanmar. Acta Trop 2010, 114, 71–75. [CrossRef]
- Kang, JM., Ju, HL., Kang, YM. Genetic polymorphism and natural selection in the C-terminal 42 kDa region of merozoite surface protein-1 among Plasmodium vivax Korean isolates. Malar J 2012, 11, 206. [CrossRef]
- Barnard-Kubow, K., Sloan, D., Galloway, L. Correlation between sequence divergence and polymorphism reveals similar evolutionary mechanisms acting across multiple timescales in a rapidly evolving plastid genome. BMC evolutionary biology 2014, 14, 1. [CrossRef]
- Vigué, L, Eyre-Walker, A. The comparative population genetics of Neisseria meningitidis and Neisseria gonorrhoeae. PeerJ 2019, 27, e7216. [Google Scholar] [CrossRef]
- Sunyaev, S., Kondrashov, F.A., Bork, P., Ramensky, V. Impact of selection, mutation rate and genetic drift on human genetic variation. Human Molecular Genetics 2003, 12, 3325–3330. [CrossRef]
- Raig, H., Nõmmsalu, H., Meripõld, H., Metlitskaja, J. Estonian Research Institute of Agriculture: Saku, Estonia, Nõmmsalu, F.G.H., Ed.; 2001; 141p.
- Andronov, E., Terefework, Z., Roumiantseva, M., Dzyubenko, N., Onichtchouk, O., Kurchak, O., Dresler-Nurmi, A., Young, J. P., Simarov, B., Lindstrom. Symbiotic and Genetic Diversity of Rhizobium galegae Isolates Collected from the Galega orientalis Gene Center in the Caucasus. Applied and Environmental Microbiology 2003, 69, 1067–1074. [CrossRef]
- Österman, J., Chizhevskaya, E., Andronov, E., Fever, D., Terefework, Z., Roumiantseva, M., Onichuk, O., Dresler-Nurumi, A., Simarov, B., Dzybenko, N., Lindstrom, K. Galega orientalis is more diverse than Galega officinalis in Caucasus—whole-genome AFLP analysis and phylogenetics of symbiosis-related genes. Mol. Ecol. 2011, 20, 4808–21. [CrossRef]
- Radeva, G., Jurgens, G., Niemi, M., Nick, G., Suominen, L., Lindström, K. Description of two biovars in the Rhizobium galegae species: biovar orientalis and biovar officinalis. System. Appl. Microbiol. 2001, 24, 192–205. [CrossRef]
- Philiptschenko, J. Varriabilität und Variation; Berlin Bornträger: Berlin, Germany, 1927. [Google Scholar]
- Goldschmidt, R. B. Phenocopies. Scientific American 1949, 181, 46–49. [Google Scholar] [CrossRef]
- Goldschmidt, R. B. The intersexual males of the beaded minute combination in Drosophila melanogaster. PNAS 1949, 35, 314–316. [Google Scholar] [CrossRef]
- Gould, S.J. Wonderful Life: The Burgess Shale and the Nature of History; W.W. Norton & Company: New York, 1989. [Google Scholar]
- Gould, S.J. & Eldredge, N. Punctuated equilibrium comes of age. Nature 1993, 366, 223–227. [CrossRef]
- Theis, K. R., Dheilly, N. M., Klassen, J. L., Brucker, R. M., Baines, J. F., Bosch, T. C., et al.. Getting the hologenome concept right: an eco-evolutionary framework for hosts and their microbiomes. mSystems. 2016, 1, e00028–e00016. [CrossRef]
- Novikova, N., Safronova, V. Transconjugants of Agrobacterium radiobacter harbouring sym genes of Rhizobium galegae can form an effective symbiosis with Medicago sativa. FEMS Microbiol. Lett. 1992, 93, 261–268. [CrossRef]
- Allen, O.N. Experiments in Soil Bacteriology; Burgess Publishing, Co.: Minneapolis, Minnesota, 1959; pp. 52–59. [Google Scholar]
- Somasegaran, P., Hoben, H.J. Isolating and Purifying Genomic DNA of Rhizobia Using a Large-Scale Method. In Handbook for Rhizobia; Garber, R.C., Ed.; Springer: New York, NY, USA, 1994; pp. 279–283. [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
| Genes* | Means ± standard errors | ||
|---|---|---|---|
| bv. orientalis | bv. officinalis | tSt (P0) | |
| p-distance | |||
| core | 0.048±0.001 | 0.010±0.001 | 106.4 (< 0.001) |
| sym | 0.028±0.008 | 0.005±0.001 | 2.84 (< 0.05) |
| tSt (P0) | 2.47 (< 0,05) | 3.57 (< 0,01) | - |
| dN/dS** | |||
| core | 1.571±0.050 (D) | 1.013±0.026 (N) | 9.91 (< 0.001) |
| sym | 1.009±0.142 (N) | 0.272±0.111 (P) | 4.09 (< 0.001) |
| tSt (P0) | 3.72 (< 0,01) | 6,50 (< 0.001) | - |
| Genes | Pearson correlation coefficients (r)* | tSt (P0) | |
|---|---|---|---|
| bv. orientalis | bv. officinalis | ||
| core | + 0,346 (P0 < 0,001) | + 0,066 (0,05 < P0 < 0,10) | 12,73 (< 0,001) |
| sym** | + 0,078 (P0 > 0,10) | – 0,991 (0,05 < P0 < 0,10) | 4,18 (< 0,001) |
| tSt (P0) | 0,99 (> 0,05) | 50,03 (< 0,001) | - |
| Numbers of GOGs in clusters contrasting for p-distance* | ||||||
|---|---|---|---|---|---|---|
| CP-I (229) | CP-II (153) |
CP-III (112) | CP-IV (288) |
Total GOGs | ||
| The same in clusters for dN/dS* | CS-I (148) | 3 | 0 | 0 | 0 | 3 |
| CS-II (190) | 5 | 3 | 1 | 0 | 9 | |
| CS-III (85) | 4 | 0 | 2 | 4 | 10 | |
| CS-IV (359) | 12 | 4 | 20 | 18 | 54 | |
| Total GOGs | 24 | 7 | 23 | 22 | 76 | |
| Items | Neorhizobium galegae* | Rhizobium leguminosarum** |
|---|---|---|
| Compared biovars (their hosts) | bv. orientalis (Galega orientalis), bv. officinalis (G. officinalis) | bv. viciae (Lathyrus, Lens, Pisum, Vavilovia, Vicia), bv. trifolii (Trifolium) |
| Taxonomic diversity of hosts of the compared biovars | Different species of the same plant genus | Various plant genera and tribes |
| Replicons harboring sym genes | Chromids (> 1600 kb) | Plasmids (200-500 kb) |
| Phylogenetic congruence of core and sym genes | High or complete | Incomplete or absents |
| Differences between biovars for the diversity parameters of sym and core genes | Highly significant for both gene groups | For sym genes are more pronounced than for core genes |
| Variation within biovars: for core genes for sym genes |
highly significant significant but lower than for core genes |
highly significant much lower than for core genes or is not revealed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





