Introduction
Known as the Oriental ratsnake (
Figure 1)[
1], Indian ratsnake or Dhaman;
Ptyas mucosa is a common non-venomous species of colubrid snakes. There are over 300 genera and 2000 species in the colubrid family, making it the largest snake family [
2]. The ratsnake, is an excitable and fast-moving snake, but it is harmless to humans, preying upon small reptiles, birds, and mammals. Therefore, in some areas, farmers will obtain and move the Oriental ratsnake from other locations to catch mice and protect their crops. Adult snakes usually prefer to subdue their prey by sitting on it instead of constricting, using their weight to suppress the prey, which is a hunting mechanism of hunting uncommon in other snake species [
3]. When they are threatened, they will inflate their necks, which can be used to imitate the king cobra or Indian cobra to scare off potential predators [
4].
In southern China, the Indian rat snake is commonly eaten by humans, and its skin is used for making the membranes of a traditional musical instrument, the erhu [
5]. In traditional Chinese medicine, its gallbladder is used make a medicinal wine to treat many diseases [
6]. In the past, due to over hunting, its numbers were greatly reduced, but with the success of artificial breeding, their numbers have gradually recovered [
6].
In this study, we present a highly continuous P. mucosa genome with a genome size of 1.74Gb by using single-tube long fragment reads (stLFR) sequencing data and combined with whole genome sequencing data for correction. Its repeat content reached 42.19%. These provide an important basis for follow-up studies elucidating the biology of P. mucosa. Taking this further high-quality reference genome and transcriptome data can provide effective help for subsequent targeted breeding.
Main content
Context
In this study we present a highly-continuous genome assembly of
P. mucosa, finding the maximum genome size of
P. mucosa is 1.74Gb. The scaffold N50 length is 9.57Mb and the maximal length of scaffold is 78.3Mb (
Table 1). Furthermore, the
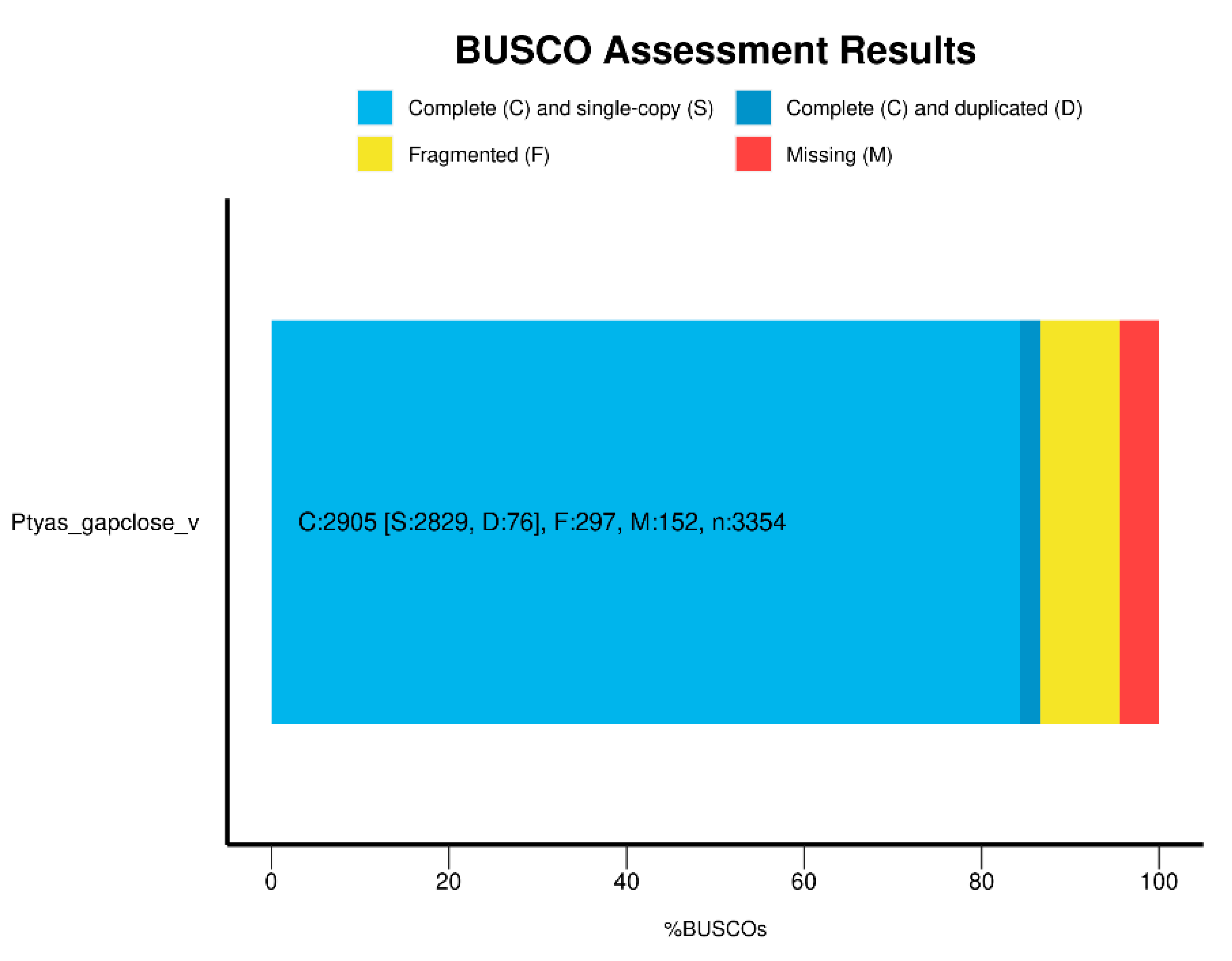
P. mucosa genome has a CG content of 37.9% and using BUSCO (
