1. Introduction
Infections caused by pathogens cause high mortality and worsen the quality of life [
1,
2,
3,
4,
5,
6,
7]. There is a development of resistant strains of bacteria that cannot be treated with classical therapy options. Among other intra-macrophage latent or "dormant" bacteria. Existing methods of early diagnosis and therapy in some severe cases may be ineffective. This is especially true of secondary atypical pneumonia (caused by
Mycoplasma pneumoniae and
Chlamydia pneumoniae), complicated bronchitis, bronchial asthma, where viral and bacterial infection is complicated by an immune response, etc. Targeted delivery to macrophages is a promising approach to improve the effectiveness of drug therapy for respiratory diseases the driver of which are macrophages: pneumonia, tuberculosis, etc. [
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23,
24]. The CD206 receptor is expressed only on activated macrophages, while there is more of it on M2 than on M1 [
2,
25]. CD206 is a transmembrane protein, a C-type lectin that recognizes mannose, fucose and
N-acetylglucosamine residues of oligosaccharides of bacteria and pathogens for subsequent absorption of this pathogen by the macrophage [
8,
14,
15,
16,
23,
26,
27]. Therefore, an effective strategy for analyzing macrophages interaction with pathogens and biomimetic systems are necessary. The perspective idea is to implement biomimetics of a polymer with carbohydrate labels as models for a pathogenic microorganism to develop and test drug delivery systems by targeting alveolar macrophages. At the same time, macrophages are interesting objects from the point of view of the diagnosis of infectious diseases: by their polarization status and phenotype, one can judge the etiology of the disease and choose the optimal treatment strategy.
Macrophages are part of innate immunity, the first frontier of defense against pathogens, so their activation is one of the key factors in a number of infectious diseases. For example, macrophages are important for the elimination of pathogens:
Mycobacterium tuberculosis, HIV and parasites such as leishmania and trypanosome, causing “sleeping sickness”, etc [
3]. At the same time, the type of macrophage polarization (M1 or M2) plays a key role in the course and features of the disease. Pro-inflammatory macrophages M1 are important for protection against bacterial infections, but bacteria can secrete substances that repolarize macrophages into anti-inflammatory M2, which is a condition for the formation of dormant resistant infection [
28,
29]. Repolarization of macrophages in tumors allows changing the microenvironment from “cold” to “hot”, thereby activating the immune response against the tumor [
30,
31,
32]. Excessive proinflammatory activity of macrophages can also be dangerous (for example, cytokine storm in COVID-19) [
10]. Thus, for medical and bioanalytical applications, it is important to create methods for express analysis of activated macrophages. On the other hand, determination of the effectiveness of binding of specific ligands to mannose receptors of macrophages is necessary for the development of effective delivery systems of targeted action to increase the effectiveness of therapy.
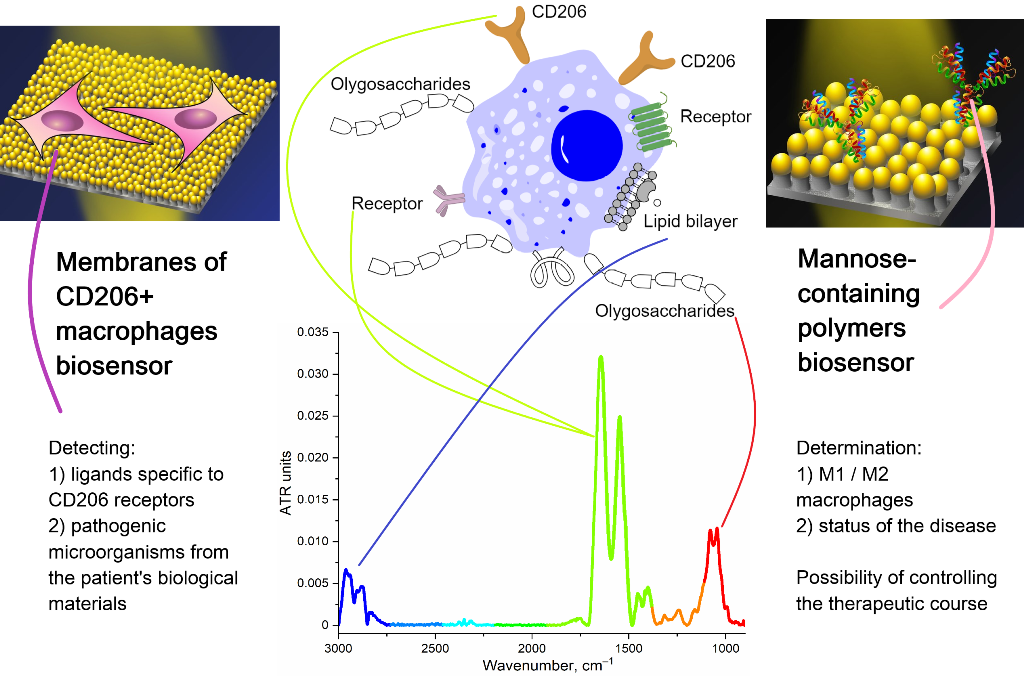
We offer 3 analytical applications of CD206 receptor interactions. The first one is targeted delivery of pharmaceutical formulations to macrophages, which we have considered in detail in a series of papers [
14,
15,
16,
33,
34,
35,
36,
37,
38]. The technique is rapid and effective for testing and optimizing high-affinity specific ligands to alveolar macrophages.
The second application is the development of biosensors based on CD206+ macrophage membranes for use them as a selective recognizer of carbohydrate fragments of pathogenic microorganisms. Macrophage-based biosensors may provide promising methods for early diagnosis of diseases, determination of the intensity of inflammation and the efficiency of the treatment. So, a sensitive and simple electrochemical sensor of mouse macrophage cells was developed for early detection of lipopolysaccharides to assess the toxicity of pathogenic bacteria [
39]. However, for such methods, significant signal distortion is possible because of use of a non-specific signal (impedance).
Presented here CD206-targeting system binds exclusively receptors that are expressed only by disease-causing activated macrophages. Thus, a biosensor with deposited CD206+ macrophage membranes have a number of unique advantages: 1) high selectivity to oligomannosidic ligands (Kd 10–7-10–9 M), 2) expressiveness of the analysis, 3) the possibility of multiple use, 4) stability during storage.
The third application is the development of biosensors based on specific polymers ligands to CD206 macrophage receptors to diagnose the status of polarization and activation of macrophages in biological fluids (for example, bronchoalveolar lavage – BAL [
40,
41,
42,
43,
44]) to monitor the course of treatment and the ability to adjust the course of therapy. Polymers with oligo- and polymannosidic fragments can be used to detect activated macrophages and determine their M1-M2 polarization. These are prerequisites for the development of personalized medicine.
Thus, the main idea of the work is to develop biosensors for analyzing the effectiveness of ligand binding with polarized macrophages. To do this, we synthesized an IR marker that specifically binds to the mannose receptors of macrophages. By displacing such an IR marker, the affinity for CD206 of various polymer oligosaccharide ligands with different molecular architecture can be determined. We tested our specific ligands on living macrophage cells and compared data on binding specificity on macrophage membranes. Our second bioanalytic system consisting in the determination of the polymers binding degree with BAL (contain activated macrophages) and NPL (which do not contain activated macrophages).
This work opens up experimental foundations for the creation of highly effective biosensors for the early diagnosis of macrophage-associated diseases, for monitoring the course of therapy and personalized medicine.
