1. Introduction
As reported by the World Health Organization, more than 17.9 million people passed away from cardiovascular sicknesses in 2016. Annually, cardiovascular disease causes approximately 31% of deaths worldwide [
12]. Cardiovascular magnetic resonance imaging (CMR) is a practical imaging modality for detecting and assessing the heart diseases (that are related to the cardiac chambers or the coronary arteries). Using CMR, we can derivate cricial metrics like left and right ventricular (LV, RV) volumes, left ventricle mass (LVM), and ejection fraction (EF) [
13]. These clinical indices are used for condition diagnosis (such as heart failure and hypertrophic cardiomyopathy ), risk stratification, and therapy decisions’ guidance.
It is worth noting that the short-axis (SAX ) cine images of the left and right ventricles are highly suited to cover the entire left and right ventricle from the base to the apex (see the section 2 and Figure 3, 2, for more information). Since the number of images per subject can be exceedingly large, the manual segmentation of LV and RV (which is done slice by slice by physicians and radiologists ) is a time consuming and error-prone task.
Despite the development of various automated/semi-automated segmentation techniques, accurate and fast LV segmentation remains a challenge. For instance, Trabeculations and the papillary muscles are located inside the LV cavity (see Figure 3) are almost indistinguishable from the myocardium using intensity. Therefore, several algorithms are struggling with the same issue of inner contours shrinking. There is a concern that the LV cavity region may appear to be too small on apical and basal slices, resulting in an imbalance between the number of pixels that belong to the LV region and the background. Additionally, variability in the shape and intensity of the LV can be considerable, particularly among those with pathology. In the past two decades, there has been significant advance in the development of fully automated algorithms, despite the handicaps that have been encountered.
The objective of this paper is to propose an improved UNET architecture to handle the automatic segmentation of the endocardium in SAX CMR images. Our enhancement consists of adding inception modules [
3] in the contraction path of the CNN. The inception modules’ goal is to enable the UNET to perform a parallel learning of the different filter kernels, and therefore, the most pertinent kernels will be chosen for representing a given set of images . Our main contributions can be summarized as follows:
We propose a UNET model with inception layers in the contracting path for capturing the endocardium of LV (i.e., the segmentation of internal cavity of LV). The motivation behind this is to enhance the accuracy of segmenting the internal cavity of LV using multiple convolution kernels at the same time.
We compare the performance of the proposed model with a baseline UNET and other existing methods using both the cross-entropy loss and the dice metric.
The rest of the paper is structured in the following way.
Section 2 presents preliminary information on the heart structure and the available types of cardiac images.
Section 3 summarizes the current methods that are devoted to left ventricle segmentation.
Section 4 presents the proposed UNET models.
Section 4 reports the experimental results on the chosen dataset.
Section 5 presents the conclusion and future works.
2. Preliminaries on Cardiac Anatomy
