Submitted:
23 August 2023
Posted:
23 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Clinical Specimen Collection
2.2. R.N.A Extraction and cDNA Synthesis
2.3. Reverse Transcriptase Polymerase Reaction(RT-PCR)
2.4. Virus Recovery and Propagation
2.5. Sequencing and sequence data Analysis of selected isolates
2.6. Development of Indirect ELISA (IELISA)
2.8. Initial Validation and Data Analysis of both Diagnostic Assays
2.9. Commercial ELISA (c-ELISA) Test
2.10. Virus Neutralization Test (V.N.T.)
2.11. Ethical considerations
3. Results
3.1. Specimen Collection
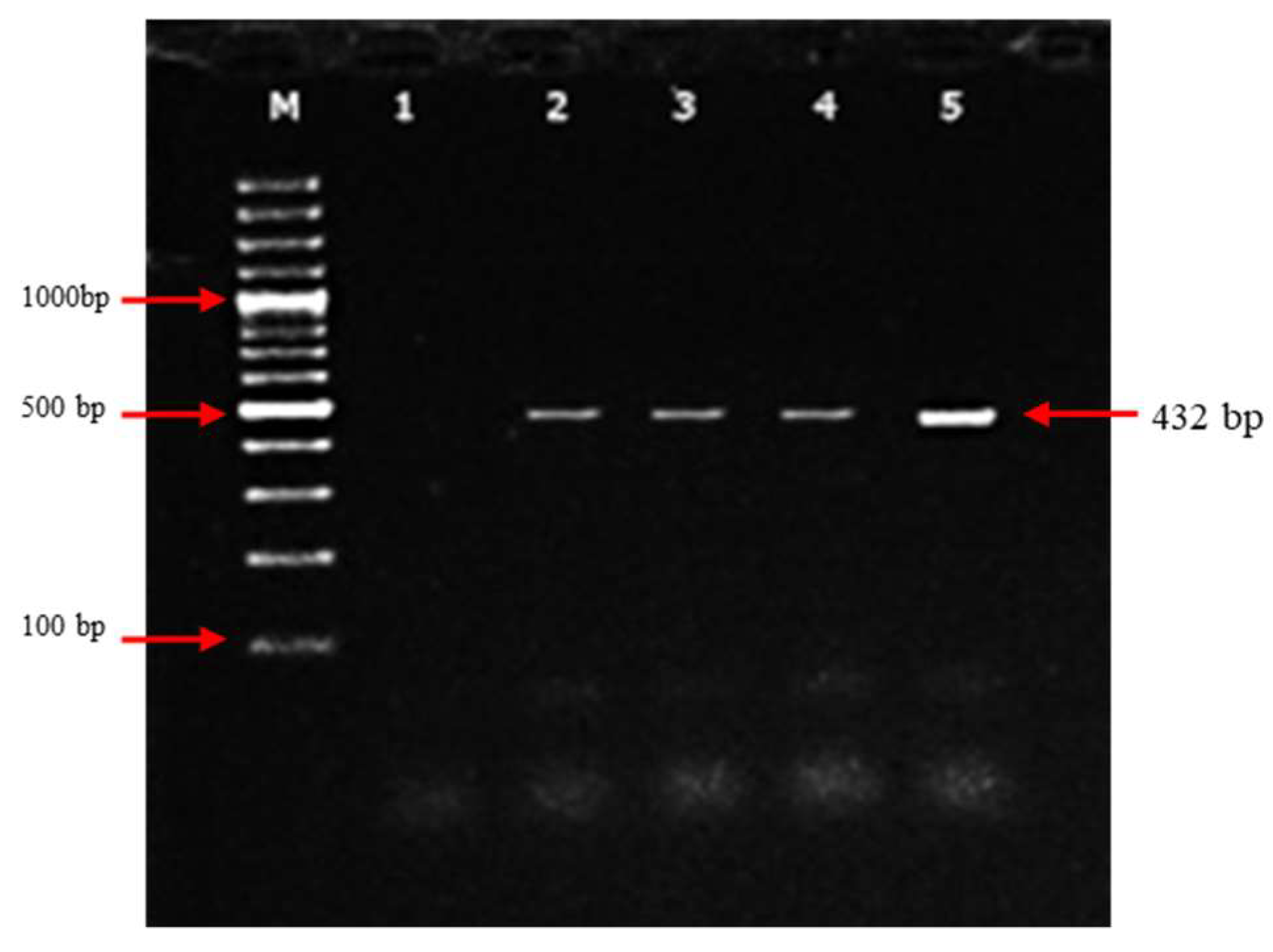
3.2. Molecular detection of PPRV based on N-genes by RT-PCR
3.3. Phylogenetic Analysis of Indigenous Peste des Petits Ruminants Isolates
3.4. PPRV Recovery and propagation:
| Isolate names | Different cytopathic effects on cells | |||||
|---|---|---|---|---|---|---|
| >Cell rounding | >Clumping of cells | >Syncytia formation | >Detachment from surface | >Cells with elongated processes | >Vacuolation | |
| ABP-NARC/Islamabad-2020/01 (Gilgit) | + | + | _ | + | + | _ |
| ABP-NARC/Islamabad-2020/02 (Fateh Jang) | + | _ | + | + | _ | + |
| ABP-NARC/Islamabad-2020/03 (Islamabad) | + | + | + | + | _ | _ |

