Submitted:
22 August 2023
Posted:
24 August 2023
You are already at the latest version
Abstract
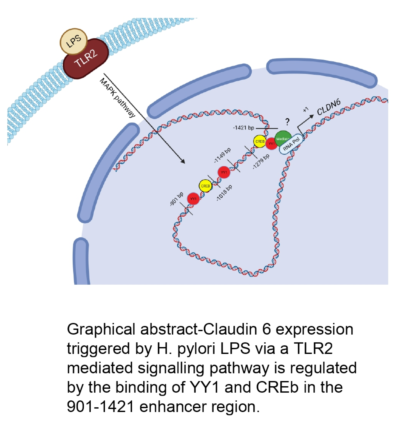
Keywords:
1. Introduction
2. Results
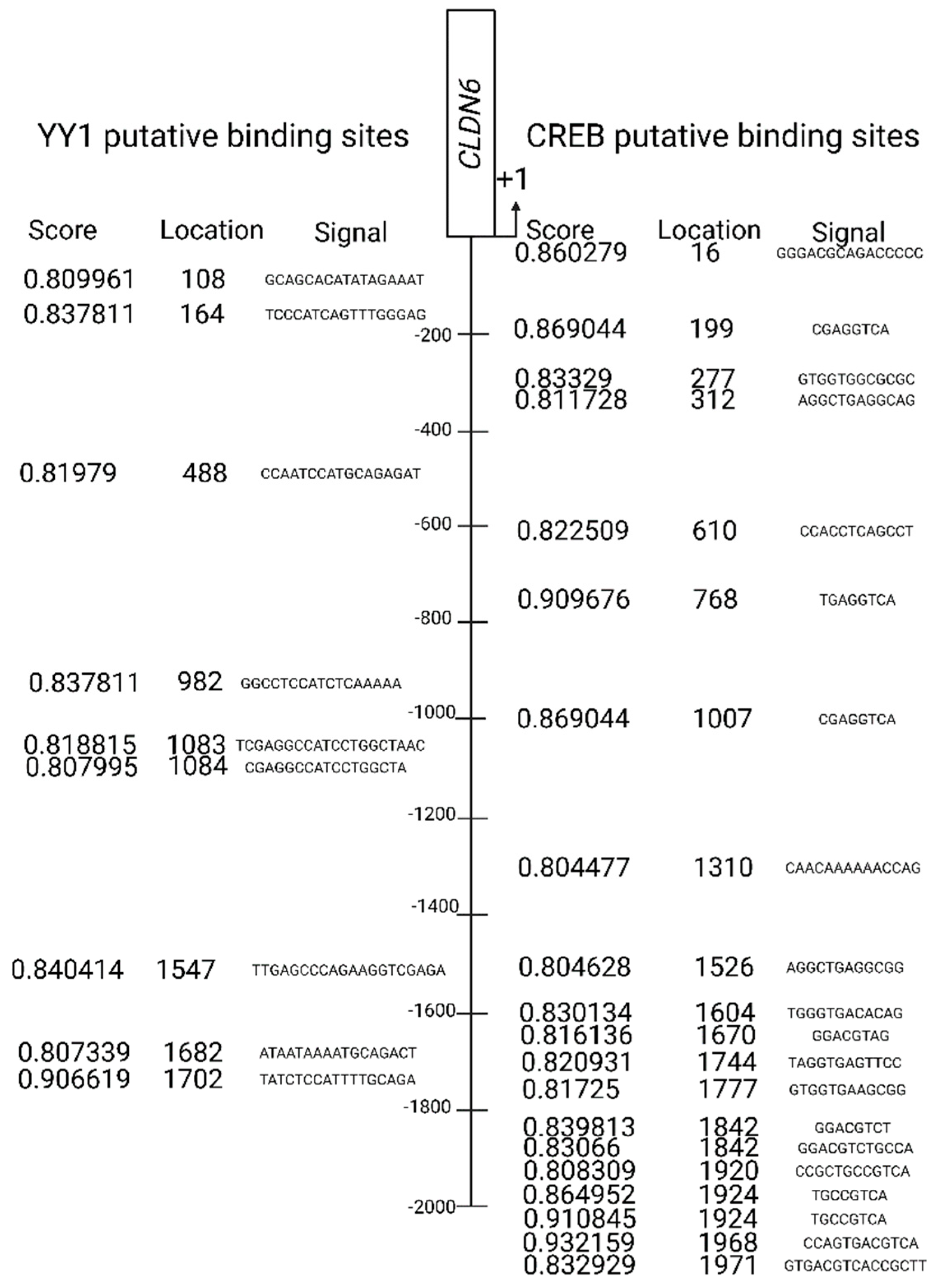
2.1. Bioinformatic analysis.
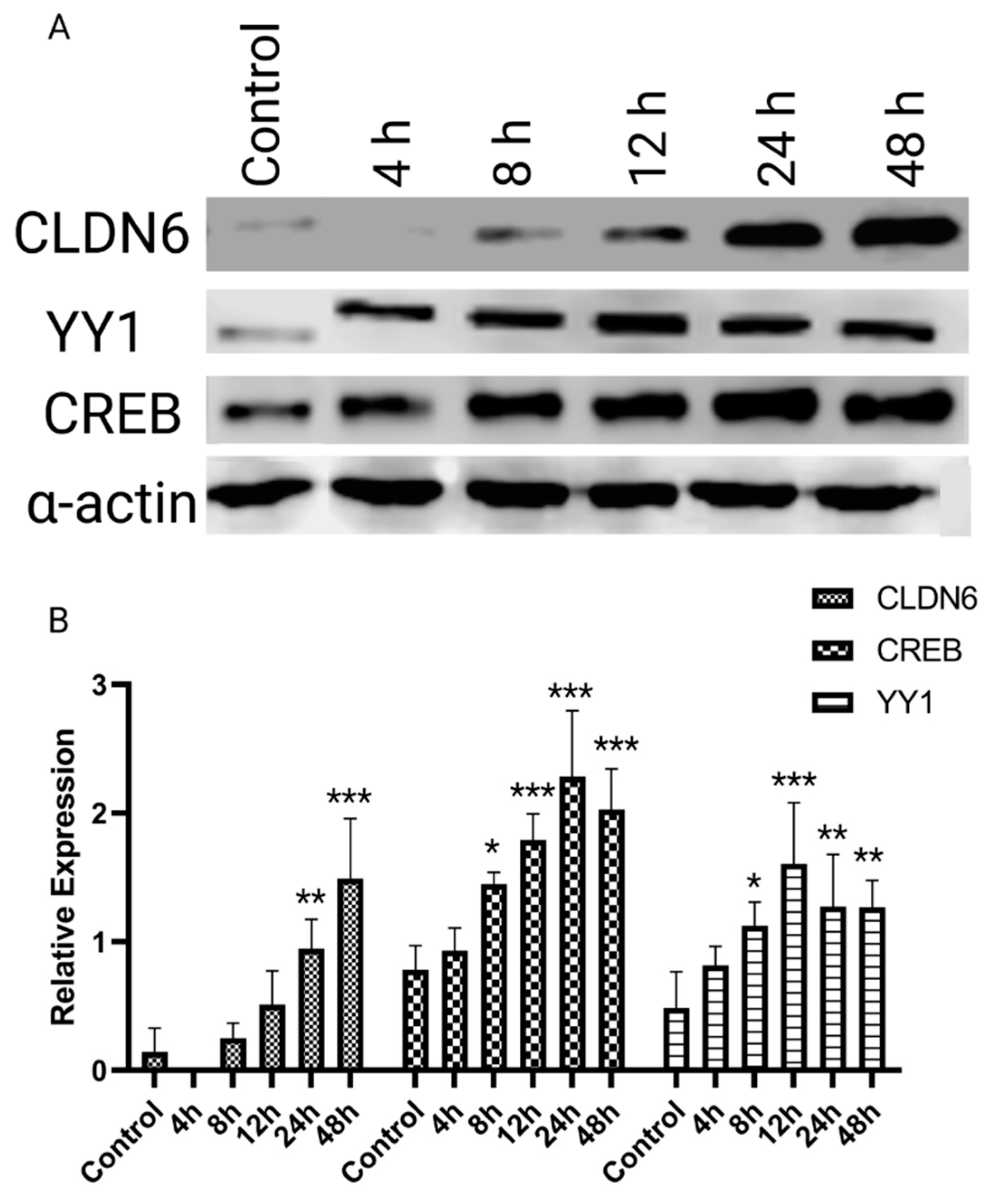
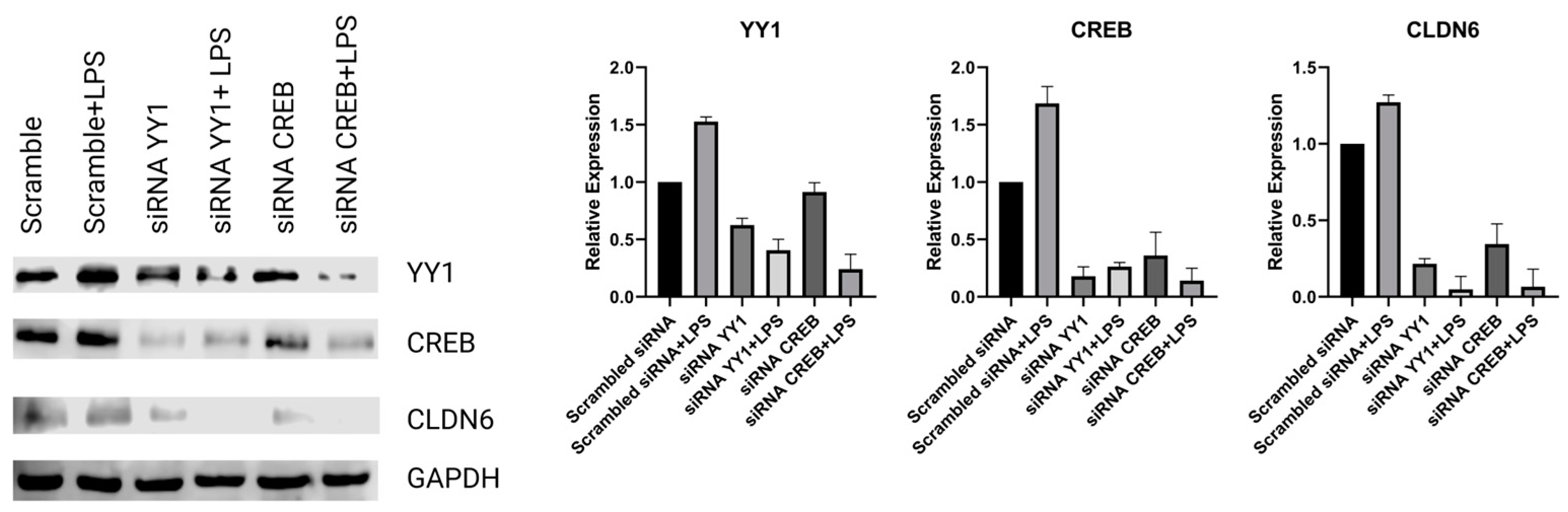