Figure 2) to measure its integrity reached 86.6%. Thus we can see from these genome assembly data that this reference is a highly contiguous genome. Here we report the draft reference genome sequence of
P. mucosa. This data will provide a valuable resource in the study of nonpoisonous snakes.
Methods
Detailed stepwise protocols are gathered in a protocols.io collection with some minor adaptations outlined below [
7] (
Figure 3).
Sample collection and sequencing
In 2021 an adult P. mucosa (NCBI:txid31142) individual from Hezhou City in Guangxi province was collected for genome assembly and RNA sequencing. The individual died of natural causes and the samples were transferred to dry ice and quickly frozen, then kept at -80℃ until further use. We isolated 8 tissues and organs for RNA sequencing, including the heart, small intestine, large intestine, lung, liver, stomach, kidney and muscle. Furthermore, genomic DNA was extracted for whole-genome sequencing utilizing the AxyPrep genomic DNA kit (AxyPrep, USA).
The total RNA was isolated utilizing the TRlzol reagent (Invitrogen, USA) following the recommended guidelines. The assessment of RNA quality, purity, and quantity was performed using the Qubit 3.0 fluorometer (Life Technologies, USA) and the Agilent 2100 Bioanalyzer System (Agilent, USA). The cDNA libraries were generated through the reverse transcription of RNA fragments ranging from 200 to 400 bp. In addition, the liver sample was used for single-tube long fragment read (stLFR) sequencing and genome survey which it refers to the means of analyzing the second generation sequencing data through k-mer to obtain genome size, heterozygosity, repeat sequence proportion, GC-content and other genomic information.
Genome survey, assembly, annotation and assessment
The stLFR sequencing data were assembled with Supernova software (v2.1.1) [
8]. NextPolish (v1.0.5) [
9] program was then used to carry out a second round of correction and third round of polishing for this assembly by using the WGS data. To get a haploid representation of the genome, duplicates were purged with Purge_Dups pipeline [
8] from the genome. The completeness of the genome was evaluated using sets of Benchmarking Universal Single-Copy Orthologs (BUSCO v5.2.2) with genome mode and lineage data from vertebrata_odb10 [
10].