2. Materials and Methods
2.1. Reagents
Mannan (46 kDa), polyethyleneimine 1.8 kDa (PEI1.8), 2-hydroxypropyl-β-cyclodextrin (HPCD), FITC, D-mannose, D-galactose, spermine, putrescine, NaBH3CN, activated PEG 5 kDa (N-succinimidyl ester of mono-methoxy poly(ethylene glycol)) were purchased from Sigma Aldrich (St. Louis, MI, USA). Mannotriose-di-(N-acetyl-D-glucosamine) (triMan) was obtained from Dayang Chem (Hangzhou) Co., Ltd. Carbonyldiimidazole (CDI) was obtained from GL Biochem Ltd. (Shanghai, China) via an intermediary Himprocess (Moscow, Russia). Other chemicals: salts and acids were from Reakhim Production (Moscow, Russia).
2.2. Polymer synthesis and characterization
Activated HPCD was obtained as described earlier [
33,
45] by reaction of HPCD OH-groups with carbonyldiimidazole in DMSO.
Mannan-sp-HPCD. A 100 mg mannan polymer was dissolved in 10 mL of 1 mM HCl, then 12 mg of potassium periodate was added. The mixture was incubated for 30 min at 40 °C followed by dialysis (cut-off 3 kDa) against water for 2 h. 10 mg of spermine and 5 mg of NaBH3CN was added to the mixture at controlled pH 4. Then mixture was incubated for 6 h at 50 °C followed by purification by dialysis against water (cut-off 3.5 kDa) for 6 h. Additionally, 35 mg (per HPCD) of activated HPCD was added to the mixture, and incubation was continued at the same temperature for 6 h. The final purification of samples was performed using 24 h dialysis (cut-off 6–8 kDa).
HPCD-PEI-X. FITC labelling of PEI was performed at first step for parts of the samples, the rest of the polymers were unlabeled.
To the aqueous solution of PEI (2 mL of 50 mg/mL in 0.01M HCl), a solution of FITC (10 mg in 1 mL DMSO) was added drop by drop with stirring; the pH was brought to 8. The mixture was incubated at 50 °C for 2 h, followed by purification by dialysis against water (cut-off 1 kDa) for 12 h. Then, to the PEI-FITC solution (or free PEI) activated HPCD was added at 6-fold molar excess. The mixture was incubated at 40 °C for 12 h at pH 7.4 (PBS) followed by dialysis against water (6-8 kDa) for 6 h. The sample was divided into three equal parts, to which an aqueous solution of: 1) mannose (15-fold times molar excess over PEI), 2) triMan (10-fold molar excess over PEI), 3) galactose (15-fold molar excess over PEI) were added, respectively. NaBH3CN has been added to each of them. The samples were incubated for a 24 h at pH 5 and 50 °C followed by purification using dialysis (cut-off 6-8 kDa) for 12 h.
The final purification of the samples was performed by HPLC gel filtration in a Knauer chromatography system (Knauer, Berlin, Germany) on Diasfer-110-C18 column (BioChemMack, Moscow, Russia) [
15].
All samples were freeze-dried for two days at –60 °C (Edwards 5, BOC Edwards, UK). Modification degree was calculated using NH2-group titration with 2,4,6-trinitrobenzenesulfonic acid.
Determination of the hydrodynamic diameter of the synthesized polymers was carried out by NTA (Nanoparticle Tracking Analysis) using a Nanosight LM10-HS device (Great Britain) [
46].
2.3. IR marker synthesis and characterization
Samples of 20 mg of triMan, 7 mg of putrescine, 5 mg of NaBH3CN are mixed and dissolved in 1 ml of 1 mM HCl. Then the mixture was incubated at 50 °C for 6 hours, followed by purification by dialysis against water (cut-off 1 kDa). 70 mg of activated PEG in PBS (pH 7.4) was added to the triMan-putrescine product, the mixture was incubated for 12 hours at 50 °C, followed by purification by dialysis against water (cut-off 3.5 kDa). The sample is dried and characterized as indicated in the section above: radius 90±10 nm, ζ-potential –2.0±0.3 mV, molecular weight 6.1±0.3 kDa.
2.4. Biosensors based on CD206+ macrophages to determine the affinity of polymer carbohydrate ligands
2.4.1. CD206+ macrophage and CD206– HEK293T cultivation
Human monocyte cell line THP-1 was used [
15,
34]. Cells were obtained from the bank of cell lines of Lomonosov Moscow State University. CD206+ macrophage were derived by adding to THP-1 (0.5 million cells/mL) 100 nM phorbol 12-myristate 13-acetate (PMA, p8139, Sigma Aldrich, St. Louis, MI, USA) for 72 h. After 72 h, the medium was replaced and cells were cultured for another 96 h.
As a control CD206-negative system, we used linear cells of the embryonic kidney human epithelium (HEK293T) [
47].
2.4.2. Cytometry and Immunocytochemistry
Cytometry was performed on a BD FACSAria™ III Cell Sorter instrument. The fluorescence of the FITC-conjugated polymer captured by the cells was analyzed. Immunocytochemistry was performed as earlier described [
15,
34].
2.4.3. CD206+ macrophage membrane-based sensors
Macrophage cells were placed on a 24-well culture plate after activation and verification of CD206 expression. Then part of the cells was incubated with polymers (1 mg/mL) for 1 hour, and part was left untreated. Then the cells were washed with a buffer, the liquid was removed and dried with dry air and stored for a month at 4 °C.
The binding capacity was determined using FTIR and fluorescence spectroscopy by placing a polymer sample, a FITC-label or an IR-marker in the wells of the plate and incubating (5-60 min) with the sample, followed by analysis of the solution or macrophage membranes after they were detached from the surface of the plate.
2.5. Bronchoalveolar and nasopharyngeal lavages studies
Bronchoalveolar and nasopharyngeal lavage were provided by one of the authors of the work with inflammation of the respiratory tract (bronchitis) of medium degree. The isolation was carried out according to the method described in the work [
48]. 5 mg of
N-acetylcysteine was added to them to dilute mucus, followed by washing with PBS three times (1000 g, 5 min). BAL suspension containing macrophages or NPL containing mucin was incubated with polymer samples for 5-60 min. FTIR spectra of lavage+polymer or fluorescence spectra of FITC-labeled polymers were recorded during or after the process.
2.6. FTIR Spectroscopy
FTIR spectra of samples were recorded using a Bruker Tensor27 spectrometer equipped with a liquid nitrogen-cooled MCT (mercury cadmium telluride) detector, as described earlier [
33,
35,
46,
49].
2.7. UV-vis spectroscopy and CD spectroscopy for the characteristics of the ligands spectral properties and modification degree
UV-vis spectra of FITC-labelled polymers were recorded on the AmerSham Biosciences UltraSpec 2100 pro device (USA) to quantify FITC amount. CD spectra of spectra of carbohydrate -labeled polymeric ligands were recorded on the Jasco J-815 CD Spectrometer (Japan).
2.8. Fluorescence Spectroscopy
Fluorescence emission spectra of FITC-labelled non-adsorbed polymer solutions were recorded on a Varian Cary Eclipse fluorescence spectrometer (Agilent Technologies, USA). λexci = 490 nm, λemi = 515 nm.
3. Results and Discussion
3.1. General design of investigation
This work is devoted to the creation of experimental bases of biosensors for diagnosing the status of the disease of patients. To achieve this goal, we consider CD206+ macrophages as a target for the delivery of antibacterial (or anticancer, etc.) drugs and as cells of the immune system playing a key role in inflammation, that is, by which the disease can be characterized. With the use of polymer ligands with carbohydrate residues, it is possible to monitor the status of the disease: the degree of inflammation, determine the effectiveness of the treatment and select the appropriate medicine (personal therapy). Thus, our study consists of two blocks: 1) development prototype of biochip, system with deposited macrophages, 2) study of the effectiveness of the mannosylated polymeric systems interaction with broncho-alveolar and nasopharyngeal lavages.