Different medical imaging modalities were used for the assessment of LV. These modalities include magnetic resonance images (MRI), echocardiography, computer tomography (CT) scan, myocardial perfusion imaging, multiple gated acquisition scanning, gated blood-pool SPECT, and fusion imaging. Magnetic resonance imaging is a widely used modality in the cardiovascular diseases diagnosis. CMR is considered as the de facto mechanism for analyzing heart problems. CMR is able to provide both anatomical and functional insights on the heart. In fact, it is appropriate to the evaluation of the heart chambers size, blood flow through major vessels, heart valves, and pericardium [
16,
17]. Echocardiography utilizes high-frequency ultrasound waves to create anatomical images of the heart (it is also termed as ultrasound (US) imaging). It is the widely used imaging mechanism for the inspection of cardiovascular diseases. Thanks to its noninvasiveness, cheapness, excellent temporal resolution, real-time imaging, it is considered as one of the preferred imaging method for evaluating the LV function. US is also considered as the main means for the assessing of LVM. As regards Computer Topography, it is known that the computed tomography (CT) scan of the heart delivers a cross section of its anatomy. It is based on the X-ray attenuation properties of tissues being scanned. CT is a widely used imaging method for the characterization of the heart anatomy and function. LV size and mass evaluation can be computed using the CT imaging. CT is also considered as a good alternative for the LV size and mass prediction for patients with CMR prohibition.
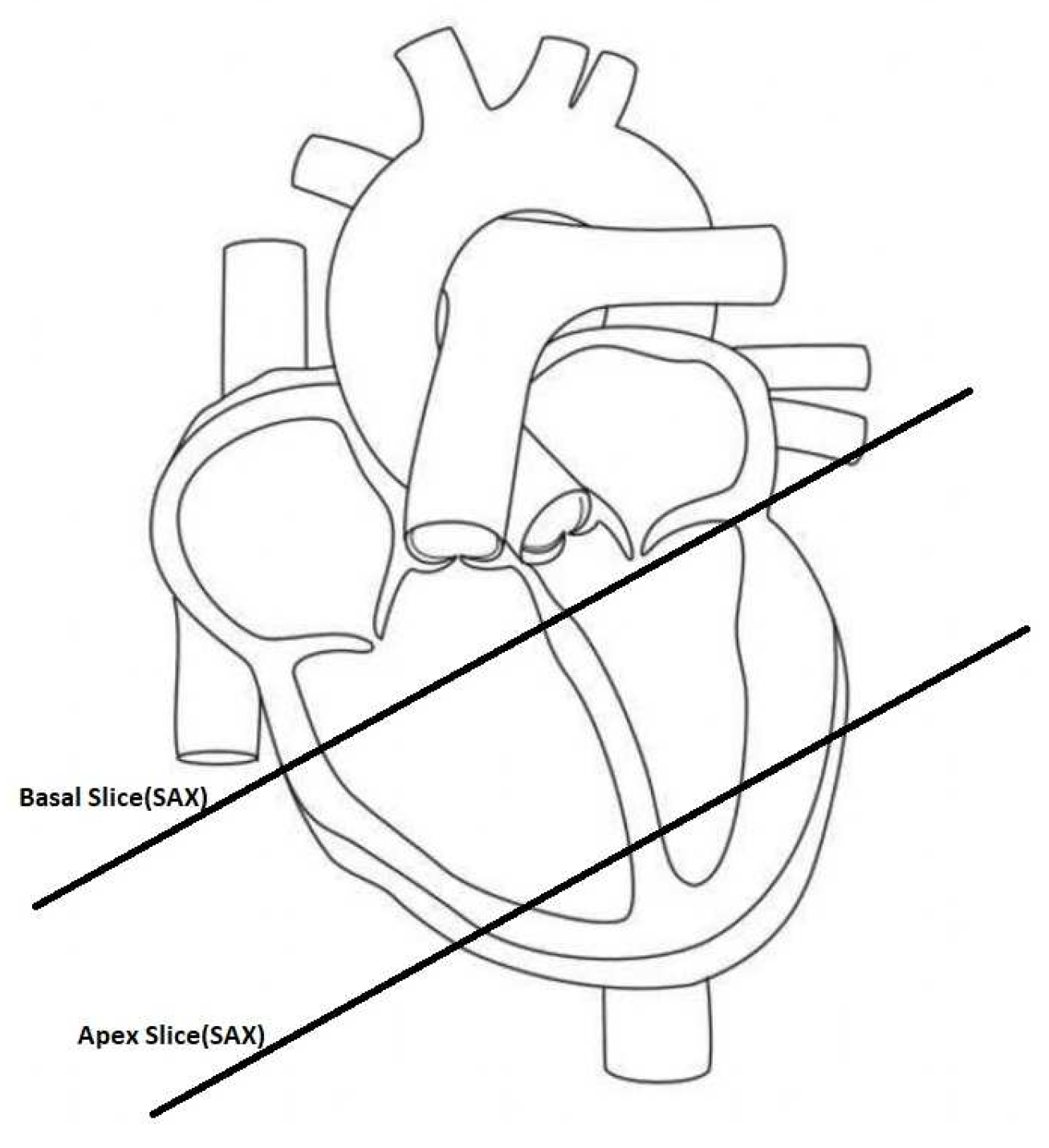
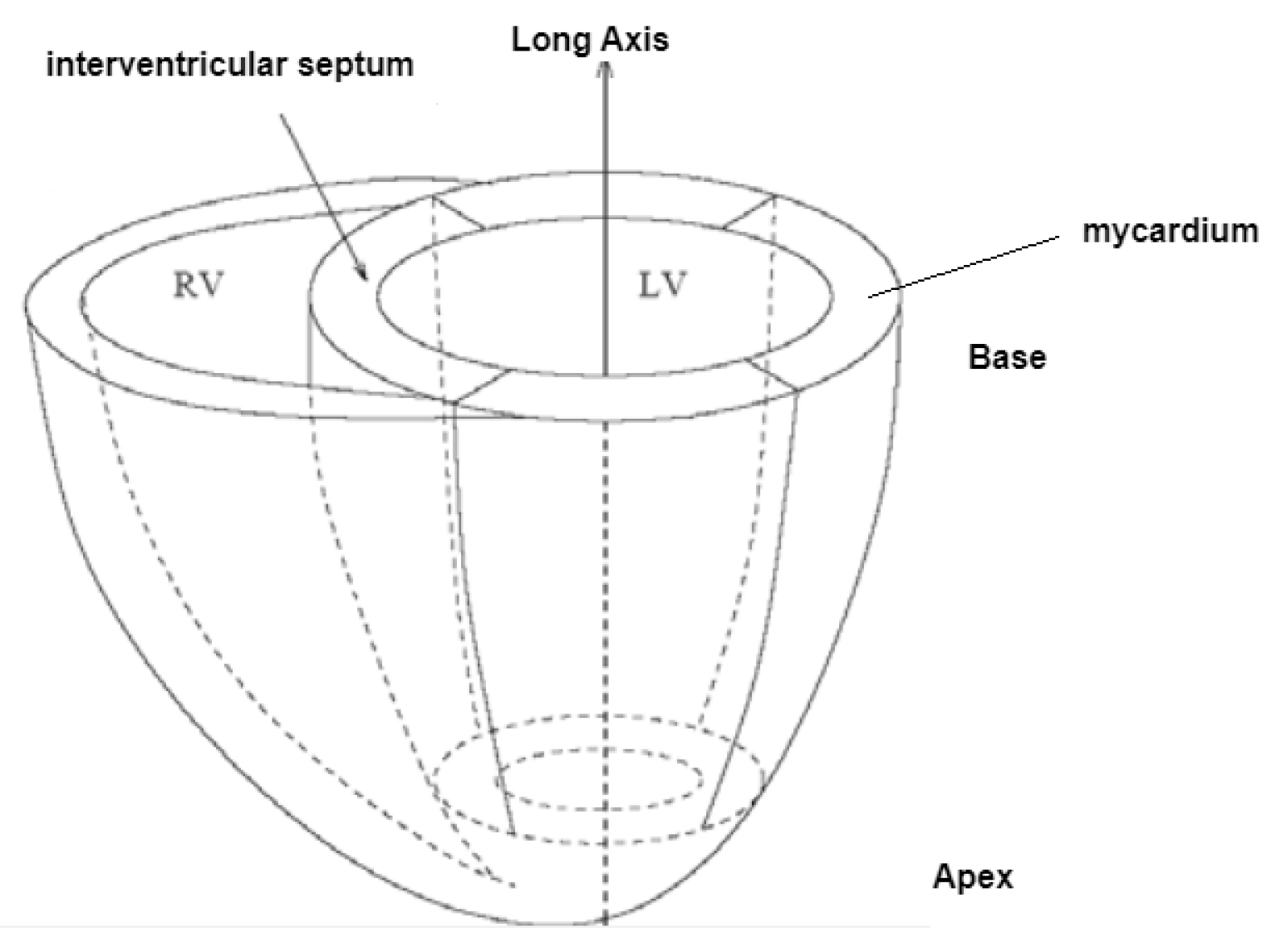
The segmentation of the heart chambers is a complex task due to the heart anatomy. It is worth noting that the left ventricle (LV) is responsible for pushing oxygen-rich blood to the aorta and ultimately to the rest of the body. Its cavity typically has an ellipsoid shape and is surrounded by myocardium, which is usually between 6 to 16 millimeters thick. In contrast, the right ventricle (RV) has a more complex crescent form and experiences lower pressure when pumping blood to the lungs, making it three to six times thinner than the LV, which can make it difficult to detect in MRI scans. For these arguments, the majority of research has been devoted to LV, as its function is more critical for the body[
15]. The magnetic resonance imaging of the heart involves the use of 8-10 short axis (SAX) slices that cover the entirety of the organ (see
Figure 1).
The SAX imaging plane is perpendicular to the apex-base axis, known as the short axis plane. Each slice is spaced with 10-20mm with the following one (see
Figure 2).
With the help of the synchronized ECG signal, dynamic MR sequences are employed to capture the heart in motion; this allows for the acquisition of 30 images per cardiac cycle, with a temporal resolution of 30 ms. With current technology, one examination can result in up to 250 images. These images, however, predominantly focus on a smaller region of interest (ROI) as the ventricles occupy a small surface area in the full-size images.
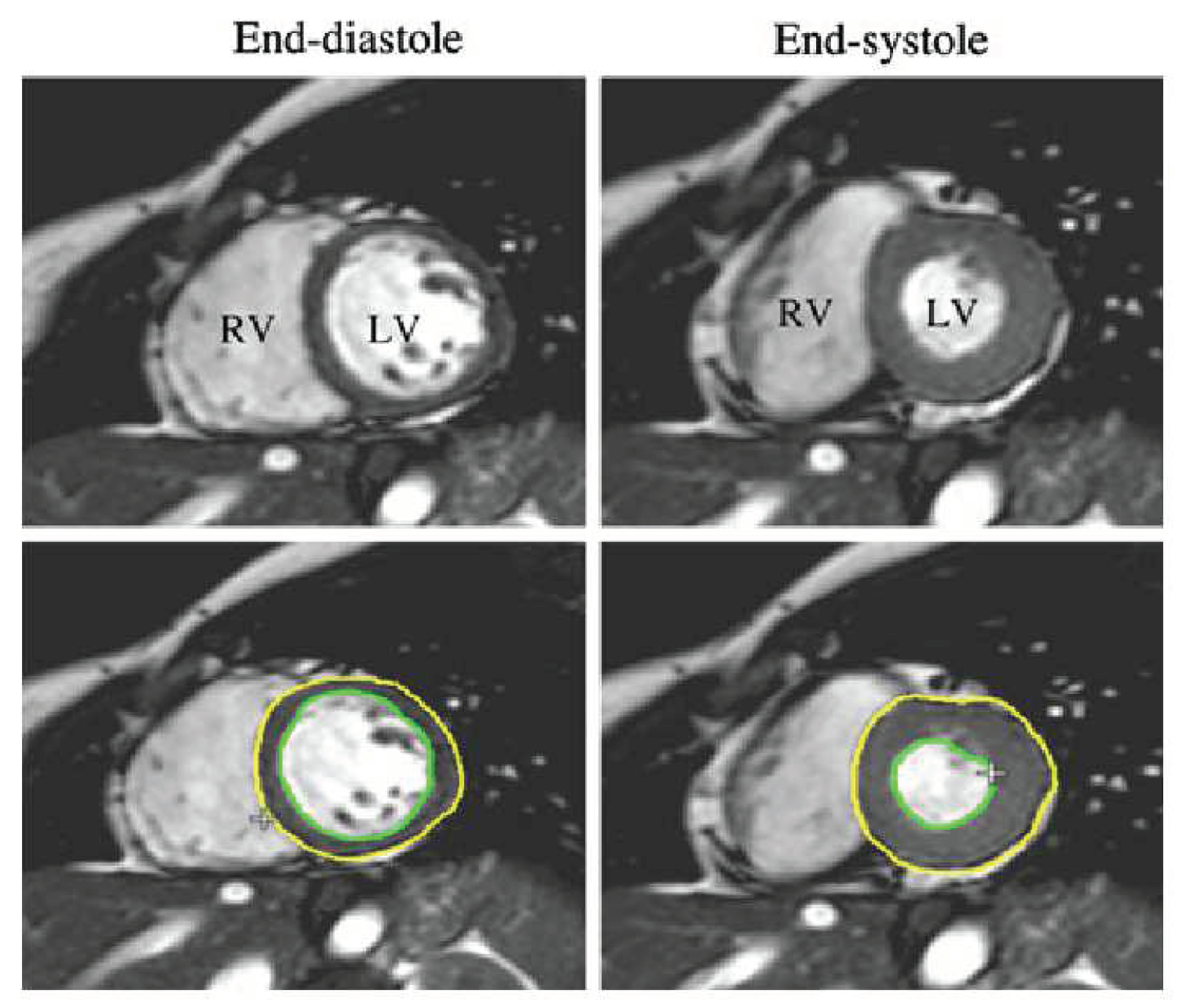
In
Figure 3, we show a short-axis cine-MRI image at end-diastole and end-systole of LV. The figure reveals that the left (and even right ventricular) cavity has a brighter gray level than that of the surrounding tissues (e.g., the myocardium, papillary muscles, trabeculae, lung parenchyma, and anterior chest wall). Moreover, we notice that the papillary muscles and trabeculae have almost the same gray value as that of the myocardium, and this constitutes a major handicap in deriving a smooth contour or a correct estimation of the LV volume. We notice that, the physicians must delineate the end-to-end epicardial ventricular edges in the end-systolic (ES) and end-diastolic (ED) stages to derive the clinical indices and orient the diagnosis of the disease.