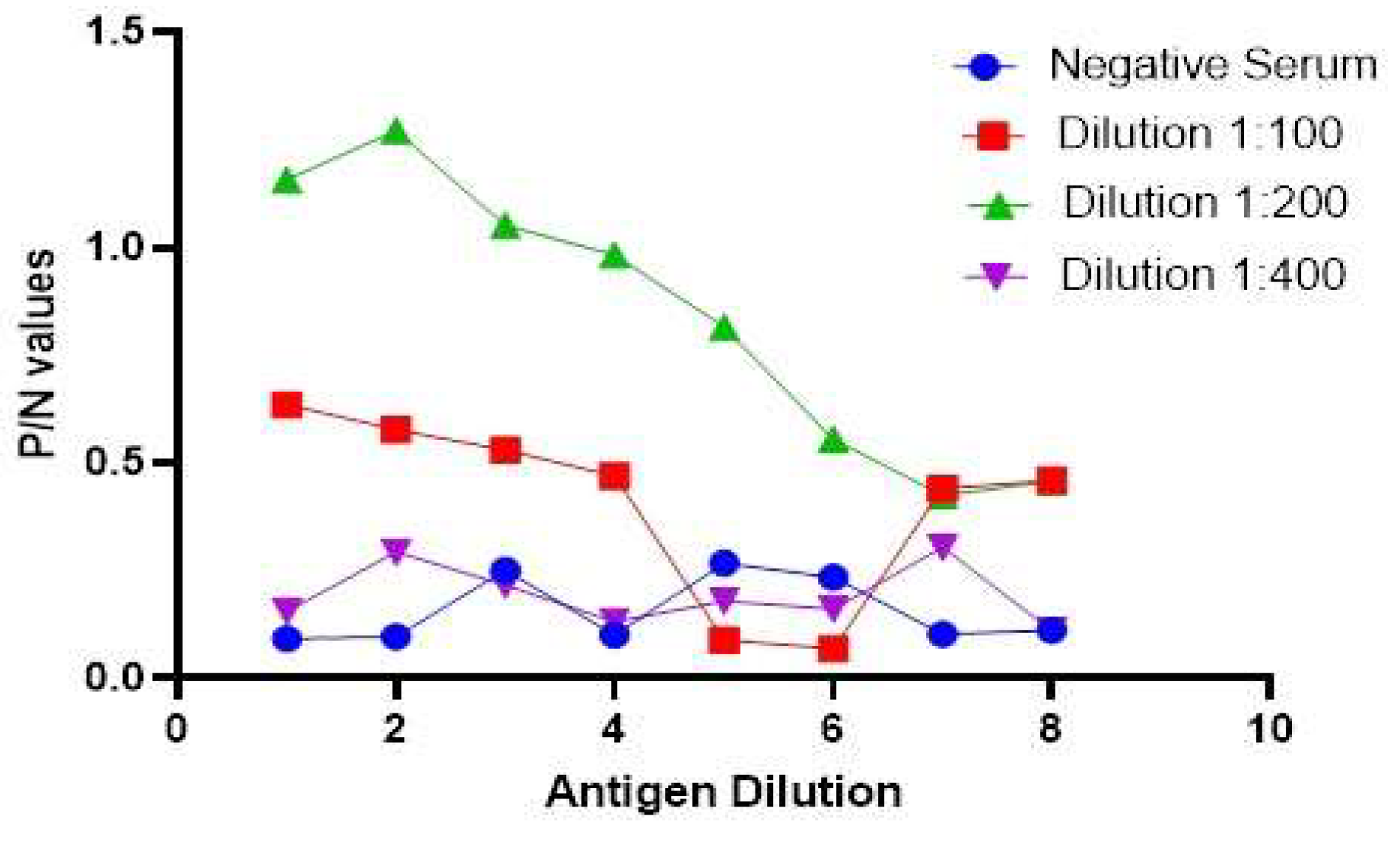
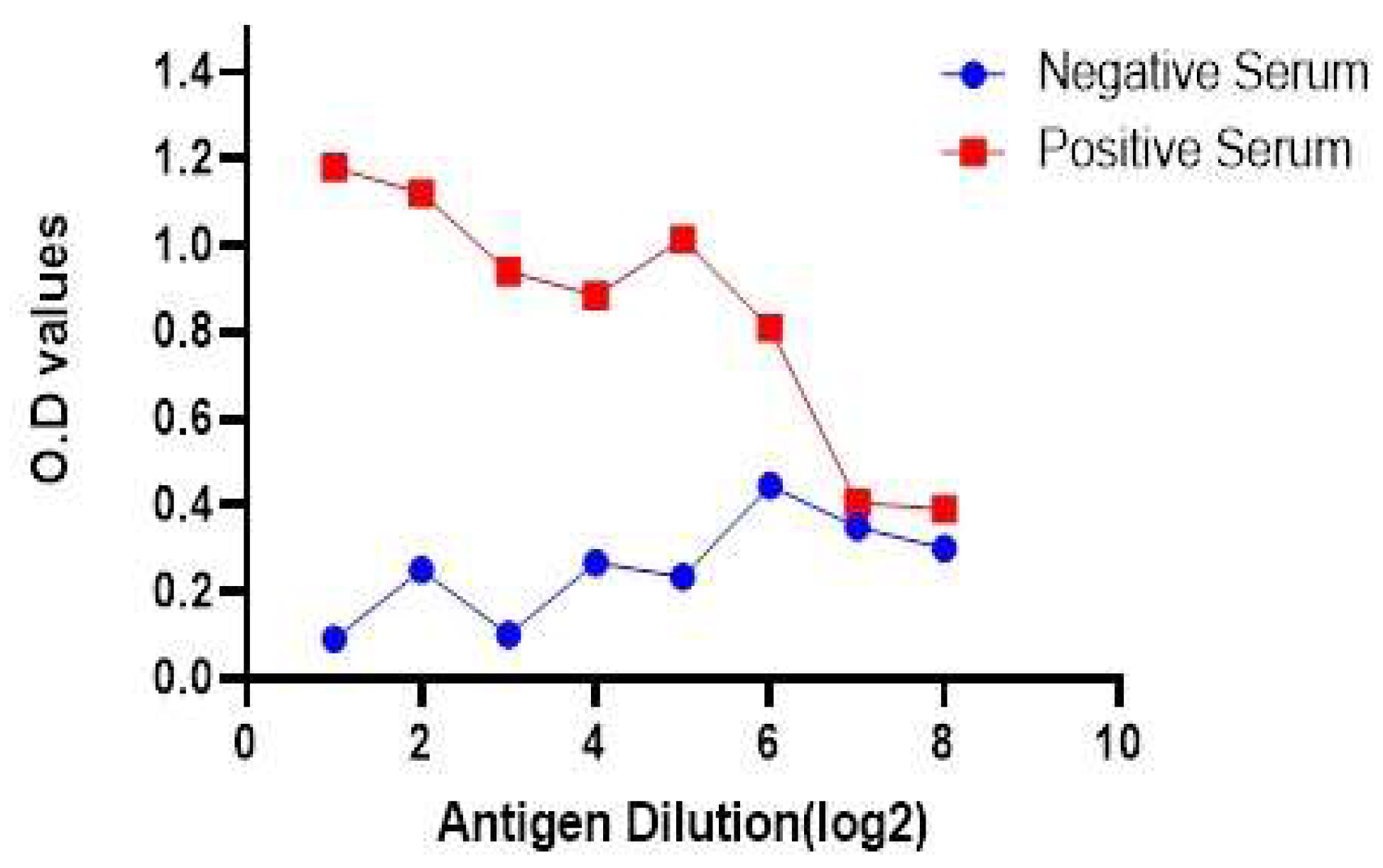
3.6. Antigen preparation and development of I-ELISA
3.7. Relative Specificity and Sensitivity of the Developed Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Regenmortel, M.H.; Fauquet, C.M.; Bishop, D.H.; Carstens, E.; Estes, M.; et al. Virus taxonomy: Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses; Academic Press, 2000. [Google Scholar]
- Radostits, O.M.; Gay, C.; Hinchcliff, K.W.; Constable, P.D. The Diseases Of Cattle, Horses, Sheep, Pigs, And Goats. Veterinary Medicine 2007, 10, 2045–2050. [Google Scholar]
- Lefèvre, P.C.; Diallo, A. Peste Des Petits Ruminants. 1990, 9, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Banyard, A.C.; Parida, S.; Batten, C.; Oura, C.; Kwiatek, O.; Libeau, G. Global distribution of peste des petits ruminants virus and prospects for improved diagnosis and control. Journal of general virology 2010, 91, 2885–2897. [Google Scholar] [CrossRef]