2.2. H. pylori LPS and YY1/CREB expression
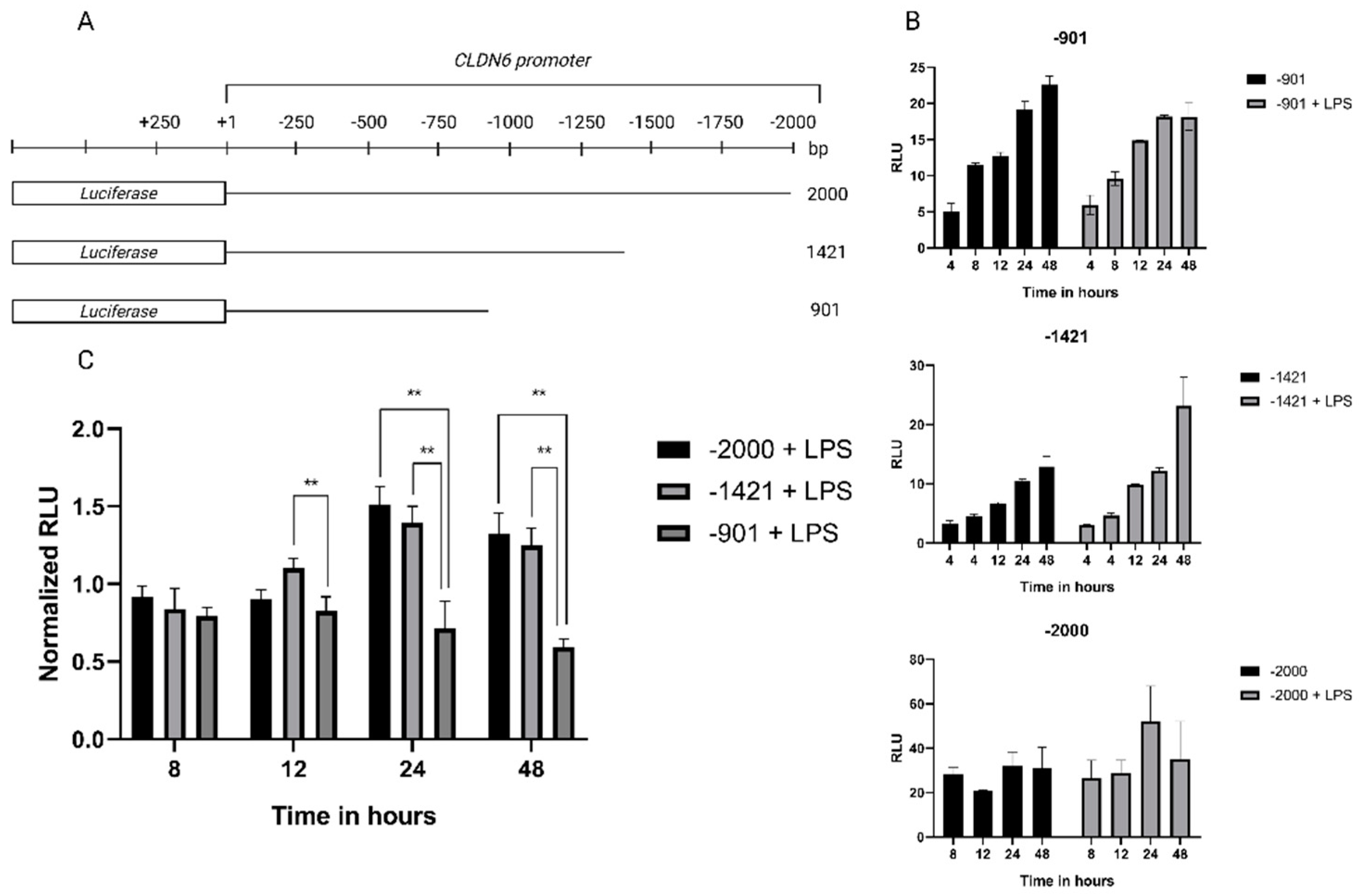
2.3. Cloning of claudin 6 promoter
2.4. Regulation of Claudin 6 expression