In order to detect the presence of known repeat elements in the genome of the many-banded P. mucosa, , the following approach was employed Repeat Finder (TRF) [
11], LTR_FINDER (RRID:SCR_015247) [
12] and RepeatModeler (v2.0.1,RRID:SCR_015027) (v1.0.8) [
13]. RepeatMasker (v3.3.0, RRID:SCR_012954) [
14] and RepeatProteinMask v3.3.0 [
15] were used to search the genome sequences for known repeat elements. BRAKER2 pipeline[
16] was used to perform gene prediction. Then the gene sets were aligned against several known databases, including SwissProt[
17], TrEMBL[
17], Kyoto encyclopedia of genes and genomes (KEGG)[
18], GO and NR [
19] database.
Results
In
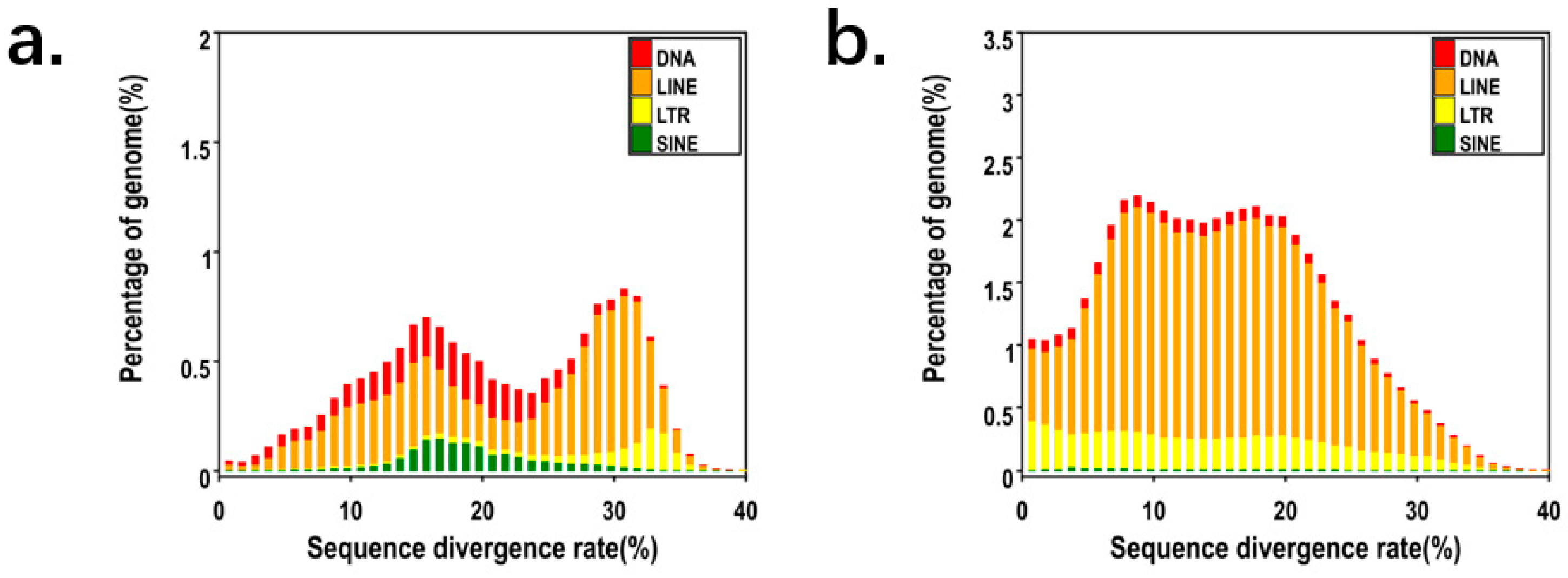
P. mucosa, the total length of the repeat sequence in the genome reached 735Mb, and its repeat content is as high as 42.16% (
Table 2 and
Table 3). We analysed the content of various repetitive elements, and several different genome families were identified within the
P. mucosa genome. We found that LINE repeat elements accounted for 35.51%, while LTR accounted for 9.15%, and DNA accounted for 4.66% (
Figure 4). Long interspersed nuclear elements were the most numerous of these repeats. Research findings suggest that despite snake species sharing similar genome sizes, they demonstrate considerable variations in TE content, with limited diversity in the types of TEs. Species with a longer evolutionary history tend to exhibit greater diversity in TE content, as indicated by research findings.