- Rahman, A.U.; Dhama, K.; Ali, Q.; Hussain, I.; Oneeb, M.; Chaudhary, U.; Wensman, J.J.; Shabbir, M.Z. Peste des petits ruminants in large ruminants, camels, and unusual hosts. Veterinary Quarterly 2020, 40, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Zohari, S.; Saeed, A.; Khan, Q.; Abubakar, M.; LeBlanc, N.; et al. Detection and phylogenetic analysis of peste des petits ruminants virus isolated from outbreaks in Punjab, Pakistan. Transboundary and emerging diseases 2012, 59, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Nafea, M.R.; Elbakry, M.; Shahein, M.; Farag, G.K.; Abdallah, F.; Ali, A.A.H. Virological and molecular studies on peste des petits ruminants virus (PPRV) in small ruminants and camels in Egypt between 2017 and 2018. Adv. Anim. Vet. Sci. 2019, 7, 12–18. [Google Scholar] [CrossRef]
- Hosny, W.; Baheeg, E.; Hassanein, S.; Mohamed, S. Preparation of a house ELISA kit for detecting Peste des petits ruminants Virus (PPRV) antibodies. BenhaVeterinary Medical Journal 2021, 41, 24–28. [Google Scholar] [CrossRef]
- Balamurugan, V.; Vinod, K.K.; Dheeraj, R.; Kurli, R.; Suresh, K.P.; Govindaraj, G.; Roy, P.J.V. Temporal and Spatial Epidemiological Analysis Of Peste Des Petits Ruminants. Outbreaks from The Past 25 Years In Sheep And Goats And Its Control In India. Viruses 2021, 13, 480. [Google Scholar] [CrossRef]