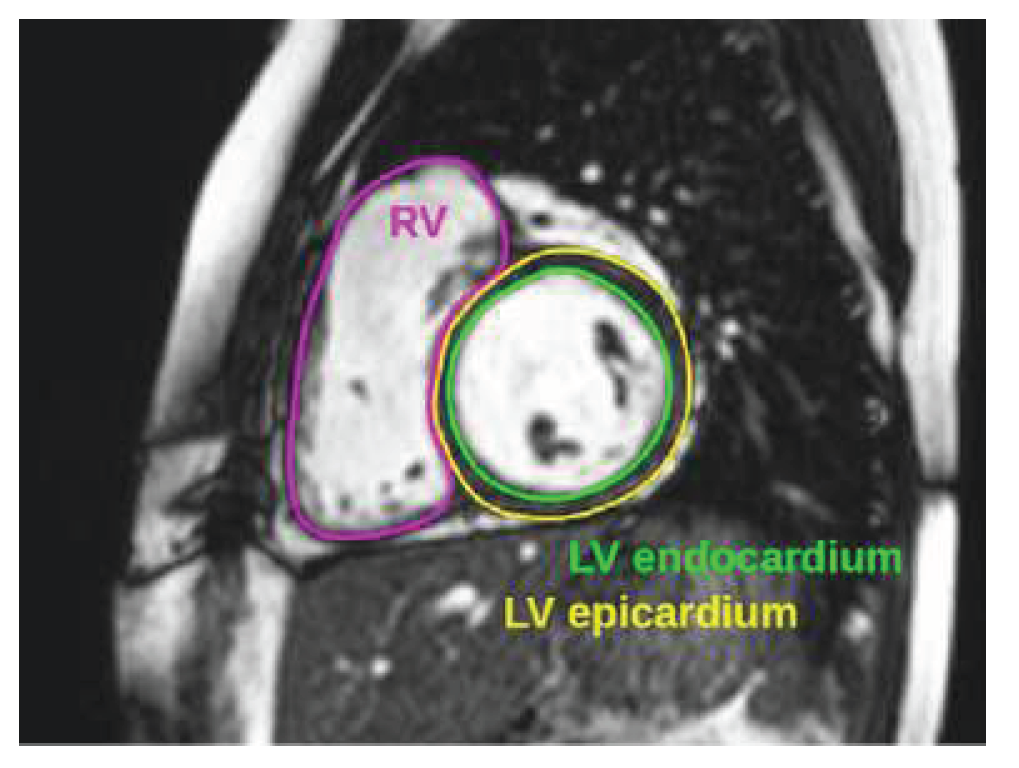
In
Figure 4, we show the endocardium of LV in green color, and the epicardium of LV in yellow, while the endocardium of RV is shown in magenta. At the end of this section, we notice that there are a few public cardiac MRI datasets which are suitable for systematic segmentation of LV, mainly, we have:
3. State of The Art
The Ventricles segmentation has gained much interest due to their involvement in evaluating cardiac indices, which are crucial for the diagnosis of cardiovascular conditions.
Many detailed reviews have been presented by the research community in the field of ventricle segmentation [
15,
19,
20,
21,
22,
23]. The current approaches of LV segmentation can be ranged into various classes, including: (1) image-based or pixel-classification techniques such as intensity thresholding and distribution modelling; (2) deformable models such as active contours; (3) statistical shape and appearance models such as truncated ellipsoids for the LV; (4) anatomical atlas-based registration;(5) deep learning such as fully convolutional networks (FCN) and UNET; and (6) combinations of these various approaches [
24,
25].
Active contour models enjoy several benefits when compared to other classical image segmentation techniques, such as thresholding, contour-detection and region-growing. First off, these models can attain sub-pixel accuracy for object borders. Additionally, these models can be structured into an energy minimization framework, which allows for the integration of prior knowledge, like shape or intensity distribution, to achieve reliable segmentation [
26]. Finally, active contour models can yield smooth and closed contours [
27,
29,
30].
In [
1], the authors introduced a method that belongs to the Active Form Model class. The authors designed a segmentation method of echographic images based on the Active Appearance and Motion Models [
2] with an adaptation of the noise model. They have taken into account both the internal texture and external texture of the endocardium, and this significantly improves the results. Broadly speaking, the approach consists of three main steps: first, the image annoation is made by the experts; then, the training of the model is performed to guide the evolution of the contour, and finally the segmentation of images is realized using binarizations and comparisons. A multi-atlas-based segmentation approach is proposed in [
32]. The approach pipeline is described as follows: first the CMR image is preprocessed to palliate the tissue intensity disparities across the scans; second, an atlas-based segmentation is performed to get the endocardium and epicardium contours LV; third, the contours are rectified using a spatio-temporal registration.