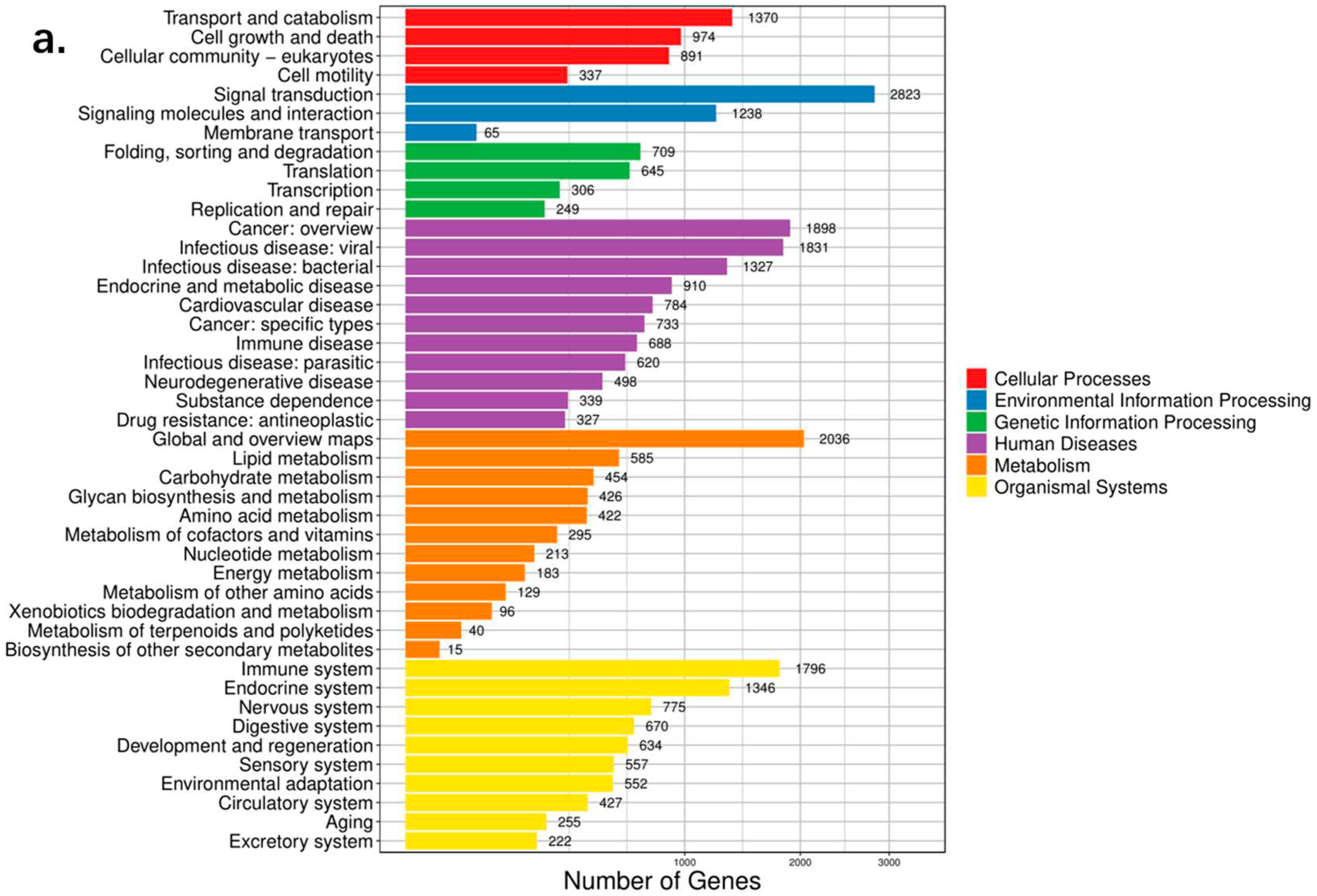
A total of 24,869 functional genes were annotated using KEGG. This showed the highest number of annotated genes in pathways related to Human Diseases, Organismal Systems and Metabolism, and the highest number of Signal Transduction genes were in Environmental Information Processing. Moreover, GO gene enrichment for
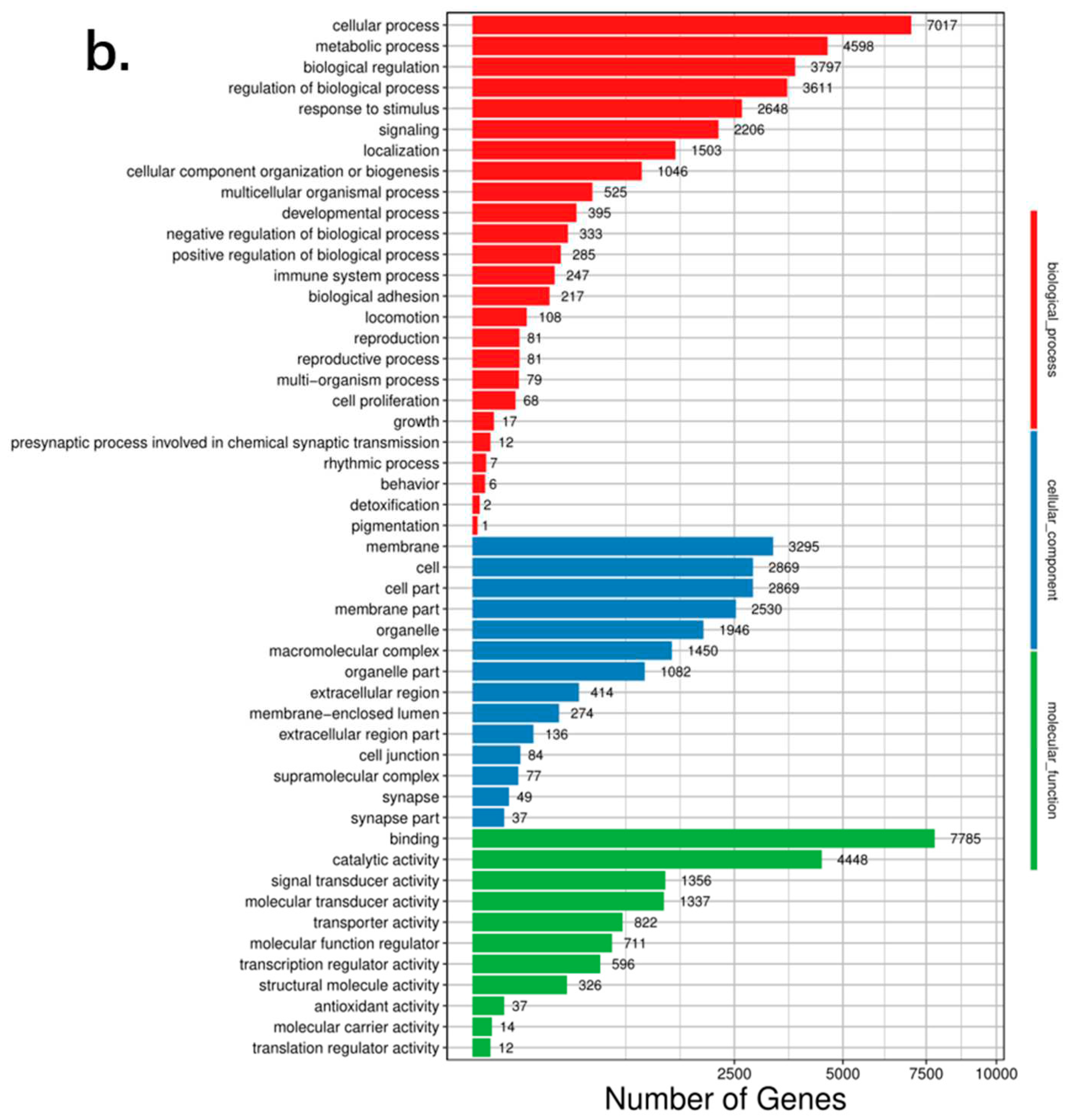
P. mucosa revealed that, among 25 biological process pathways, 247 genes were related to immune system processes, and 2 genes were related to detoxification (
Figure 5)
Reuse Potential
P. mucosa is a species of snake belonging to the species rich Colubrid family. Therefore, assembling the genome of the P. mucosa genome helps to understand the development process and origin of the Colubrids. Alongside this, as an economically important species, understanding the genome of the P. mucosa can potentially helpful for the breeding and breeding of the mouse snake in the future and can provide guidance for its breeding.