- Gargadennec, L.; Lalanne, A. La peste des petits ruminants. Bull. Serv. Zoo. AOF 1942, 5, 15–21. [Google Scholar]
- Taylor, W. The Distribution And Epidemiology Of Peste Des Petits Ruminants. Preventive Veterinary Medicine 1984, 2, 157–166. [Google Scholar] [CrossRef]
- Athar, M.; Muhammad, G.; Azim, F.; Shakoor, A. An Outbreak Of Peste Des Petits Ruminants-Like Disease Among Goats In Punjab (Pakistan). Pakistan Veterinary Journal 1995, 15, 140–140. [Google Scholar]
- Zahur, A.B.; Ullah, A.; Hussain, M.; Irshad, H.; Hameed, A.; Jahangir, M.; Farooq, M.S. Sero-Epidemiology Of Peste Des Petits Ruminants (PPR) In Pakistan. Preventive Veterinary Medicine 2011, 102, 87–92. [Google Scholar] [CrossRef]
- Balamurugan, V.; Vinod, K.K.; Dheeraj, R.; Kurli, R.; Suresh, K.P.; Govindaraj, G.; Roy, P.J.V. Temporal and Spatial Epidemiological Analysis Of Peste Des Petits Ruminants Outbreaks From The Past 25 Years In Sheep And Goats And Its Control In India. Viruses 2021, 13, 480. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization Of The United Nations. Peste Des Petitis Ruminant(Ppr)In Moroco. 2008. Available online: Https://Www.Fao.Org/Fileadmin/User_Upload/Newsroom/Docs/Aj120e00.Pdf.