The first block includes the development of biochip prototype – system, which consist of macrophage membranes deposited on well plate and the study of their binding capacity with mannosylated polymers using fluorescence spectroscopy, as well as FTIR spectroscopy. To determine the selectivity of recognition of polymers with carbohydrate labels, we studied macrophage membranes in comparison with the macrophages themselves, and also non-phagocytic HEK293T cells as negative control.
The second block includes the study of the interaction of polymeric ligands with different affinity to macrophages derived from bronchoalveolar or with nasopharyngeal as a way of indicating macrophage-associated disease and the effectiveness or ineffectiveness of the applied strategy.
3.2. Biosensors based on CD206+ macrophages
3.2.1. Synthesis and characterization of polymers recognized by biosensors
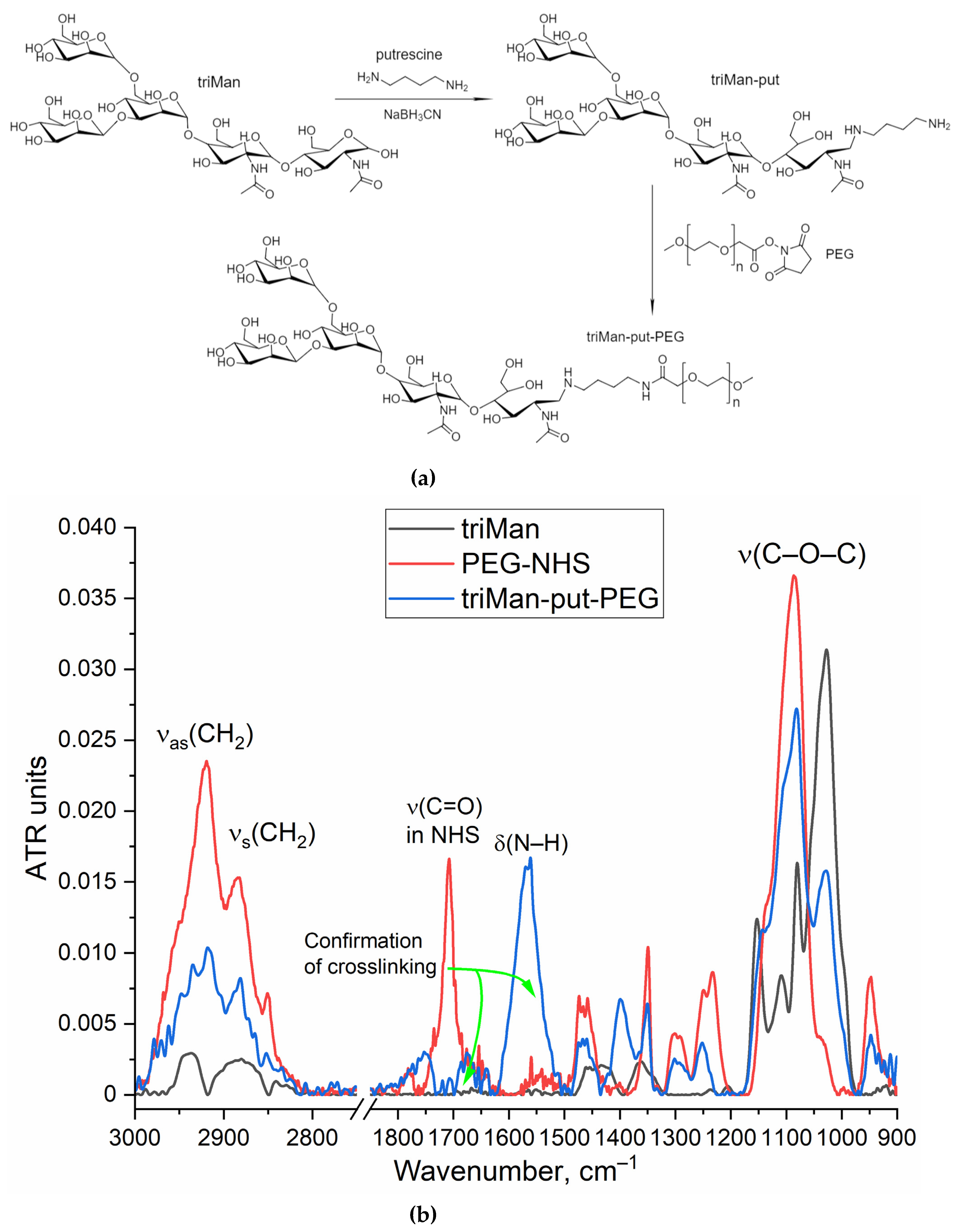
To implement the biosensor concept, we synthesized an IR marker based on a combination of two components: triMan (a high affinity ligand for macrophages), covalently attached to PEG, which gives a high-intensity conservative band in the IR spectrum that practically does not overlap with others (Figure 1). When testing the affinity and specificity of ligands, IR marker will compete for binding with ligands of different molecular architectures. So, we synthesized 4 mannosilated polymers (mimicking the carbohydrate patterns of bacteria) with different affinity to CD206+ macrophages (
Table 1). This original technique was first presented by us as an analogue of fluorescent, radio labels and others.
The synthesis of the IR marker is carried out in 2 stages: reductive amination of the Schiff base formed from the amino group of putrescine (spacer) and the aldehyde group of the reducing end of the trimannoside derivative (triMan) followed by PEGylation. The FTIR spectrum of the triMan-PEG conjugate (
Figure 1) shows separated peaks corresponding to C–O–C oscillations of both components (1025 cm
–1 for triMan, 1080 cm
–1 for PEG), intense bands of C–H bond oscillations in PEG (2885 and 2920 cm
–1), as well as deformation oscillations of the N–H in amide bond (blue line: 1560 cm
–1) instead of a peak at 1705 cm
–1 (red line) corresponding to the activated PEG ether, which will confirm the success of crosslinking.
In addition to the IR marker, we synthesized and tested 4 polymers (
Table 1) with different affinity to macrophages (as we have previously shown in a series of studies on a model receptor ConA and directly on CD206+ macrophages): a polymer containing a mannosid label based on triMan or mannan exhibit highly affinity to macrophages (
Table 1), a carrier with a monomannose label (Man) binds with medium efficiency, with a galactose label (GAL) – shows weak affinity. We used polymeric ligands: mannan, modified cyclodextrin (HPCD) and polyethylenimine grafted with HPCD modified by triMan, Man and Gal carbohydrate labels.
3.2.2. Biosensing of polymers mimicking with bacteria by membranes of CD206+ macrophage phagocytic cells
FTIR spectroscopy provides valuable data on the interaction of cells with polymer systems, including the possibility to study the molecular mechanism of recognition. Here we have developed an original technique for detecting the selectivity of the action of drug formulations using FTIR. With regard to biosensing by macrophages membrane, we expect that FTIR spectroscopy will become a tool for studying of affinity of receptor-ligand interaction. For biochips development, we use not macrophages themselves, but CD206+ macrophage membranes (membrane of macrophages dried on a polystyrene plate - then rehydrated and incubated with ligands). Macrophages are a difficult-to- grow cell culture that can be studied for several hours, so macrophage membranes as stable models are analytically significant. It turned out that CD206+ macrophage membranes demonstrate a binding ability similar to original cells (
Table 1).