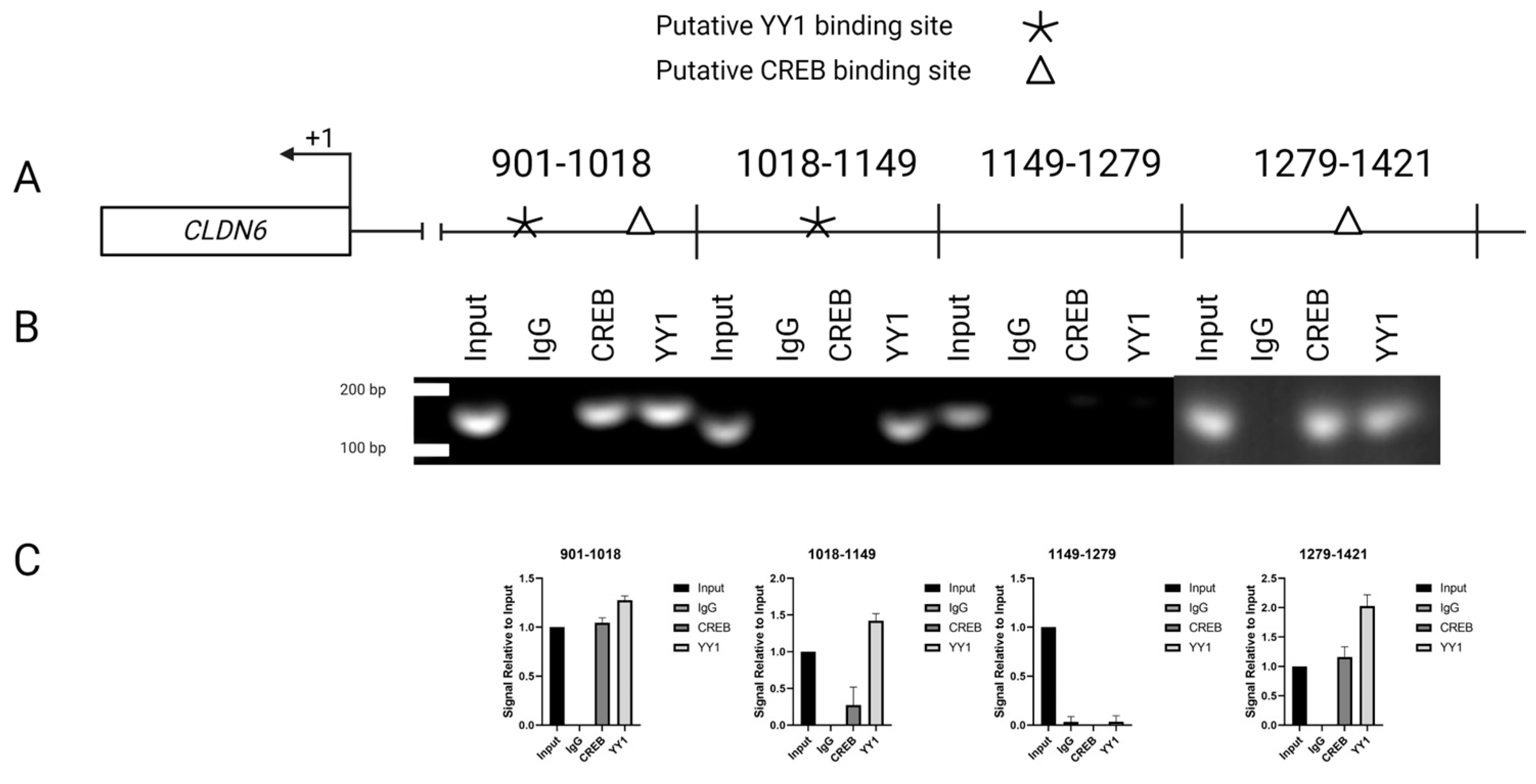
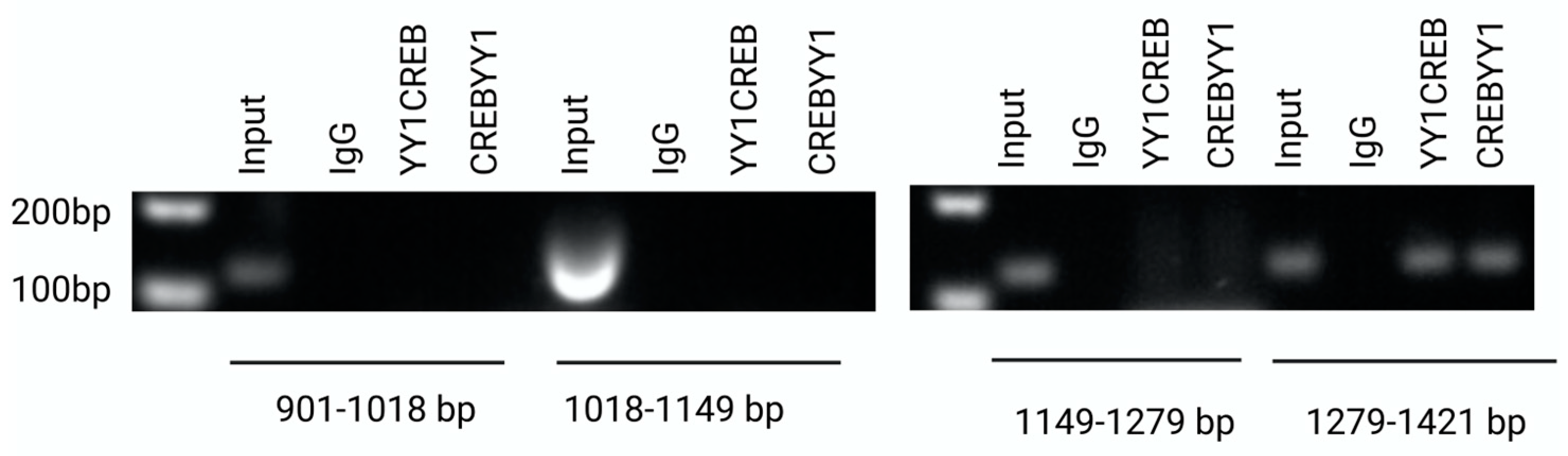
2.5. Interaction of YY1/CREB in Claudin 6 promoter
3. Discussion
4. Materials and Methods
Reagents
Cell culture
Bioinformatic analysis
YY1 and CREB silencing
Preparation of LPS Helicobacter pylori and exposure to AGS cells.