- Kabir, A.; Mirani, A.H.; Kashif, J.; Manzoor, S.; Iqbal, A.; Khan, I.U.; Abubakar, M. Serological detection and confirmation of PPR among sheep and goats were kept under different production systems. Pakistan Journal of Zoology 2020, 52, 1137. [Google Scholar] [CrossRef]
- Abubakar, M.; Irfan, M.; Manzoor, S. Peste Des Petits Ruminants In Pakistan; Past, Present And Future Perspectives. Journal Of Animal Science And Technology 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Couacy-Hymann, E.; Roger, F.; Hurard, C.; Guillou, J.P.; Libeau, G.; Diallo, A. Rapid and sensitive detection of peste des petits ruminants virus by a polymerase chain reaction assay. J. Virol. Methods 2002, 100, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Kinimi, E.; Odongo, S.; Muyldermans, S.; Kock, R.; Misinzo, G. A paradigm shift in the diagnosis of peste des petits ruminants: A scoping review. Acta Vet Scand. 2020, 62, 7. [Google Scholar] [CrossRef]
- Balamurugan, V.; Sen, A.; Saravanan, P.; Singh, R.P.; Singh, R.K.; Rasool, T.J.; Bandyopadhyay, S.K. One-step multiplex RT-PCR for detection of PPR virus in clinical samples. Vet. Res. Commun. 2006, 30, 655–666. [Google Scholar] [CrossRef]
- Balamurugan, V.; Singh, R.; Saravanan, P.; Sen, A.; Sarkar, J.; Sahay, B.; et al. Development of an indirect ELISA for the detection of antibodies against the Peste-des-petits-ruminants virus in small ruminants. Veterinary research communications 2007, 31, 355–364. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of R.N.A. isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical biochemistry 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Forster, P.; Torroni, A.; Renfrew, C.; Röhl, A. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Molecular Biology and Evolution 2001, 18, 1864–1881. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Kumar, N.; Chaubey, K.K.; Chaudhary, K.; Singh, S.V.; Sharma, D.K.; Gupta, V.K.; Mishra, A.K.; Sharma, S. Isolation, identification and characterization of a Peste des Petits Ruminants virus from an outbreak in Nanakpur, India. Journal of virological methods 2013, 189, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.-U.-D.; Burgess, G.W. Production and characterization of monoclonal antibodies to fowl adenoviruses. Avian Pathology 2001, 30, 457–463. [Google Scholar] [CrossRef]
- Singh, R.P.; Sreenivasa, B.P.; Dhar, P.; Roy, R.N.; Bandyopadhyay, S.K. Development and evaluation of a monoclonal antibody-based competitive enzyme-linked immunosorbent assay for the detection of rinderpest virus antibodies. Revue Scientifique et Technique-Office International des Epizooties. Revue Scientifique et Technique-Office International des Epizooties 2000, 19, 754–763. [Google Scholar] [CrossRef]