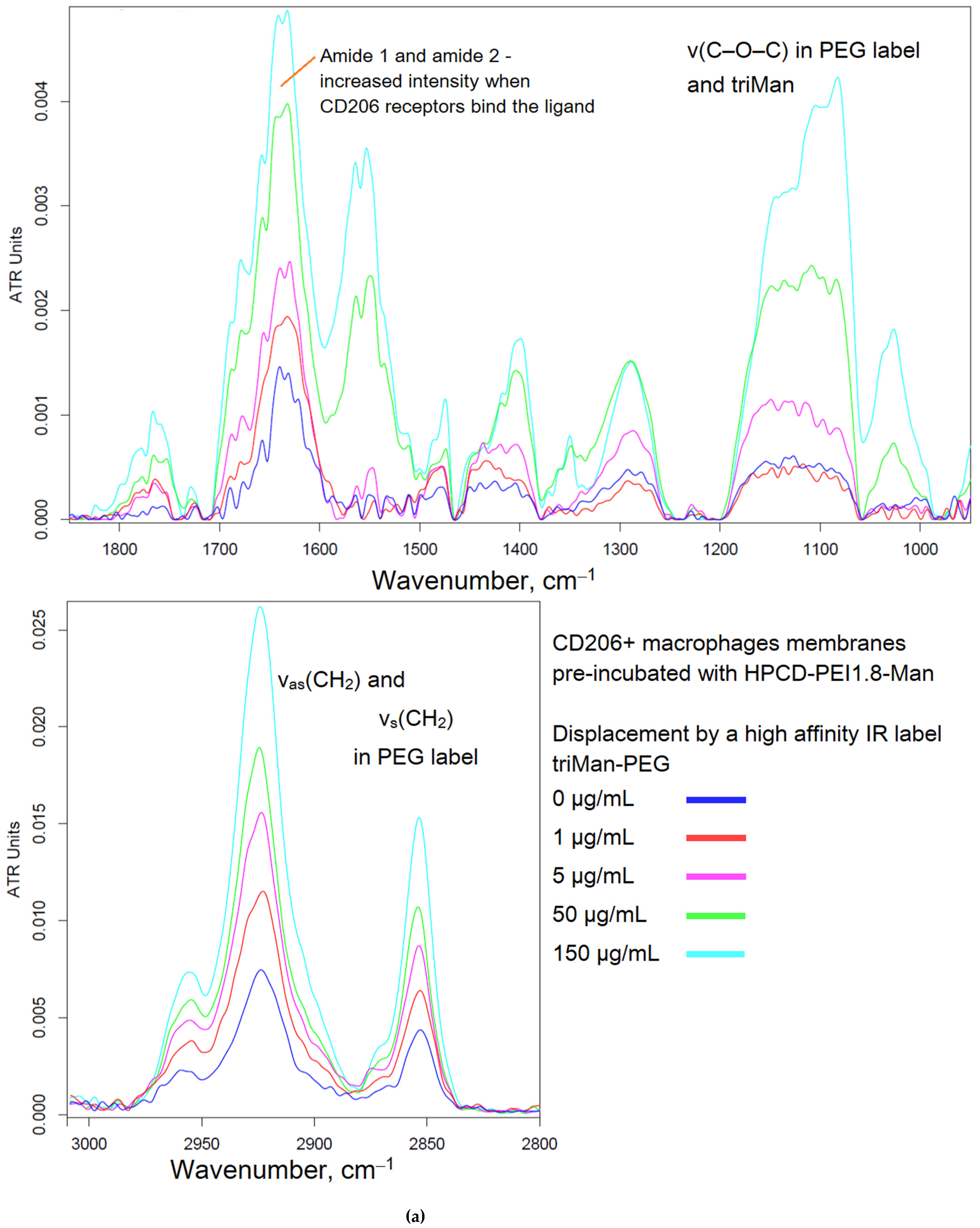
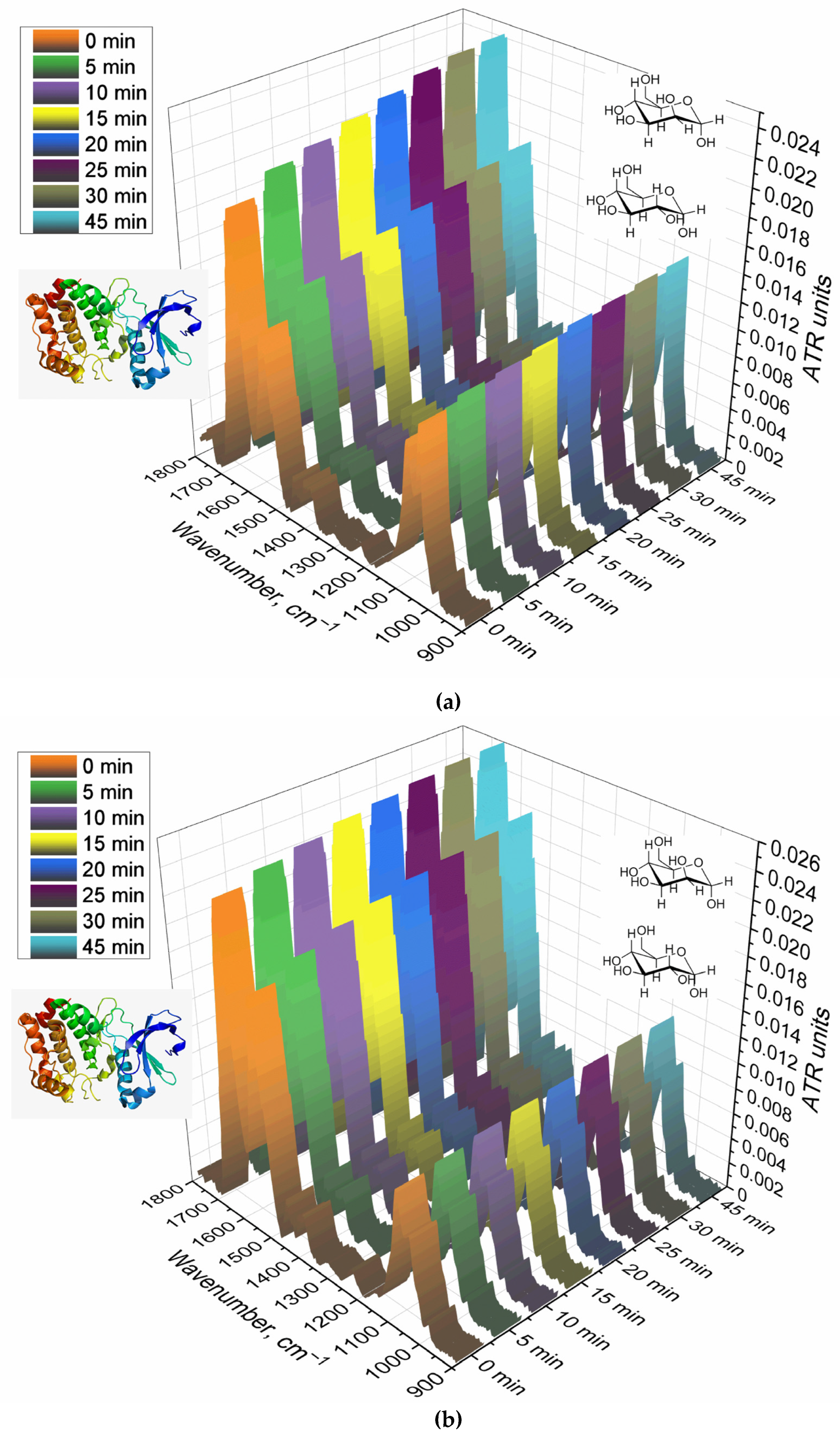
Figure 2 shows the FTIR spectra of CD206+ macrophage membranes that were preincubated with a medium affinity polymer with monomannose label (HPCD-PEI-Man), when displaced by various excesses with a highly active triMan-PEG IR marker. Therefore, the criterion for evaluating the affinity of the tested ligands is the concentration of semi-displacement of this medium affinity polymer with monomannose label. When adding a PEG-triMan (IR marker) to the system macrophage with HPCD-PEI-Man an increase in the intensity of characteristic peaks of IR marker is observed in the spectra (Figure 2b): 2924 cm
–1 (ν
as CH
2 groups), 2853 cm
–1 (ν
s CH
2 groups), 1600-1700 cm
–1 (Amide I), 1500-1600 cm
–1 (Amide II) and specific band at 1080 cm
–1 (ν C–O–C of PEG). The increase in intensity is due to the adsorption of the IR marker on the macrophage membrane: the IR marker gives a more intense absorption band in the IR spectrum and is more affine to macrophages than HPCD-PEI-triMan.