In what follows we will principally focus on deep learning-based segmentation methods since they have achieved dazzling success in medical image analysis. It is known that FCN models [
33] are largely used in semantic segmentation. In the same perspective, U-net models [
34] further enhance FCN and are largely applicable to medical images. Obtaining a large amount of labeled LV images is difficult in practice, due to the expense and slow speed of manual segmentation of the ventricles. In this regard, semi-supervised approaches are much more suitable for clinical applications, as they do not require large quantities of labeled data while still achieving excellent results. Consequently, the use of semi-supervised approaches in image segmentation is exceedingly growing . One basic hypothesis in semisupervised learning is that the data points are likely to preserve the topology, and this means that if two examples x1 and x2 are close in a dense area, then the corresponding outputs y1 and y2 must be also close. A similar assumption can be extrapolated for examples in a cluster. In this context, generative adversarial networks (GAN) have gained promising popularity in image segmentation [
35,
36]. In [
35], the authors proposed a semisupervised segmentation approach that uses FCN based discriminators to create real segmentation maps and predicted segmentation maps. In [
37], the authors used the feature adjustment that matches the properties’ distributions between labeled and unlabeled data. The framework proposed in [
14] handles the segmentation of the LV ( endocordium and epicardium) as regression problem. More specifically a set of control points belonging to the LV (resp RV) contour that are previously marked by the experts must be predicted by deep neural networks. The model involves two steps: first, it locates the region of the ventricle in a short-axis MRI image using a residual network backbone; then, it predicts the contours of the endocardium and epicardium using varied residual networks. In [
19], a fully convolutional network (FCN) is introduced to segment the LV using short-axis MRI images (without using the concatenation from the contraction path). The authors perform preprocessing on the images by normalizing the pixel values and balancing the weights of both the target and the background classes. The FCN is fine-tuned by choosing the best values for the learning algorithm and the batch size. In [
5], a two-step approach is proposed to segment the endocardium and the epicardium of LV. Firstly, a UNET-based network is trained to capture the central-line of LV (endocardium). Secondly, the level set approach is used to delineate the myocardium region; more precisely, the result of the UNET step is used in the level set energy expression as a constraint term. In [
9], an FCN architecture (without the expanding path) is proposed for segmenting the endicardium and epicardium of LV. The model contains 15 stacked convolution layers and three layers of overlapping, two-pixel strided max pooling. It shows a higher performance in delineating the the epicardium. To handle the great variability of the ventricles, the framework presented in [
38] firstly extracts a multiscale pyramid of descriptors of bi-ventricle. Then a strategy for adjusting the weighted pyramid is performed to get smooth properties for both labeled and unlabeled data. More specifically, the weighting task is performed using adversarial learning. In [
39], the segmentation of LV is defined in terms of radial distances between the LV walls (the endo- and epicardial contours) and the center of the blood pool rather than a classification task. To this end, two convolutional neural networks perform a regression task to infer these scores. The work by [
23] proposed a UNET architecture with multiscale decision heads. In particular, each layer of the expansion path derives an upsampled final output that is used in an complex weighted loss. In addition, the model uses a special weighting scheme to palliate imbalanced classes. A strengthened Residual UNET is proposed in [
40] to enhance the accuracy of the endocardium /epicardium segmentation of CMR images. The model mainly introduces medium skip connections in the contracting path and short skip connections in the residual blocks.
4. Proposed Approach
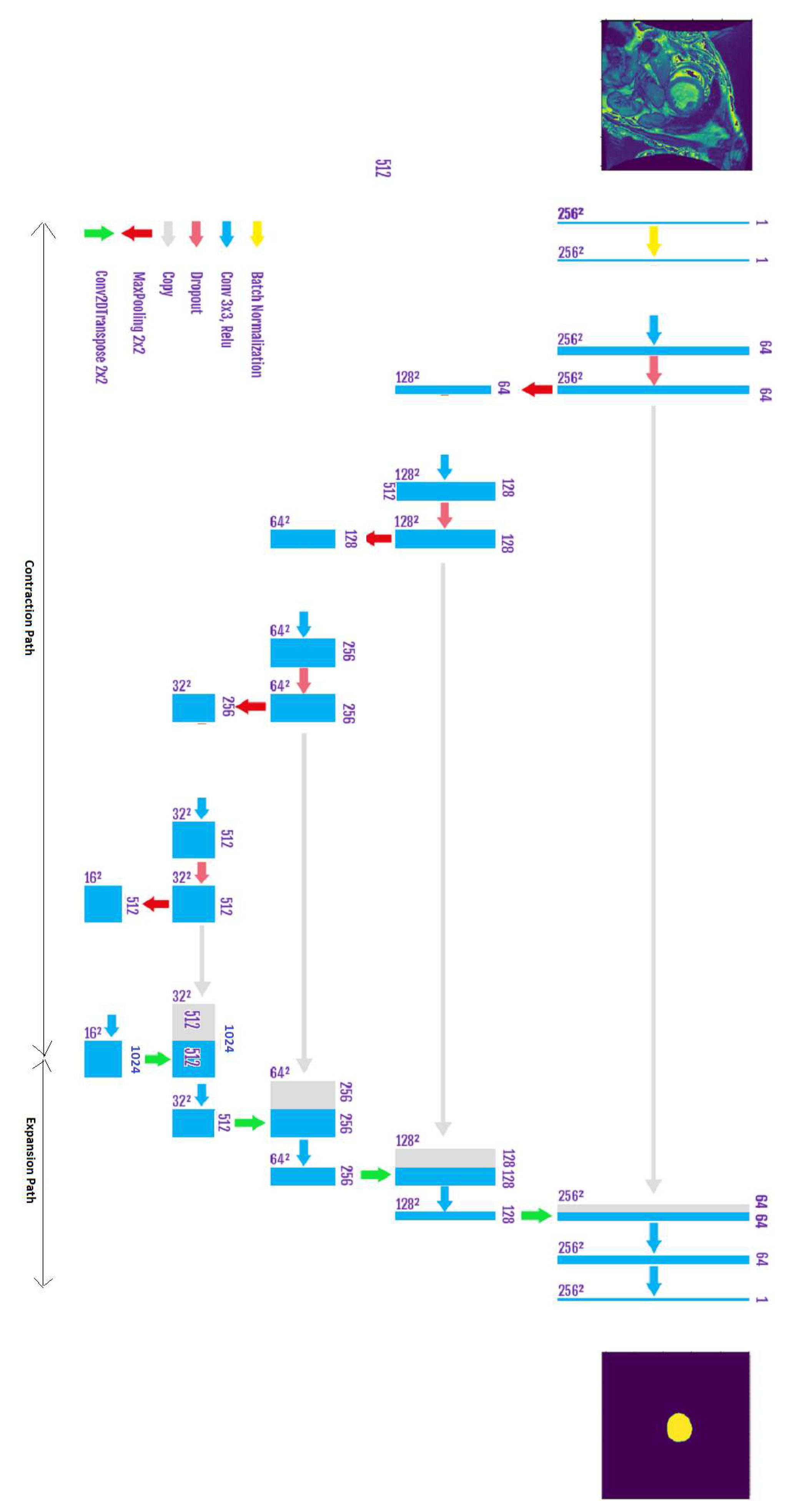
To handle the segmentation of LV endocardium (from SAX CMR images) we propose a baseline UNET network and an improved UNET (that uses additional inception modules). It is worth noting that the UNET architecture is well suited to image segmentation due to the following reasons:
The contraction path of the the model allows for a correct classification of the current pixel (of an object) since its context (neighboring pixels) is large, thanks to the length of the contraction network, but on the other hand, the edges of that object can be erroneous.
To palliate the above-mentioned issue, the expansion path tends to re-establish the correct edges of the detected objects using up-sampling and concatenated layers from the contraction path.