Protein extraction
Western blot
Plasmid construction and luciferase assay
Chromatin immunoprecipitation
Statistical analysis.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Moonwiriyakit, A.; Pathomthongtaweechai, N.; Steinhagen, P.R.; Chantawichitwong, P.; Satianrapapong, W.; Pongkorpsakol, P. Tight junctions: from molecules to gastrointestinal diseases. Tissue Barriers 2023, 11, 2077620. [Google Scholar] [CrossRef]
- Morita, K.; Furuse, M.; Fujimoto, K.; Tsukita, S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA 1999, 96, 511–516. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Claudins and epithelial paracellular transport. Annu Rev Physiol 2006, 68, 403–429. [Google Scholar] [CrossRef]
- Krause, G.; Winkler, L.; Mueller, S.L.; Haseloff, R.F.; Piontek, J.; Blasig, I.E. Structure and function of claudins. Biochim Biophys Acta 2008, 1778, 631–645. [Google Scholar] [CrossRef]
- Colegio, O.R.; Van Itallie, C.; Rahner, C.; Anderson, J.M. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol 2003, 284, C1346–C1354. [Google Scholar] [CrossRef] [PubMed]
- Turksen, K.; Troy, T.C. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev Dyn 2001, 222, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Nudel, N.; Benvenisty, N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat Commun 2013, 4, 1992. [Google Scholar] [CrossRef]
- Du, H.; Yang, X.; Fan, J.; Du, X. Claudin 6: Therapeutic prospects for tumours, and mechanisms of expression and regulation (Review). Mol Med Rep 2021, 24. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Uppada, S.; Achkar, I.W.; Hashem, S.; Yadav, S.K.; Shanmugakonar, M.; Al-Naemi, H.A.; Haris, M.; Uddin, S. Tight Junction Proteins and Signaling Pathways in Cancer and Inflammation: A Functional Crosstalk. Front Physiol 2018, 9, 1942. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Zhang, T.; Han, Z.G.; Shan, L. Low claudin-6 expression correlates with poor prognosis in patients with non-small cell lung cancer. Onco Targets Ther 2015, 8, 1971–1977. [Google Scholar] [CrossRef]
- Ito, Y.; Takasawa, A.; Takasawa, K.; Murakami, T.; Akimoto, T.; Kyuno, D.; Kawata, Y.; Shano, K.; Kirisawa, K.; Ota, M.; et al. Aberrant expression of claudin-6 contributes to malignant potentials and drug resistance of cervical adenocarcinoma. Cancer Sci 2022, 113, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jin, X.; Lin, D.; Liu, Z.; Zhang, X.; Lu, Y.; Liu, Y.; Wang, M.; Yang, M.; Li, J.; et al. Clinicopathologic significance of claudin-6, occludin, and matrix metalloproteinases -2 expression in ovarian carcinoma. Diagn Pathol 2013, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Xu, X.; Liu, Z.; Zhang, T.; Zhang, X.; Wang, L.; Wang, M.; Liu, Y.; Lu, Y.; Liu, Y.; et al. Apoptosis signal-regulating kinase 1 is associated with the effect of claudin-6 in breast cancer. Diagn Pathol 2012, 7, 111. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, X.; Li, Y.; Ruan, Y.; Lu, Y.; Yang, M.; Lin, D.; Song, P.; Guo, Y.; Zhao, S.; et al. DNA methylation of claudin-6 promotes breast cancer cell migration and invasion by recruiting MeCP2 and deacetylating H3Ac and H4Ac. J Exp Clin Cancer Res 2016, 35, 120. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, Y.; Shen, X.; Ruan, Y.; Lu, Y.; Jin, X.; Song, P.; Guo, Y.; Zhang, X.; Qu, H.; et al. CLDN6 promotes chemoresistance through GSTP1 in human breast cancer. J Exp Clin Cancer Res 2017, 36, 157. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, X.; Liu, Z.; Liu, Q.; Wang, L.; Lu, Y.; Liu, Y.; Wang, M.; Yang, M.; Jin, X.; et al. The distinct expression patterns of claudin-2, -6, and -11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol 2013, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Kohmoto, T.; Masuda, K.; Shoda, K.; Takahashi, R.; Ujiro, S.; Tange, S.; Ichikawa, D.; Otsuji, E.; Imoto, I. Claudin-6 is a single prognostic marker and functions as a tumor-promoting gene in a subgroup of intestinal type gastric cancer. Gastric Cancer 2020, 23, 403–417. [Google Scholar] [CrossRef]
- Zavala-Zendejas, V.E.; Torres-Martinez, A.C.; Salas-Morales, B.; Fortoul, T.I.; Montano, L.F.; Rendon-Huerta, E.P. Claudin-6, 7, or 9 overexpression in the human gastric adenocarcinoma cell line AGS increases its invasiveness, migration, and proliferation rate. Cancer Invest 2011, 29, 1–11. [Google Scholar] [CrossRef]