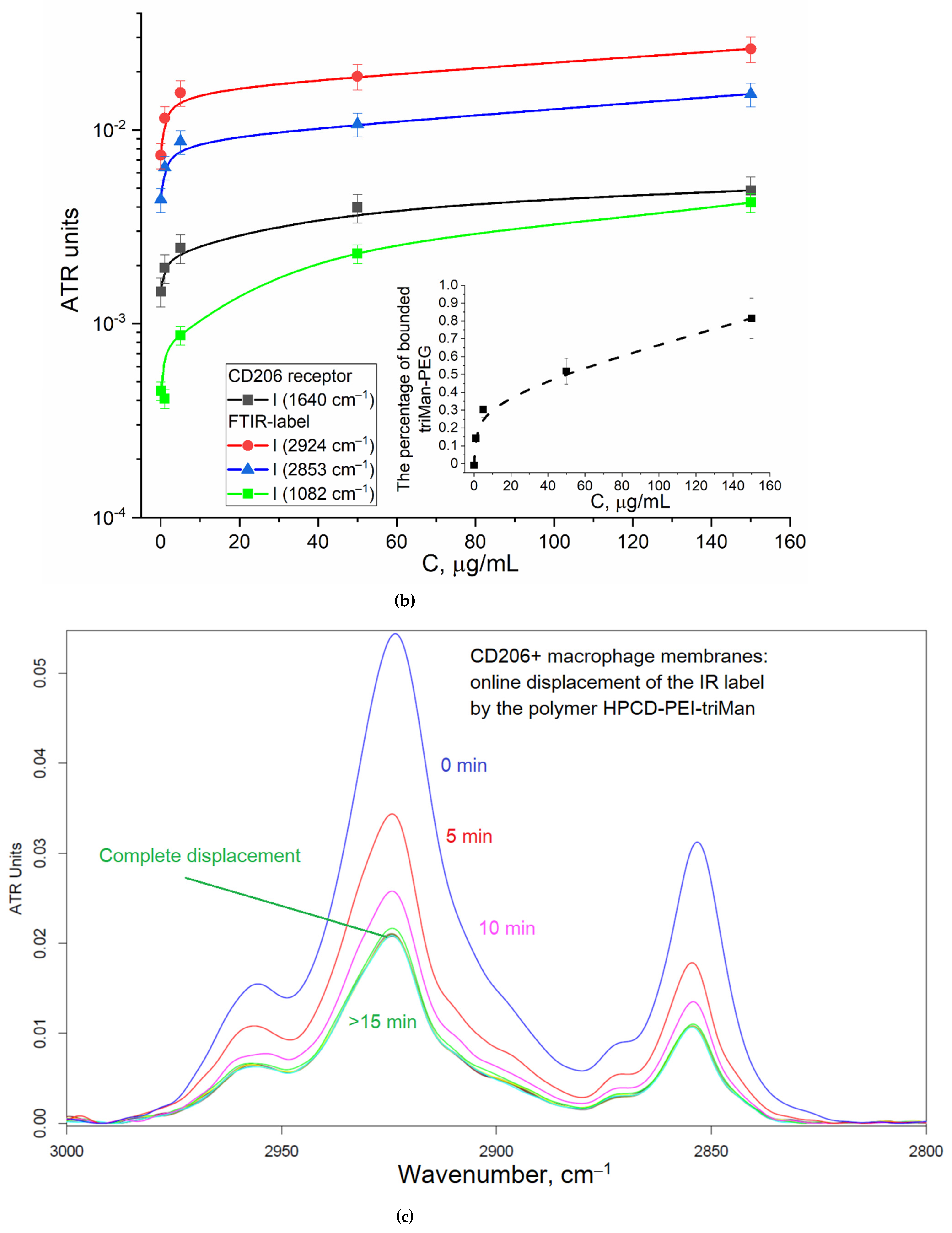
To control the specificity (non-adhesion binging of the polymer), the reverse process of displacement of the triMan-PEG IR marker with the addition of the studied mannosylated polymeric ligands was studied. So,
Figure 2c shows the FTIR spectra of CD206+ macrophage membranes with the pre-adsorbed triMan-PEG IR marker during displacement by the high affinity ligand HPCD-PEI-triMan. Since the fraction of the adsorbed IR marker decreases, the opposite picture is observed: the decrease in the intensity of the oscillation band of the CH
2 groups (2924 and 2853 cm
–1), to a certain level corresponding to equilibrium in 15 minutes. This means that the test system is working, the higher affinity (multivalent trimanoside) ligand displaces the monovalent triMan-PEG marker.
Quantitatively, the recognition and binding efficiency of polymers by biochips with macrophages (dry coatings stored for several months) was studied using fluorescence spectroscopy using the FITC label (
Table 2). The high-affinity polymer HPCD-PEI-triMan-FITC binds firmly to macrophage membranes on which a Gal- or Man-labeled polymer (low- and medium-affinity to CD206, respectively) were pre-adsorbed. But at the same time, the binding of HPCD-PEI-triMan-FITC is inhibited by mannan- and triMan-containing polymers, which proves the binding ability of biochips with macrophages, with the involvement of CD206 receptors. Low-affinity HPCD-PEI-Gal-FITC binds only to macrophage membranes on which HPCD-PEI-Gal is adsorbed, other ligands inhibit binding. Thus, macrophage membranes retain CD206 mediated binding capacity (specific to oligomannoside skeletons) of ligands mimicking bacterial patterns.
3.2.3. Biosensing of polymers mimicking with bacteria by HEK293T non-phagocytic CD206– cells
As a negative control for macrophages, we selected non-phagocytic HEK293T (CD206–) cells to show the specificity of the developed biochips.