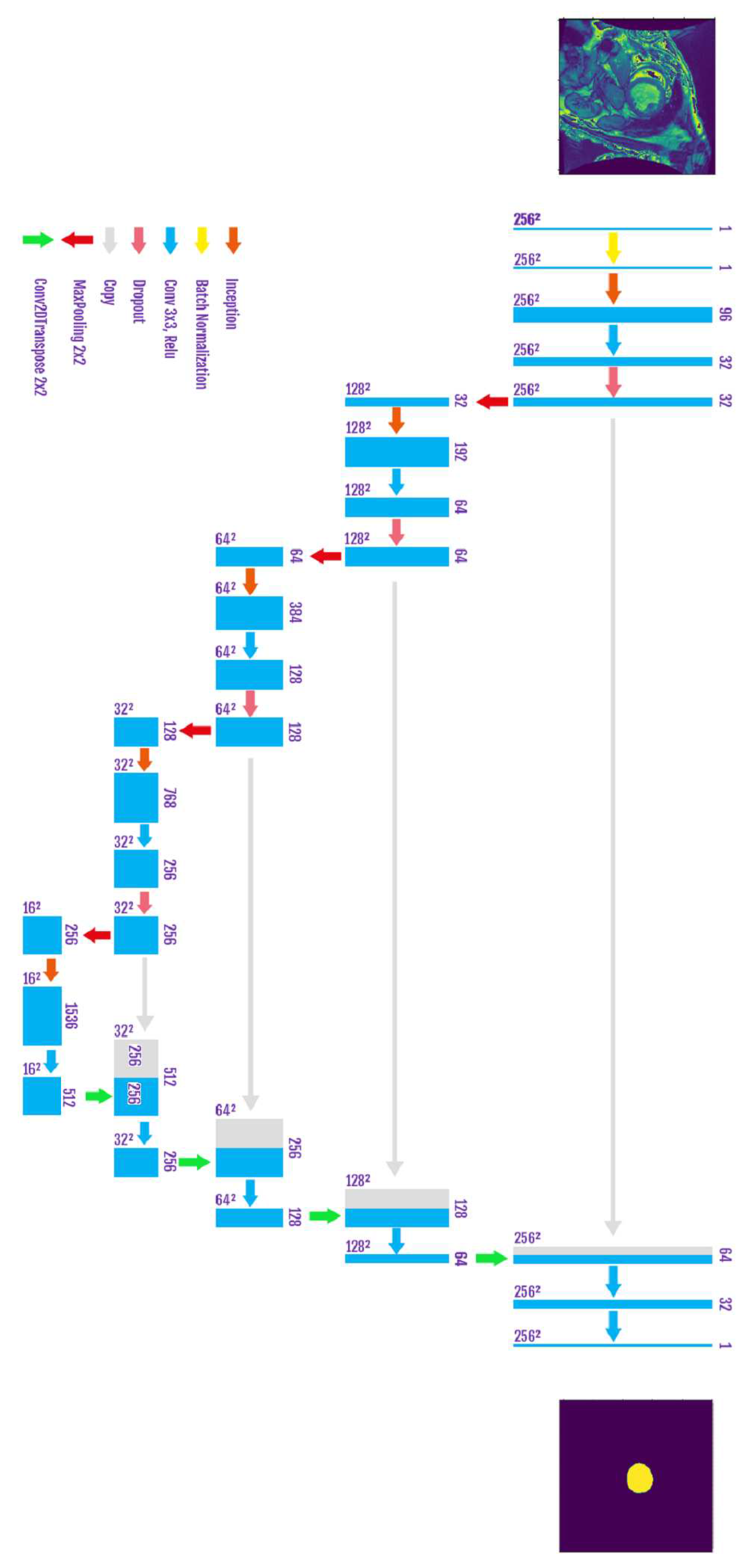
Figure 5 demonstrates the architecture of our Baseline UNET. To enhance the accuracy of the segmented cavity of LV, we add inception layers in each stair of the contraction path. The contraction path is shown in
Figure 5, while the inception module is shown
Figure 6.
The combination of the two elements is shown in
Figure 7.
The motivation behind the inception modules [
3] is to perform a kind of ensemble learning within the CNN, by doing so, the network will derive the right filters that cope with the current images and give better representations.
5. Experimental Study
The accuracy of pixel classification is not a useful metric in our segmentation task since most of the pixels in the input image belong to the background. In other words, if we have an algorithm that classifies all the background pixels as target object, we can achieve an accuracy of 90% up to 95% in many images. The reason behind this is that the left ventricle occupies a very small region with respect to the entire image (in average, it is less than 5%, in the SAX image case). To palliate this issue, we use the Dice similarity coefficient as an evaluation metric, in addition to the cross entropy dissimilarity. The specifications of the cross entropy dissimilarity, dice similarity, and hybrid loss are given in Equations
1,
2, and
3.
Label stands for the ground truth image (given by the expert), while P represents the image that is predicted by our model. To improve the results of our training, we replace the standard cross-entropy loss by a hybrid loss that mixes both the cross-entropy and the dice similarity measure (see Equation3).
The MICCAI 2009 dataset [
4] (also termed Sunnybrook Cardiac Data) consists of 45 CMR images (1.6 GB) belonging to different patients (male and female). These subjects have multiple pathologies: healthy, hypertrophy, heart failure with infarction and heart failure without infarction. The MRI images are in DICOM (Digital Imaging and Communications in Medicine) image format, which consists of several patient and image metadata parameters. For each patient record, there is a set of hand-drawn contours for the End-Systole and End-Dystole slices. The contours were drawn by the experts of Sunnybrook Health Science Center. The contours were available in text files consisting of the contour points, which had to be converted to an image.
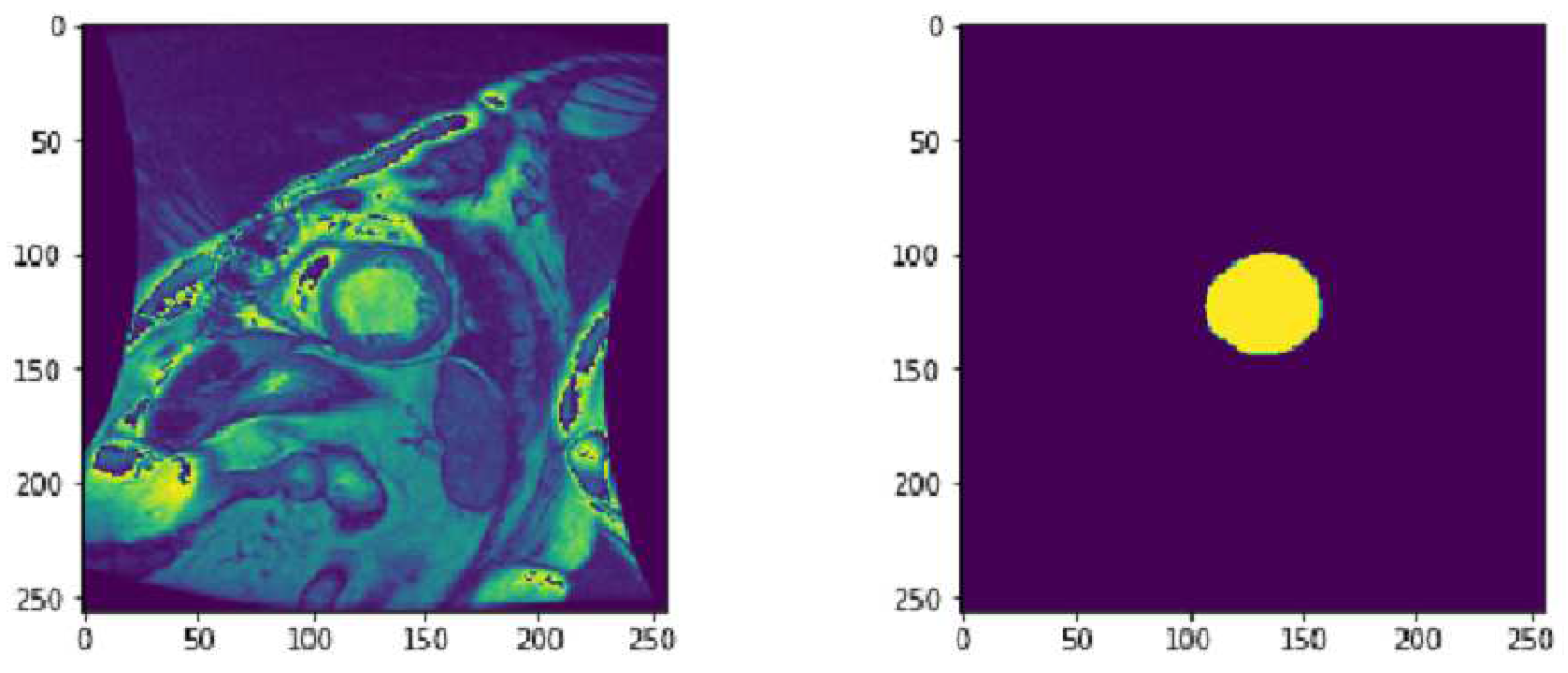
Figure 8 shows a SAX CMR Image and the corresponding LV contour provided in the data set. The labeled data provided in the dataset represent approximately 20 image instances for each patient. This results in 805 labeled DICOM images.