- Spearman, C. The method of right and wrong cases (constant stimuli) without Gauss’s formulae. British Journal of Psychology 1908, 2, 227. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Samad, A.; Awaz, K.B.; Sarkate, L. Diagnosis of Bovine Traumatic Reticulo-Peritonitis I: Strength of Clinical Signs in Predicting Correct Diagnosis. Journal of Applied Animal Research 1994, 6, 13–18. [Google Scholar] [CrossRef]
- Libeau, G.; Prehaud, C.; Lancelot, R.; Colas, F.; Guerre, L.; Bishop, D.H.L.; Diallo, A. Development of a competitive ELISA for detecting antibodies to the peste des petits ruminants virus using a recombinant nucleoprotein. Research in Veterinary Science 1995, 58, 50–55. [Google Scholar] [CrossRef]
- OIE. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; World Organization for Animal Health: Paris, France, 2013. [Google Scholar]
- Elhaig, M.M.; Selim, A.; Mandour, A.S.; Schulz, C.; Hoffmann, B. Prevalence and molecular characterization of peste des petits ruminants virus from Ismailia and Suez, Northeastern Egypt, 2014–2016. Small ruminant research 2018, 169, 94–98. [Google Scholar] [CrossRef]
- Mahapatra, M.; Neto, M.M.; Khunti, A.; Njeumi, F.; Parida, S. Development and Evaluation of a Nested PCR for Improved Diagnosis and Genetic Analysis of Peste des Petits Ruminants Virus (PPRV) for Future Use in Nascent PPR Eradication Programme. Animals 2021, 11, 3170. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Ahsan, A.; Rasheed, T.; Farooq, U.; Ameen, M.K.; Zahur, A.B. Genetic characterization of peste des petits ruminants virus circulating in different regions of Pakistan based on nucleocapsid gene sequence. Japanese Journal of Veterinary Research 2019, 67, 139–144. [Google Scholar]
- Francki, R.I.; Fauquet, C.M.; Knudson, D.L.; Brown, F. (Eds.) Classification and nomenclature of viruses: Fifth report of the international committee on taxonomy of viruses. Virology division of the international union of microbiological societies; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Shaila, M.S.; Shamaki, D.; Forsyth, M.A.; Diallo, A.; Goatley, L.; Kitching, R.P.; Barrett, T. Geographic distribution and epidemiology of peste des petits ruminants viruses. Virus research 1996, 43, 149–153. [Google Scholar] [CrossRef]
- Anees, M.; Shabbir, M.Z.; Muhammad, K.; Nazir, J.; Shabbir, M.A.; Wensman, J.J.; Munir, M. Genetic analysis of peste des petits ruminants virus from Pakistan. BMC Veterinary Research 2013, 9, 1–5. [Google Scholar] [CrossRef]