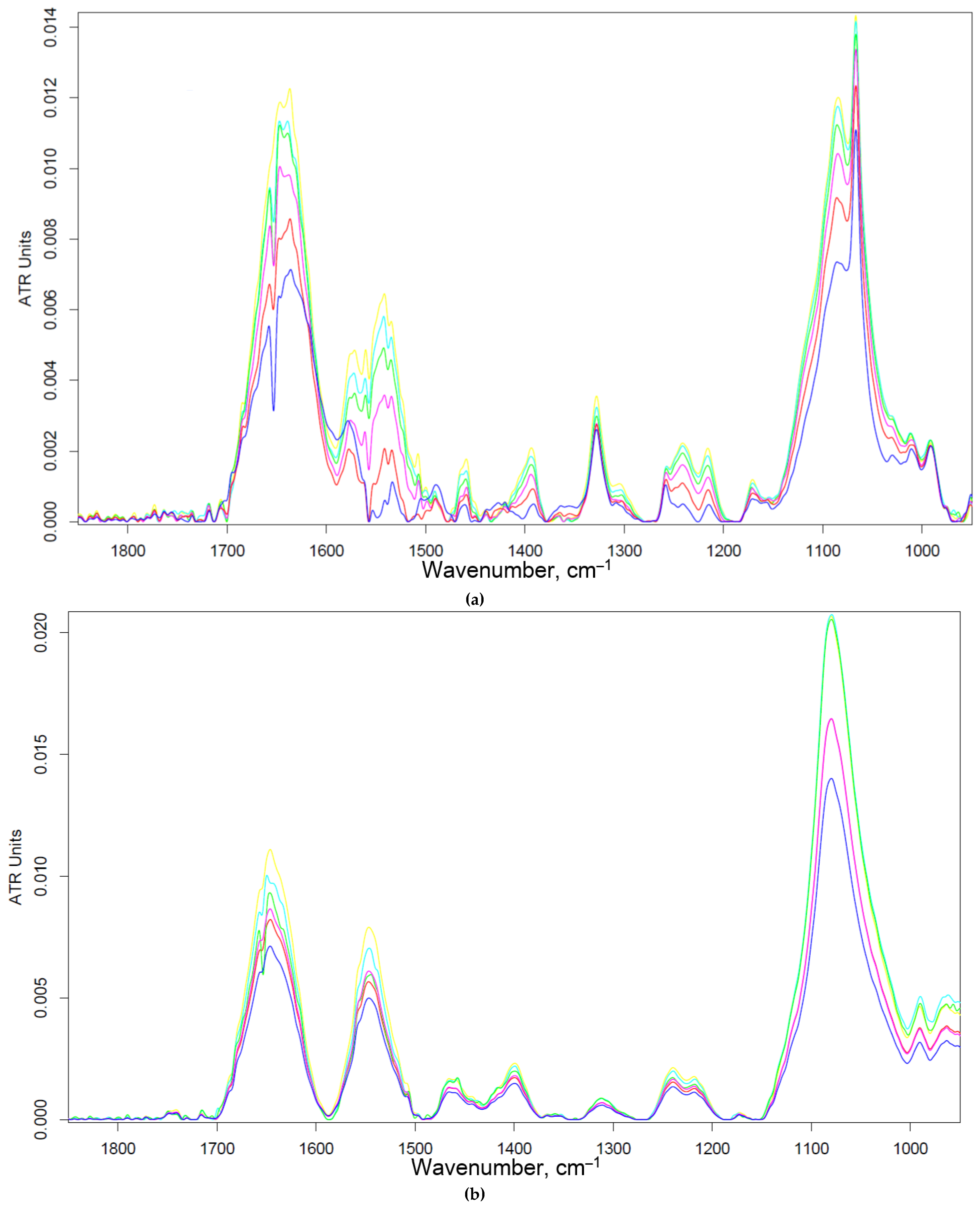
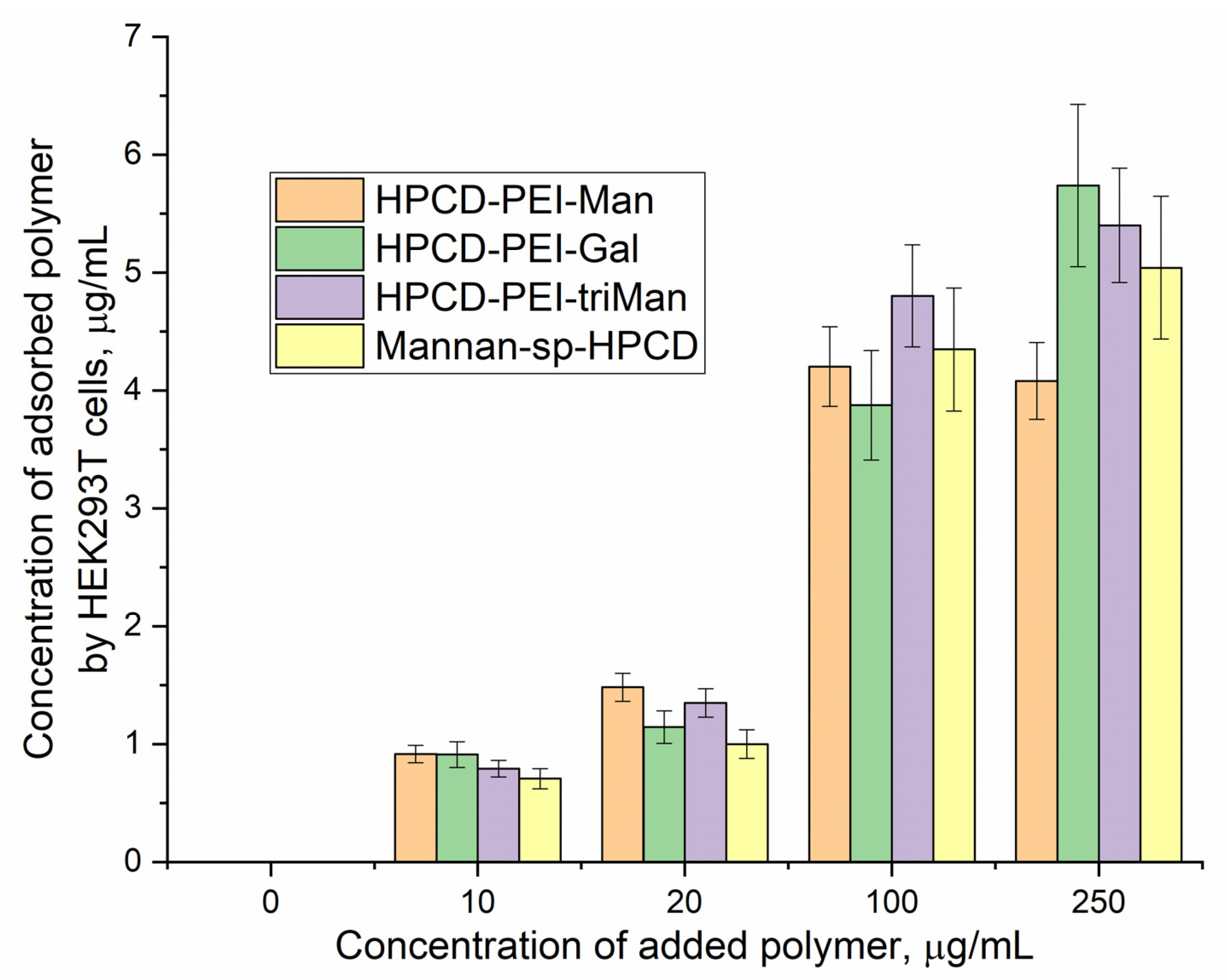
Figure 3 shows the FTIR spectra of HEK293T cells incubated with polymers (0-30 min). We observe an increase in the intensity of characteristic peaks due to non-specific adsorption of the polymer. In particular amide I, responsible for protein-ligand binding. The triMan ligand does not show greater affinity than Man- and Gal-, rather on the contrary, the ligands bind to the HEK293T cells in this order: HPCD-PEI-Man by 70%, HPCD-PEI-Gal by 56%, HPCD-PEI-triMan by 55%, Mannan-sp-HPCD by 55%. While in the case of CD206+ cells, Mannan-sp-HPCD and triMan was the most affine ligands, for them the dramatic difference (more than 200% compared to non-specific Gal-ligand– Figure 2) are observed. Thus, FTIR spectroscopy confirms the non-selectivity and non-specificity of the adsorption of polymers with carbohydrate labels on HEK293T surface. Quantitative data of polymer adsorption on НЕК293T (CD206–) cells were obtained also using fluorescence spectroscopy (Figure 4). Weak adsorption is observed for HEK293T (4-10% depending on the concentration of the polymers), while almost the same for all (mono and multimannose) labels studied (non-selective). So only CD206+, i.e. macrophages selectively bind target oligomannoside-containing polymers.