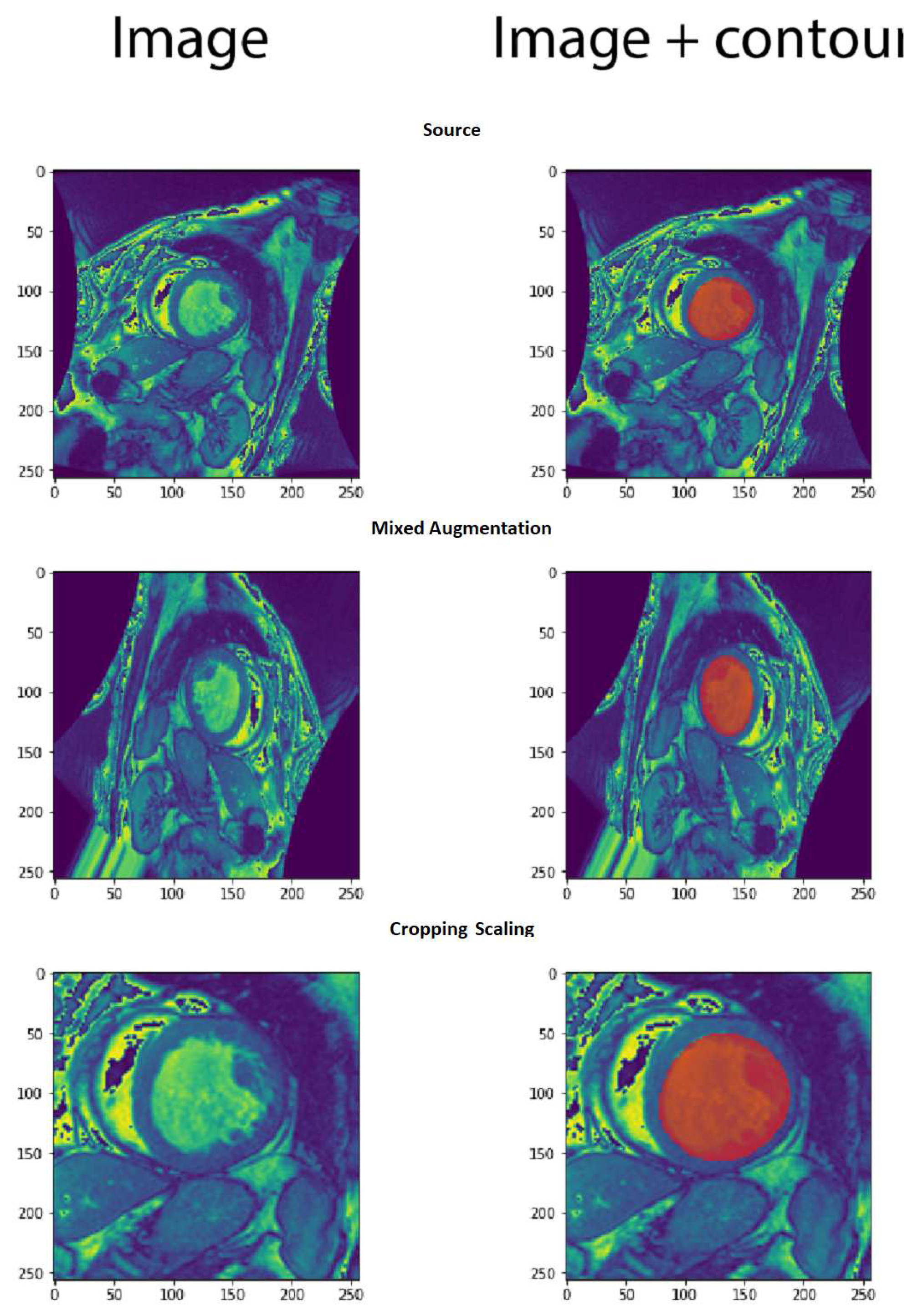
Since the size of dataset is not large, we performed a data augmentation in order to avoid the overfitting. First, we have retained 80% of data (644 images) for training and the remaining examples are devoted ( 20% or 161 images) to testing. Second, The training set is augmented the following operators: rotation with less than 90 degree, flipping, cropping and scaling. Each of the mentioned methods was applied to the training images, and this allowed us to increase the dataset 3 times its original size (1932 images).
Figure 9 shows an example of augmentation.
We have used the hardware configuration given by Google Colaboratory:
CPU : Intel(R) Xeon(R) CPU @ 2.30GHz
GPU : Tesla P100-PCIE 16GB
RAM : 32 GB
OS : Ubuntu 18.04.3 LTS (Bionic Beaver)
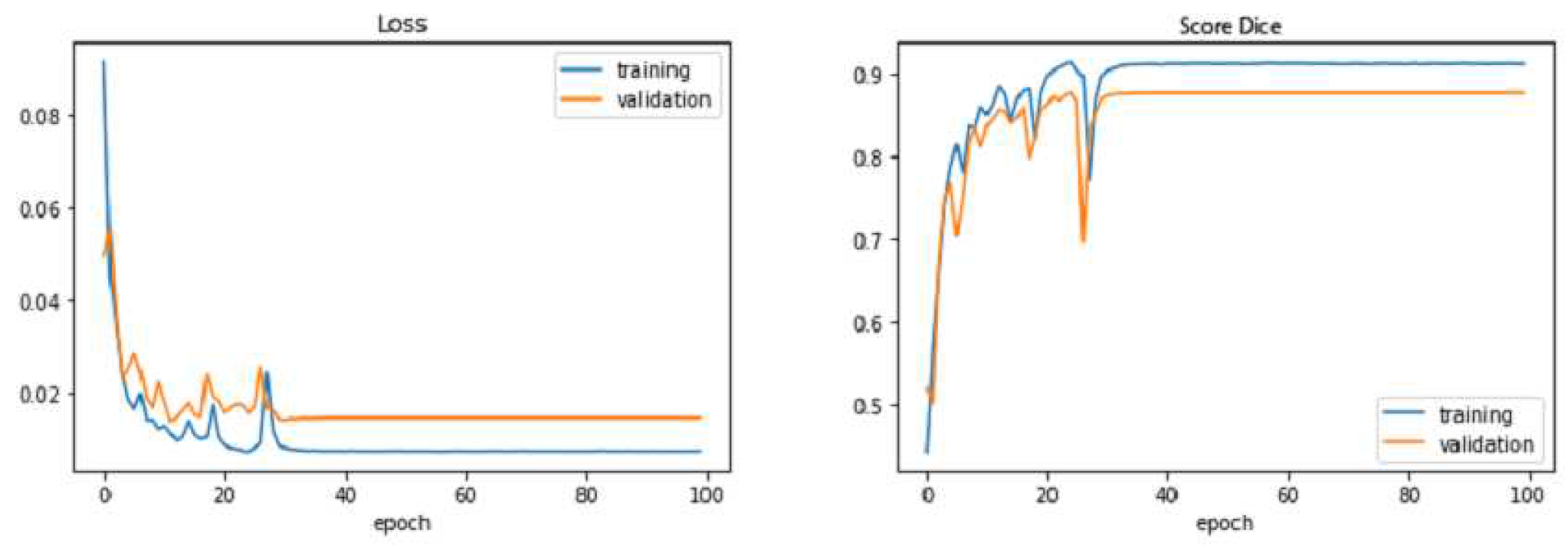
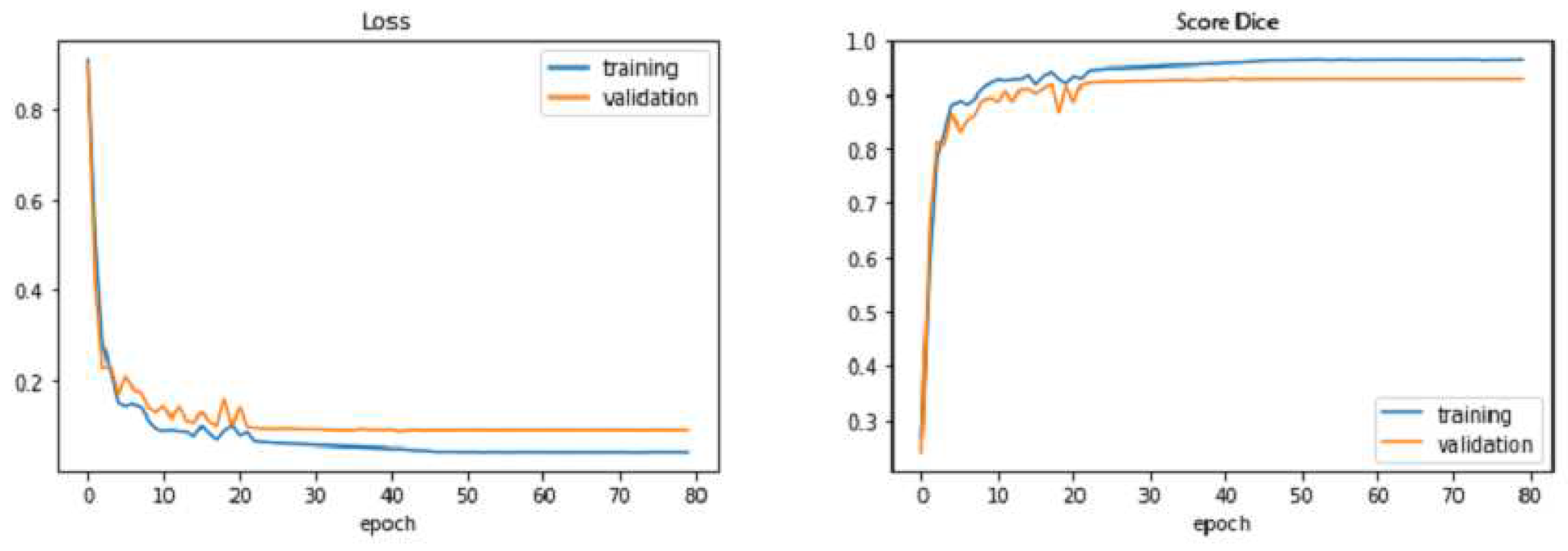
In
Figure 10,
Figure 11, we observe that the improved UNET outperforms the baseline UNET for almost all epochs, both of models reach their stability point (or overfitting threshold) around the 40
th epoch. we also notice that the loss curve of the the improved UNET is smoother than that of the baseline UNET; moreover, the improved UNET experiences less overfitting as compared with the baseline UNET, since the gap between the training and the testing losses is less than that of the baseline model.
Table 1 shows a comparison between the model performance expressed in terms of dice score and the hybrid loss. It is clearly shown that the training time of both models is reasonable, especially if we take into account the number of trainable parameters. By observing the time it took for each model to finish the training and taking into account the number of parameters that could be trained, we can say that they both took a reasonable amount of time. We can clearly see that even when the original model had much more trainable parameters (around 6 million more than the proposed model), meaning more flexibility during training, it did not perform better. And this proved that the architecture and the way the layers were linked had a crucial effect on the results. In our case, the Inception layers implemented in the improved UNET gave it more paths to take (and choose from), and this results in better performance (see the dice column in
Table 1) and less training time.
In
Table 2, we give a comparison between our improved UNET and some existing state-of-the-art models on the MICCAI 2009 dataset [
4] .
It is clearly shown that our proposition outperforms the majority of them and prove that the inception layers and batch normalization are highly effective in improving the performance of UNET models.
6. Conclusion