- Torres-Martinez, A.C.; Gallardo-Vera, J.F.; Lara-Holguin, A.N.; Montano, L.F.; Rendon-Huerta, E.P. Claudin-6 enhances cell invasiveness through claudin-1 in AGS human adenocarcinoma gastric cancer cells. Exp Cell Res 2017, 350, 226–235. [Google Scholar] [CrossRef]
- Medrano-Gonzalezl, P.A.; Cruz-Villegas, F.; Alarcon Del Carmen, A.; Montano, L.F.; Rendon-Huerta, E.P. Claudin-6 increases SNAI1, NANOG and SOX2 gene expression in human gastric adenocarcinoma AGS cells. Mol Biol Rep 2022, 49, 11663–11674. [Google Scholar] [CrossRef]
- Chavarria-Velazquez, C.O.; Torres-Martinez, A.C.; Montano, L.F.; Rendon-Huerta, E.P. TLR2 activation induced by H. pylori LPS promotes the differential expression of claudin-4, -6, -7 and -9 via either STAT3 and ERK1/2 in AGS cells. Immunobiology 2018, 223, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Lu, H.; Wu, J.Y.; Ohno, T.; Wu, M.J.; Genta, R.M.; Graham, D.Y.; Yamaoka, Y. Pattern of transcription factor activation in Helicobacter pylori-infected Mongolian gerbils. Gastroenterology 2007, 132, 1024–1038. [Google Scholar] [CrossRef] [PubMed]
- Andrisani, O.M. CREB-mediated transcriptional control. Crit Rev Eukaryot Gene Expr 1999, 9, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Guo, Y.; Li, Y.; Ruan, Y.; Zhang, M.; Jin, X.; Yang, M.; Lu, Y.; Song, P.; Zhao, S.; et al. Bioinformatic analysis reveals potential properties of human Claudin-6 regulation and functions. Oncol Rep 2017, 38, 875–885. [Google Scholar] [CrossRef]
- Resende, C.; Regalo, G.; Duraes, C.; Pinto, M.T.; Wen, X.; Figueiredo, C.; Carneiro, F.; Machado, J.C. Interleukin-1B signalling leads to increased survival of gastric carcinoma cells through a CREB-C/EBPbeta-associated mechanism. Gastric Cancer 2016, 19, 74–84. [Google Scholar] [CrossRef]
- Kim, S.; Yu, N.K.; Kaang, B.K. CTCF as a multifunctional protein in genome regulation and gene expression. Exp Mol Med 2015, 47, e166. [Google Scholar] [CrossRef]
- Yao, Y.L.; Yang, W.M.; Seto, E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol 2001, 21, 5979–5991. [Google Scholar] [CrossRef]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588. [Google Scholar] [CrossRef]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front Oncol 2019, 9, 1230. [Google Scholar] [CrossRef]
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription 2010, 1, 81–84. [Google Scholar] [CrossRef]
- Chen, F.; Sun, H.; Zhao, Y.; Wang, H. YY1 in Cell Differentiation and Tissue Development. Crit Rev Oncog 2017, 22, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Bonavida, B.; Baritaki, S. The novel role of Yin Yang 1 in the regulation of epithelial to mesenchymal transition in cancer via the dysregulated NF-kappaB/Snail/YY1/RKIP/PTEN Circuitry. Crit Rev Oncog 2011, 16, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Engel, D.A. Adenovirus E1A243 disrupts the ATF/CREB-YY1 complex at the mouse c-fos promoter. J Virol 1995, 69, 7402–7409. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, J.; Xue, C.; Zhou, X.; Chen, S.; Zheng, L.; Duan, Y.; Deng, H.; Xiong, W.; Tang, F.; et al. Dissecting the roles and clinical potential of YY1 in the tumor microenvironment. Front Oncol 2023, 13, 1122110. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.H.; Mai, R.T.; Lee, Y.H. Transcription factor YY1 and its associated acetyltransferases CBP and p300 interact with hepatitis delta antigens and modulate hepatitis delta virus RNA replication. J Virol 2008, 82, 7313–7324. [Google Scholar] [CrossRef]
- Bhaskar Rao, D.; Panneerpandian, P.; Balakrishnan, K.; Ganesan, K. YY1 regulated transcription-based stratification of gastric tumors and identification of potential therapeutic candidates. J Cell Commun Signal 2021, 15, 251–267. [Google Scholar] [CrossRef]
- Tsunoda, T.; Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef]
- Gunzel, D.; Fromm, M. Claudins and other tight junction proteins. Compr Physiol 2012, 2, 1819–1852. [Google Scholar] [CrossRef]
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem Sci 2019, 44, 141–152. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. Phosphorylation of tight junction transmembrane proteins: Many sites, much to do. Tissue Barriers 2018, 6, e1382671. [Google Scholar] [CrossRef]