Author Contributions
Tianming Lan designed and initiated the project. Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences collected the samples. Jiangang Wang performed the DNA extraction, library construction and data analysis. Jiangang Wang wrote the manuscript. All authors read and approved the final manuscript.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0004141. Raw reads are in the SRA [accession] and additional data is in the GigaDB repository[
20].
Acknowledgments
Our project was financially supported by the Guangdong Provincial Key Laboratory of Genome Read and Write (grant no. 2017B030301011). This work was also supported by China National GeneBank (CNGB).
Conflicts of Interest
The authors declare no conflict financial interests.
References
- 2002. A Photographic Guide to Snakes and Other Reptiles of India. Ralph Curtis Books. Sanibel Island, Florida. 144 pp. ISBN 0-88359-056-5.
- Colubridae. Science Direct. https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/colubridae.
- "Ptyas mucosa - Dhaman (Oriental) Ratsnake". Snakesoftaiwan.com. Retrieved 25 November 2021.
- Young, B.A., Solomon, J., Abishahin, G. 1999. "How many ways can a snake growl? The morphology of sound production in Ptyas mucosus and its potential mimicry of Ophiophagus". Herpetological Journal 9 (3):89–94.
- 滑鼠蛇 Ptyas mucosus -专题库 国家动物标本资源共享平台[引用日期2022-12-09]. 2022.
- Wang Zhang,Mingxing Hu,Qunying Tan,PeiPeng Li,Zhenghong Qin. Investigation and suggestions for the development of the pharmaceutical farm-raised snake industry [J]. 蛇志, 2021, 33 (04): 369-374.
- Liu B, Cui L, Deng Z, Ma Y, Yang D, Gong Y, et al. Protocols for the assembly and annotation of snake genomes. 2023. Protocols.io. [CrossRef]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y and Durbin R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36 9:2896-8. [CrossRef]
- Guan D, McCarthy SA, Wood J, Howe K, Wang Y and Durbin R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36 9:2896-8. [CrossRef]
- Wick RR, Holt KE. Benchmarking of long-read assemblers for prokaryote whole genome sequencing.F1000Research, 2019; 8: 2138. [CrossRef]
- Benson and G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 27 2:573-80. [CrossRef]
- Zhao X and Hao W. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Research. 2007; suppl_2:suppl_2. [CrossRef]
- Smit A, Hubley R and Green P. RepeatModeler Open-1.0. 2008–2015. Seattle, USA: Institute for Systems Biology Available from: httpwww repeatmasker org, Last Accessed May. 2015;1:2018.
- Tarailo-Graovac M and Chen N. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Current protocols in bioinformatics / editoral board, Andreas D Baxevanis [et al]. 2009;Chapter 4 Unit 4:Unit 4.10. [CrossRef]
- Tempel S. Using and understanding RepeatMasker. Mobile Genetic Elements. Springer; 2012. p. 29-51.
- Bruna T, Hoff KJ, Lomsadze A, Stanke M and Borodovsky M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics and Bioinformatics. 2021;3 1:lqaa108. [CrossRef]
- Amos B and Rolf A. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research. 2000; 1:45. [CrossRef]
- Pitk E. KEGG database. Novartis Foundation Symposium. 2006;247:91-103.
- Jian Z. Species-based distribution of BLASTX matches for unigenes against NCBI NR database. 2015.
- Insert GigaDB dataset DOI here when completed.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).