We have presented in this paper an improved UNET model that allows for segmenting the endocardium of LV. The addition of inception modules achieves better results with respect to the baseline UNET architecture and many existing methods.
In future works, we plan to segment both the endocardium and the epicardium of the LV and RV. In addition, we aim to estimate the myocardium indices such as LV volume , RV volume, septum width, and ejection fraction.
References
- Bosch, J.G. , Mitchell, S.C., Lelieveldt, B.P., Nijland, F., Kamp, O., Sonka, M. and Reiber, J.H. 2002. Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE transactions on medical imaging, 21, no. 11 (2002), pp.1374-1383. [CrossRef]
- Lebossé, J. , Lecellier, F., Revenu, M. and Saloux, E. 2005. Segmentation du contour de l’endocarde sur des séquences d’images d’échographie cardiaque. In GRETSI 2005 (pp. 537-540).
- Szegedy, C. , Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Erhan, D., Vanhoucke, V. and Rabinovich, A. 2015. Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition (2015). pp. 1–9.
- RADAU, Perry, LU, Yingli, CONNELLY, Kim, et al. Evaluation framework for algorithms segmenting short axis cardiac MRI. The MIDAS Journal-Cardiac MR Left Ventricle Segmentation Challenge 2009, 49, 4. [Google Scholar]
- Xie, Lipeng, Yi Song, and Qiang Chen. 2020. Automatic left ventricle segmentation in short-axis MRI using deep convolutional neural networks and central-line guided level set approach." Computers in Biology and Medicine 122. (2020): 103877. [CrossRef]
- AVENDI, Michael R., KHERADVAR, Arash, et JAFARKHANI, Hamid. 2016. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Medical image analysis, vol. 30,(2016), p. 108-119. [CrossRef]
- NGO, Tuan Anh et CARNEIRO, Gustavo. 2013. Left ventricle segmentation from cardiac MRI combining level set methods with deep belief networks. In : 2013 IEEE International Conference on Image Processing. IEEE, (2013). p. 695-699. [CrossRef]
- HU, Huaifei, GAO, Zhiyong, LIU, Liman, et al. 2014. Automatic segmentation of the left ventricle in cardiac MRI using local binary fitting model and dynamic programming techniques. PloS one, vol. 9, no 12, 2014. p. e114760. [CrossRef]
- TRAN, Phi Vu. A fully convolutional neural network for cardiac segmentation in short-axis MRI. 2016. arXiv:1604.00494, 2016. [CrossRef]
- YUAN, Tianchen, TONG, Qianqian, LIAO, Xiangyun, et al. 2018. Fully automatic segmentation of the left ventricle using multi-scale fusion learning. In: 2018 24th International Conference on Pattern Recognition (ICPR). IEEE, 2018. p. 3838-3843. [CrossRef]
- WIJNHOUT, J. , HENDRIKSEN, Dennis, ASSEN, H., et al. 2009. LV challenge LKEB contribution: fully automated myocardial contour detection. The MIDAS journal, vol. 43,2009, p. 2.