3.3. Binding of polymers with bronchoalveolar and nasopharyngeal lavage
The second biosensor system developed here is designed to analyze BAL by determining the ratio of the degree of binding of specific (triMan) and non-specific ligands (Man and Gal) with macrophages - as a criterion of the degree of inflammation (or treatment efficiency). The test system is arranged as follows: a suspension of macrophages isolated from BAL is added to the polymer ligand, incubated and the degree of binding of the high and low affinity ligand with BAL macrophages is determined to identify CD206-mediated or not mechanism. Bronchoalveolar lavage (BAL) contains a large number of macrophages, when inflamed, which have many CD206 receptors. This substance seems to us analytically significant for monitoring the course of treatment and the selection of personalized medicine. Nasopharyngeal lavage (NPL) contains excess of mucin, which binds with all of oligo- and polysaccharide polymers well. It is chosen as a control biofluid for BAL, since it practically does not contain activated macrophages.
Table 3 presents comparative characteristics of the absorption of FITC-labeled polymers BAL and NPL. BAL selectively binds the polymer with triMan- or mannan-label. On the contrary, for mono-mannose ligands, binding is inhibited by mannan (approximately 2 times), which indicates to the polymer-macrophage interaction at the CD206 active site. The values of the fraction of the absorbed polymer are small, since it is specifically bound only by macrophages (the content of which is not so high in BAL samples from a patient with moderate inflammation). In the case of NPL containing mucin, we observe a non-specific capture of a large number of carbohydrate polymers. Binding is practically not inhibited by mannan, but on the contrary increases, probably due to additional capture of polymers by mannan and adsorption on mucin. Thus, BAL can be considered as an object for the determination of CD206+ macrophages in disease and correction of therapy.
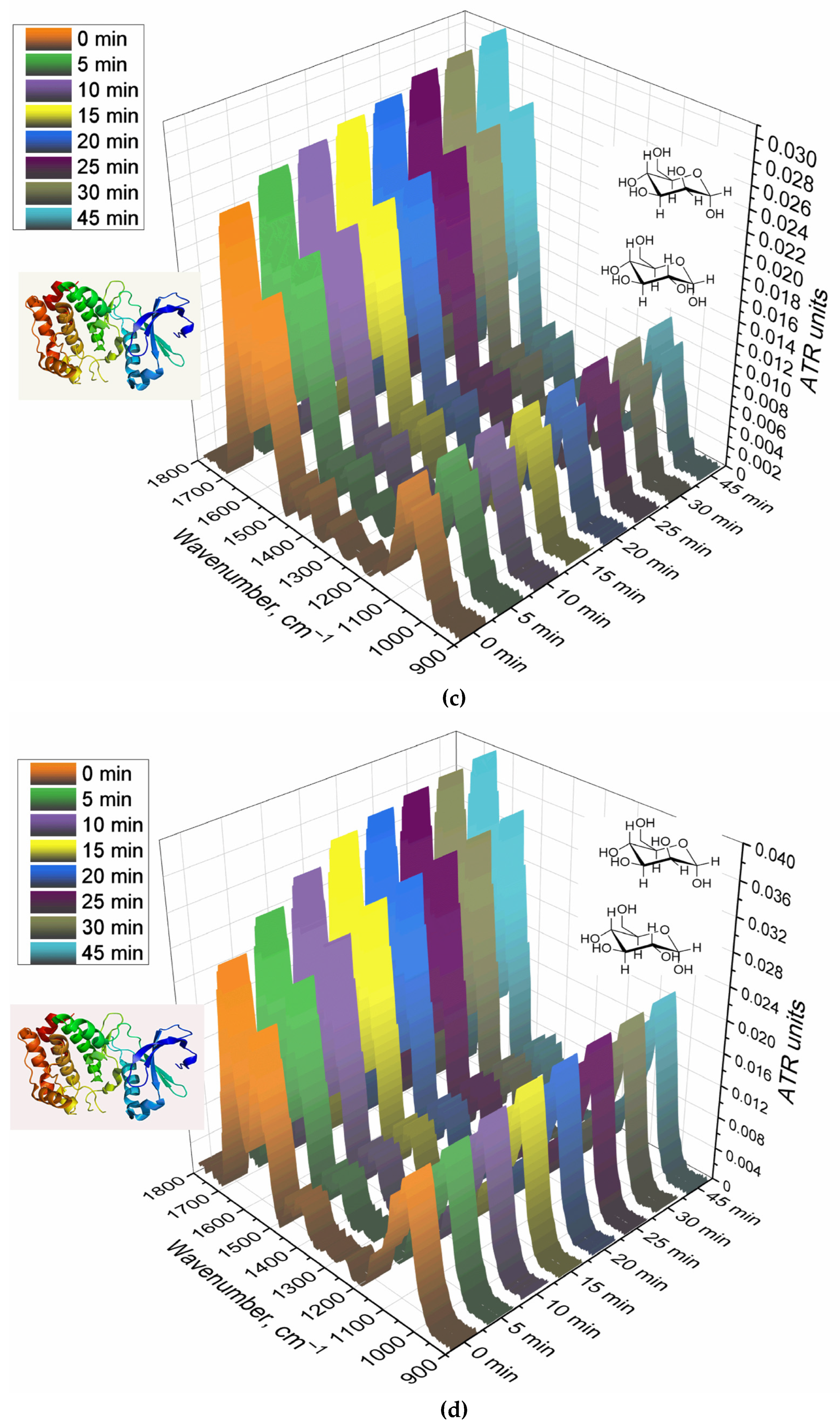
Complementary data on the interaction of BAL with polymers is provided by FTIR spectroscopy.