- Jimenez, F.R.; Lewis, J.B.; Belgique, S.T.; Wood, T.T.; Reynolds, P.R. Developmental lung expression and transcriptional regulation of claudin-6 by TTF-1, Gata-6, and FoxA2. Respir Res 2014, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Guo, Y.; Jin, Q.; Qu, H.; Qi, D.; Song, P.; Zhang, X.; Wang, X.; Xu, W.; Dong, Y.; et al. A SUMOylation-dependent HIF-1alpha/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis. J Exp Clin Cancer Res 2020, 39, 42. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, F.R.; Lewis, J.B.; Belgique, S.T.; Milner, D.C.; Lewis, A.L.; Dunaway, T.M.; Egbert, K.M.; Winden, D.R.; Arroyo, J.A.; Reynolds, P.R. Cigarette smoke and decreased oxygen tension inhibit pulmonary claudin-6 expression. Exp Lung Res 2016, 42, 440–452. [Google Scholar] [CrossRef]
- Qu, H.; Jin, Q.; Quan, C. CLDN6: From Traditional Barrier Function to Emerging Roles in Cancers. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Li, M.; Xiang, R.; Zhou, X.; Zhu, L.; Zhai, Y. Expression of CLDN6 in tissues of gastric cancer patients: Association with clinical pathology and prognosis. Oncol Lett 2019, 17, 4621–4625. [Google Scholar] [CrossRef]
- Shi, Y.; Seto, E.; Chang, L.S.; Shenk, T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 1991, 67, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Seto, E.; Shi, Y.; Shenk, T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature 1991, 354, 241–245. [Google Scholar] [CrossRef]
- Khachigian, L.M. The Yin and Yang of YY1 in tumor growth and suppression. Int J Cancer 2018, 143, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Kwok, R.P.; Lundblad, J.R.; Chrivia, J.C.; Richards, J.P.; Bachinger, H.P.; Brennan, R.G.; Roberts, S.G.; Green, M.R.; Goodman, R.H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 1994, 370, 223–226. [Google Scholar] [CrossRef]
- Steven, A.; Seliger, B. Control of CREB expression in tumors: from molecular mechanisms and signal transduction pathways to therapeutic target. Oncotarget 2016, 7, 35454–35465. [Google Scholar] [CrossRef]
- Kang, W.; Tong, J.H.; Chan, A.W.; Zhao, J.; Dong, Y.; Wang, S.; Yang, W.; Sin, F.M.; Ng, S.S.; Yu, J.; et al. Yin Yang 1 contributes to gastric carcinogenesis and its nuclear expression correlates with shorter survival in patients with early stage gastric adenocarcinoma. J Transl Med 2014, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Kobayashi, R.; Iwai, Y.; Noda, K.; Yamazaki, M.; Kurita-Ochiai, T.; Yoshimura, A.; Ganss, B.; Ogata, Y. C/EBPbeta and YY1 bind and interact with Smad3 to modulate lipopolysaccharide-induced amelotin gene transcription in mouse gingival epithelial cells. FEBS Open Bio 2019, 9, 276–290. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Xu, Y.; Wang, Y.; Wang, F.; Zhang, Q.; Ni, C.; Zhen, Y.; Xu, R.; Liu, Q.; et al. Enterobacterial LPS-inducible LINC00152 is regulated by histone lactylation and promotes cancer cells invasion and migration. Front Cell Infect Microbiol 2022, 12, 913815. [Google Scholar] [CrossRef] [PubMed]
- Noda, K.; Yamazaki, M.; Iwai, Y.; Matsui, S.; Kato, A.; Takai, H.; Nakayama, Y.; Ogata, Y. IL-1beta and TNF-alpha regulate mouse amelotin gene transcription in gingival epithelial cells. J Oral Sci 2018, 60, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lu, K.; Hou, Y.; You, Z.; Shu, C.; Wei, X.; Wu, T.; Shi, N.; Zhang, G.; Wu, J.; et al. YY1 complex in M2 macrophage promotes prostate cancer progression by upregulating IL-6. J Immunother Cancer 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Furlong, E.E. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012, 13, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Mercola, M.; Wang, X.F.; Olsen, J.; Calame, K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science 1983, 221, 663–665. [Google Scholar] [CrossRef]
- Panigrahi, A.; O'Malley, B.W. Mechanisms of enhancer action: the known and the unknown. Genome Biol 2021, 22, 108. [Google Scholar] [CrossRef]
- Wicks, K.; Knight, J.C. Transcriptional repression and DNA looping associated with a novel regulatory element in the final exon of the lymphotoxin-beta gene. Genes Immun 2011, 12, 126–135. [Google Scholar] [CrossRef]
- Lee, J.S.; Galvin, K.M.; See, R.H.; Eckner, R.; Livingston, D.; Moran, E.; Shi, Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev 1995, 9, 1188–1198. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