- Buettner, Ricardo, and Marc Schunter. 2019.Efficient machine learning based detection of heart disease. In 2019 IEEE international conference on E-health networking, application and services (HealthCom), 2019. pp. 1-6. [CrossRef]
- Grothues, Frank, James C. Moon, Nicholas G. Bellenger, Gillian S. Smith, Helmut U. Klein, and Dudley J. Pennell. 2004. "Interstudy reproducibility of right ventricular volumes, function, and mass with cardiovascular magnetic resonance. American heart journal 147, no. 2 (2004). 218-223. [CrossRef]
- BUDAI, Adam, SUHAI, Ferenc I., CSORBA, Kristof, et al. 2020. Fully automatic segmentation of right and left ventricle on short-axis cardiac MRI images. Computerized Medical Imaging and Graphics vol. 85,2020, p. 101786. [CrossRef]
- PETITJEAN, Caroline et DACHER, Jean-Nicolas. A review of segmentation methods in short axis cardiac MR images.2011. Medical image analysis, vol. 15, no 2,2011, p. 169-184. [CrossRef]
- Weingärtner, Sebastian, Mehmet Akçakaya, Sébastien Roujol, Tamer Basha, Cory Tschabrunn, Sophie Berg, Elad Anter, and Reza Nezafat. 2015. Free-breathing combined three-dimensional phase sensitive late gadolinium enhancement and T1 mapping for myocardial tissue characterization. Magnetic resonance in medicine 74, no. 4 (2015): 1032-1041. [CrossRef]
- Bustamante, Mariana, Sven Petersson, Jonatan Eriksson, Urban Alehagen, Petter Dyverfeldt, Carl-Johan Carlhäll, and Tino Ebbers. 2015. Atlas-based analysis of 4D flow CMR: automated vessel segmentation and flow quantification.Journal of Cardiovascular Magnetic Resonance 17, no. 1 (2015): 1-12. [CrossRef]
- Andreopoulos, Alexander, and John K. Tsotsos. 2008. Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI. Medical image analysis 12, no. 3 (2008). 335-357. [CrossRef]
- Shaaf, Zakarya Farea, Muhammad Mahadi Abdul Jamil, Radzi Ambar, Ahmed Abdu Alattab, Anwar Ali Yahya, and Yousef Asiri. 2022. Automatic left ventricle segmentation from short-axis cardiac mri images based on fully convolutional neural network. Diagnostics 12, no. 2 (2022). 414. [CrossRef]
- Krasnobaev, Arseny, and Andrey Sozykin. 2016. An overview of techniques for cardiac left ventricle segmentation on short-axis MRI. (2016).
- Ammari, Asma, Ramzi Mahmoudi, Badii Hmida, Rachida Saouli, and Mohamed Hédi Bedoui. 2021. A review of approaches investigated for right ventricular segmentation using short-axis cardiac MRI. IET Image Processing 15, no. 9 (2021): 1845-1868. [CrossRef]
- Shoaib, Muhammad Ali, Joon Huang Chuah, Raza Ali, Khairunnisa Hasikin, Azira Khalil, Yan Chai Hum,
Yee Kai Tee, Samiappan Dhanalakshmi, and Khin Wee Lai. 2023. An Overview of Deep Learning Methods
for Left Ventricle Segmentation. Computational intelligence and neuroscience 2023 (2023). 2023.
- Wang, Zhongrong, Lipeng Xie, and Jin Qi. 2020. Dynamic pixel-wise weighting-based fully convolutional neural networks for left ventricle segmentation in short-axis MRI. Magnetic Resonance Imaging 66 (2020). 131-140. [CrossRef]
- Suinesiaputra, Avan, Brett R. Cowan, Ahmed O. Al-Agamy, Mustafa A. Elattar, Nicholas Ayache, Ahmed S. Fahmy, Ayman M. Khalifa et al. 2014. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Medical image analysis 18, no. 1 (2014). 50-62. [CrossRef]
- Tavakoli, Vahid, and Amir A. Amini. 2013.A survey of shaped-based registration and segmentation techniques for cardiac images. Computer Vision and Image Understanding 117, no. 9 (2013): 966-989. [CrossRef]
- Chen, Yunmei, Hemant D. Tagare, Sheshadri Thiruvenkadam, Feng Huang, David Wilson, Kaundinya S. Gopinath, Richard W. Briggs, and Edward A. Geiser. 2002. Using prior shapes in geometric active contours in a variational framework. International Journal of Computer Vision 50 (2002). 315-328. [CrossRef]
- Liu, Yu, Gabriella Captur, James C. Moon, Shuxu Guo, Xiaoping Yang, Shaoxiang Zhang, and Chunming Li. 2016. Distance regularized two level sets for segmentation of left and right ventricles from cine-MRI. Magnetic resonance imaging 34, no. 5 (2016). 699-706. [CrossRef]
- Li, Chunming, Chenyang Xu, Changfeng Gui, and Martin D. Fox. 2010. Distance regularized level set evolution and its application to image segmentation. IEEE transactions on image processing 19, no. 12 (2010). 3243-3254. [CrossRef]
- Jia Wu, Katharine G Brigham, Marc A Simon, and John C Brigham. 2014. An implementation of independent component analysis for 3d statistical shape analysis. Biomedical Signal Processing and Control. (2014). 13:345–356. [CrossRef]
- Miao, Chuang, and Hengyong Yu. 2015. A General-Thresholding Solution for l p (0. [CrossRef]
- Zheng, Y. , Georgescu, B., Vega-Higuera, F. and Comaniciu, D., 2009, March. Left ventricle endocardium segmentation for cardiac CT volumes using an optimal smooth surface. In Medical Imaging 2009: Image Processing (Vol. 7259, pp. 1249-1259). SPIE. [CrossRef]
- Shahzad, R. , Tao, Q., Dzyubachyk, O., Staring, M., Lelieveldt, B.P. and van der Geest, R.J. 2017. Fully-automatic left ventricular segmentation from long-axis cardiac cine MR scans. Medical image analysis, 39, (2017). pp.44-55. [CrossRef]
- E. Shelhamer, J. Long, and T. Darrell. Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O. , Fischer, P. and Brox, T. 2015. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, -9, 2015, Proceedings, Part III 18 (pp. 234-241). Springer International Publishing. 5 October. [CrossRef]
- Hung, Wei-Chih, Yi-Hsuan Tsai, Yan-Ting Liou, Yen-Yu Lin, and Ming-Hsuan Yang. 2018. Adversarial learning for semi-supervised semantic segmentation. arXiv:1802.07934.(2018). [CrossRef]
- Souly, Nassim., Spampinato, Concetto., Shah, Mubarak. 2017. Semi supervised semantic segmentation using generative adversarial network. IEEE International Conference onComputer Vision, ICCV 2017, Venice, Italy, –29, 2017, 5689–5697. 22 October.
- Kamnitsas, Konstantinos, Christian Baumgartner, Christian Ledig, Virginia Newcombe, Joanna Simpson, Andrew Kane, David Menon et al. 2017. Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In Information Processing in Medical Imaging: 25th International Conference, IPMI 2017, Boone, NC, USA, -30, 2017, Proceedings 25, pp. 597-609. Springer International Publishing, 2017. 25 June.
- Yu C, Yan Y, Zhao S, Zhang Y. 2020. Pyramid feature adaptation for semi-supervised cardiac bi-ventricle segmentation. Computerized Medical Imaging and Graphics. 2020 Apr 1;81:101697. [CrossRef]
- TAN, Li Kuo, LIEW, Yih Miin, LIM, Einly, et al. 2017. Convolutional neural network regression for short-axis left ventricle segmentation in cardiac cine MR sequences. Medical image analysis, vol. 39. 2017. p. 78-86. [CrossRef]
- XU, Shengzhou, LU, Haoran, CHENG, Shiyu, et al. 2022. Left Ventricle Segmentation in Cardiac MR Images via an Improved ResUnet. International Journal of Biomedical Imaging, 2022, vol. 2022. [CrossRef]
- H. Wu, X. H. Wu, X. Lu, B. Lei, and Z. Wen. 2021. Automated left ventricular segmentation from cardiac magnetic resonance images via adversarial learning with multi-stage pose estimation network and co-discriminator,” Medical Image Analysis, vol. 68, no. 11, article 101891, 2021. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).