- Sharma, K.K.; Kshirsagar, D.P.; Kalyani, I.H.; Patel, D.R.; Vihol, P.D.; Patel, J.M. Diagnosis of peste des petits ruminants infection in small ruminants through in-house developed indirect ELISA: Practical considerations. Veterinary world 2015, 8, 443. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Parvin, R.; Bhuiyan, A.R.; Giasuddin, M.; Chowdhury, S.M.Z.H.; Islam, M.R.; Chowdhury, E.H. Genetic characterization of Peste des petits ruminants virus circulating in Bangladesh. British Journal of Virology 2016, 3, 115–122. [Google Scholar] [CrossRef]
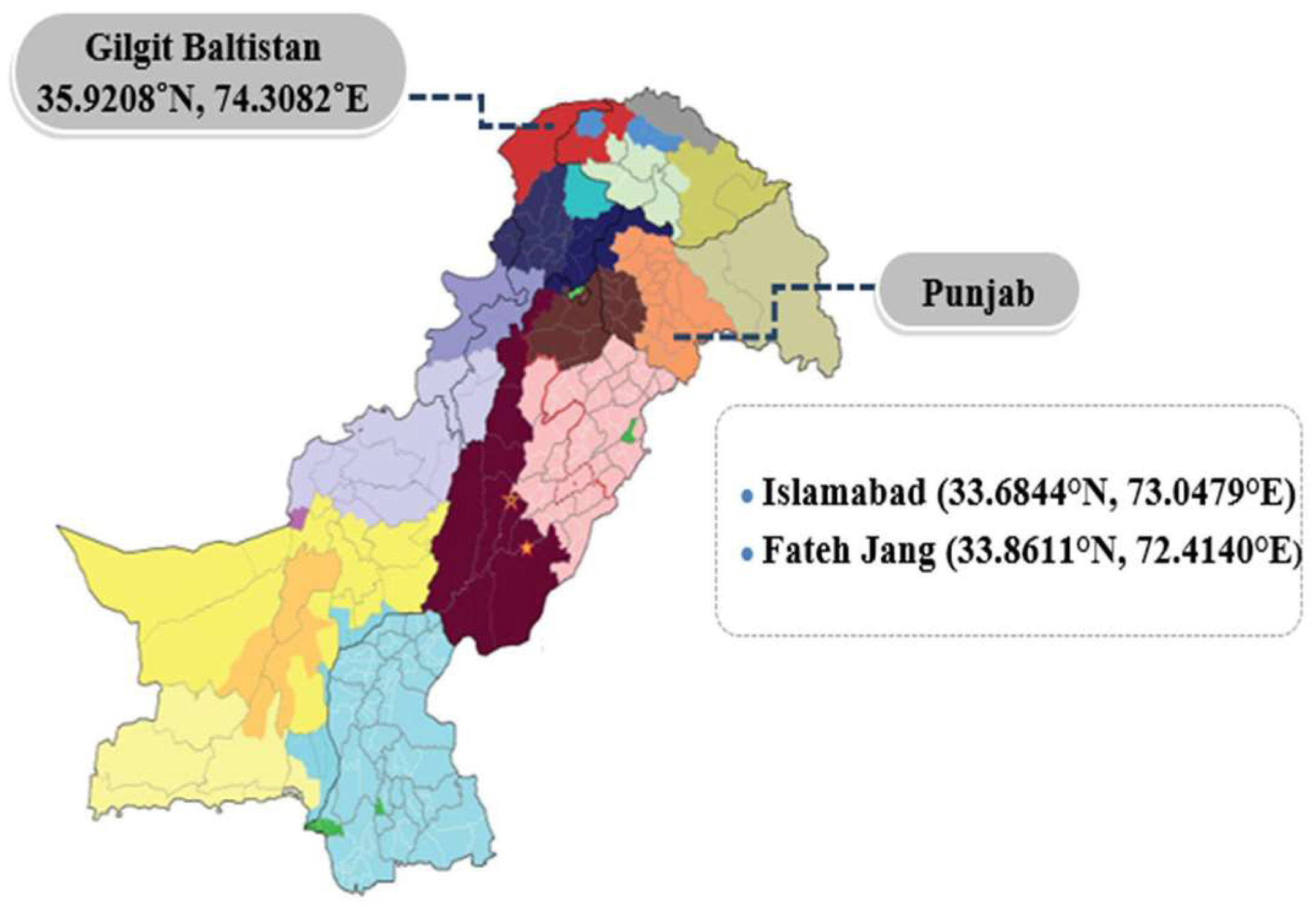
| Sr. No. | Location | No. of serum samples collected from goats | No. of serum samples collected from sheep | Total no. of serum samples |
|---|---|---|---|---|
| 1 | Fateh Jang | 52 | 42 | 94 |
| 2 | Islamabad (I.C.T.) | 109 | 57 | 166 |
| 3 | Gilgit | 44 | 21 | 65 |
| Total | 205 | 120 | 325 |
| Sample sources | Total samples | Positive samples | Negative samples |
|---|---|---|---|
| Swabs | 19 | 2 | 17 |
| Tissue | 6 | 2 | 4 |
| Sr. No. | Sample I.D. | Date of collection | Area | Source | Nature of Sample | Farm Name | Apparent Animal status | Virus Isolate |
|---|---|---|---|---|---|---|---|---|
| 1 | Pak-ICT-1350/NARC | 22-10-2020 | Islamabad | Goat | Tissue | Pakrozgar Goat Farm | Apparently Healthy | No |
| 2 | Pak-ICT-1349/NARC | 23-10-2020 | Islamabad | Goat | Tissue | Madina Goat Farms | Apparently Healthy | No |
| 3 | Pak-ICT-1346/NARC | 23-10-2020 | Islamabad | Goat | Tissue | Madina Goat Farms | Diseased | yes |
| 4 | Pak-ICT-1347/NARC | 10/11/2020 | Islamabad | Sheep | Tissue | Pure breed Farms | Diseased | No |
| 5 | Pak-ICT-1344/NARC | 10/11/2020 | Islamabad | Sheep | Tissue | Pure breed Farms | Apparently Healthy | No |
| 6 | Pak-ICT-1348/NARC | 10/11/2020 | Islamabad | Goat | Tissue | Pakrozgar Goat Farm | Apparently Healthy | No |
| 7 | Pak-GIL-1326/NARC | 15-10-2020 | Gilgit Baltistan | Goat | Swab | Hunza Farms | Diseased | yes |
| 8 | Pak-FJ-1336/NARC | 5/10/2020 | Fateh Jang | Goat | Swab | AlBarka Farms | Diseased | yes |
| I-ELISA | Commercial kit | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 135(a) | 26(d) | 161 |
| Negative | 14(b) | 150(c) | 164 |
| Total | 149 | 176 | 325 |
| I-ELISA | V.N.T. Assay | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 161(a) | 0(d) | 161 |
| Negative | 35(b) | 129(c) | 164 |
| Total | 196 | 129 | 325 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