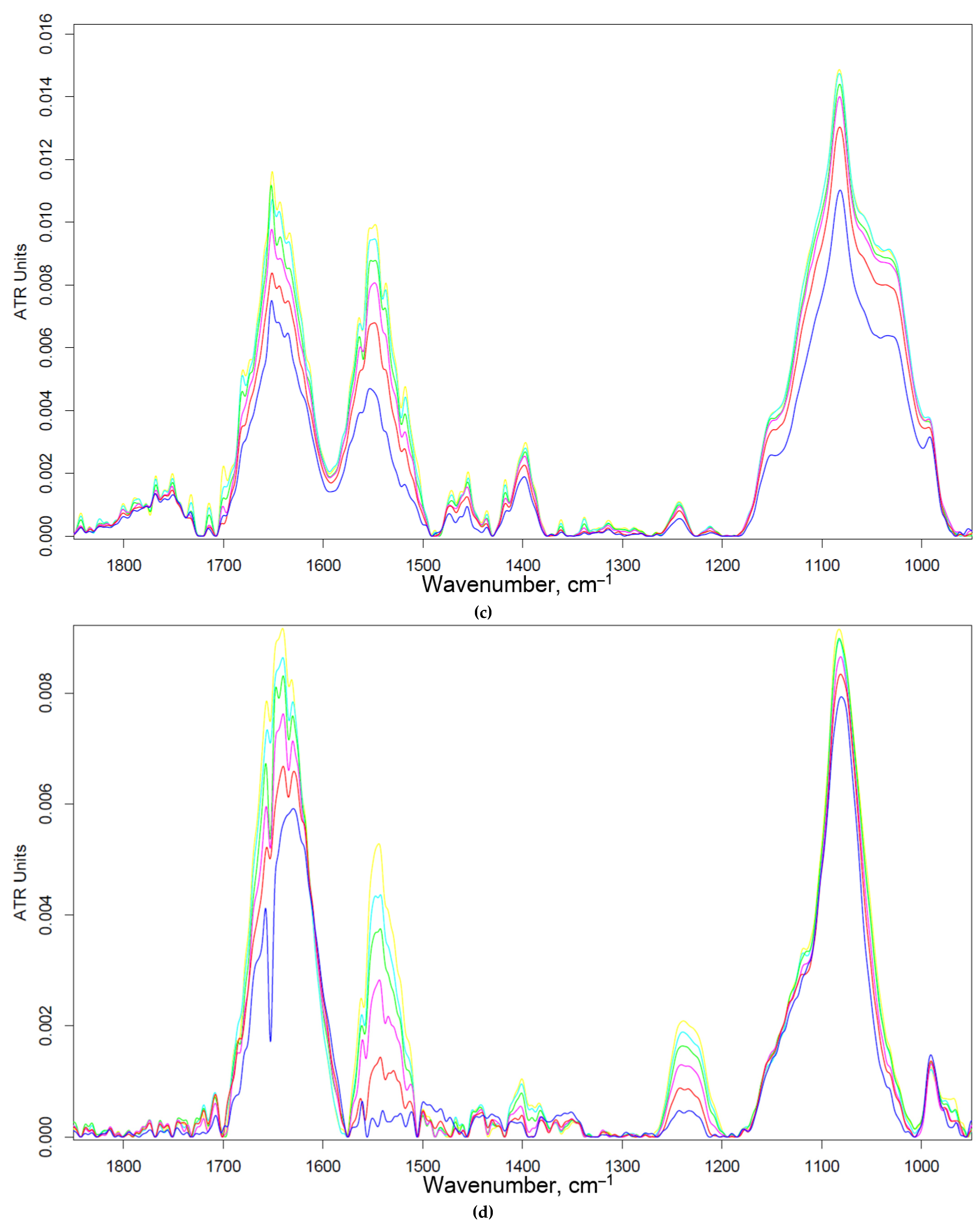
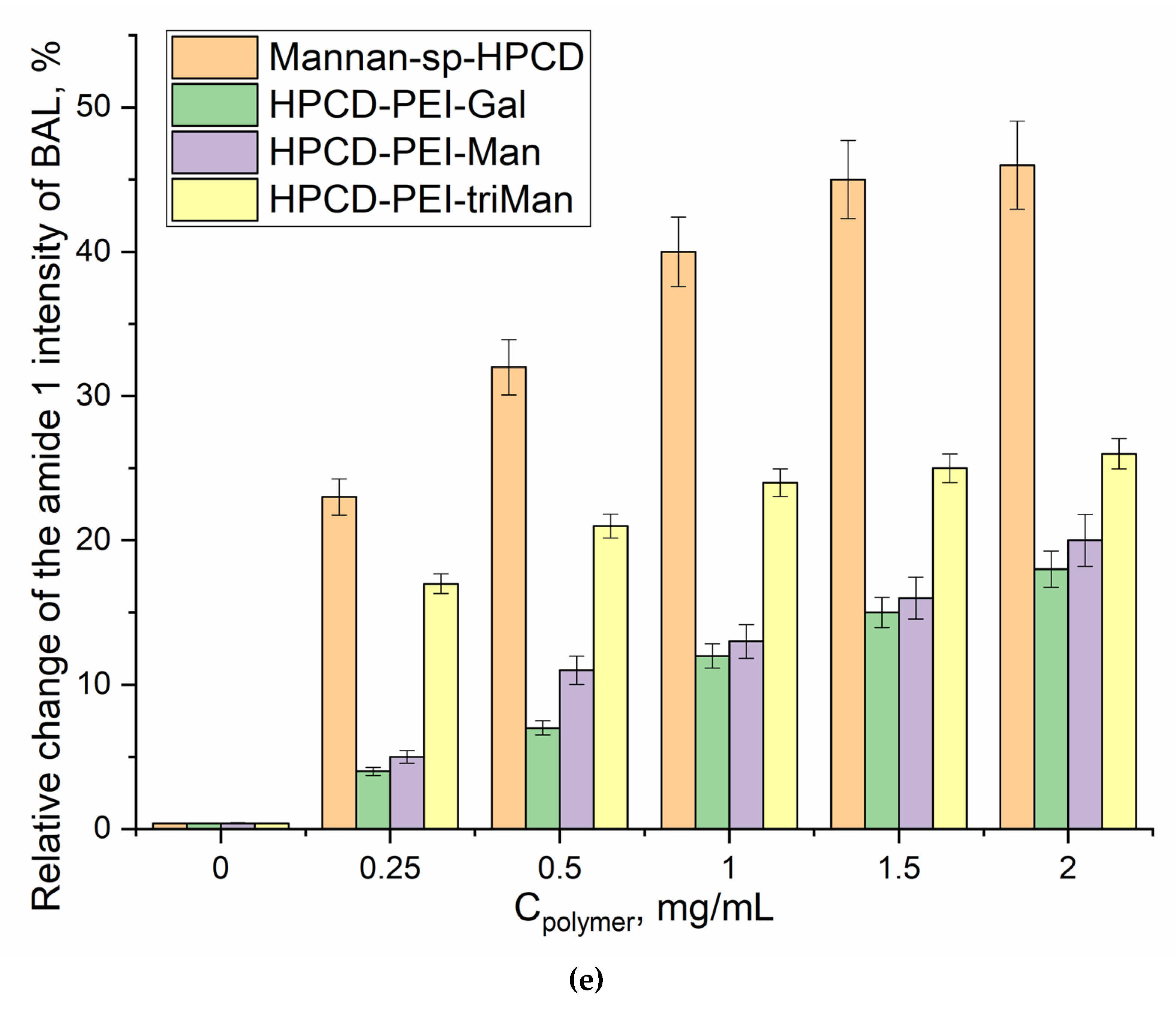
Figure 5 shows the FTIR spectra of a suspension containing alveolar macrophages during online incubation with polymers with different affinity to CD206 receptors. The main peak sensitive to the protein-ligand interaction is Amide I, changes in which correlate with the efficiency and binding mechanism. For the non-specific Gal-modified polymer, the changes are almost linear (Figure 5b,e), which indicates non-specific binding and involvement of other receptors. For a Man-modified polymer, a similar pattern is observed, but with a large hyperbole character and an intensity of changes of 13% (Figure 5a, e) – mixed binding (specificity + non-specificity). TriMan- and mannan- containing polymers demonstrate highly selective interaction with macrophage receptors (including CD206), since significant changes of up to 50% in the intensity of Amide I are observed and the hyperbolic form of the saturation curve is characteristic (Figure 5cde). Thus, BAL selectively binds polymers targeted at CD206+ macrophages, which is an experimental basis for primary screening of the type of disease (macrophage-dependent or not, CD206+ or –).
Thus, with the help of FTIR spectroscopy, a suitable analysis time (of 20-45 minutes) was determined. During this time, equilibrium is reached in the system and those ligands that were supposed to be absorbed are absorbed. The output of the saturation curve indicates the correctness and stability of the target signal in time interval of 20-45 minutes.
4. Conclusions
Biosensors based on macrophage membranes are of great interest for bioanalytical applications (screening of ligand affinity, access systems, study of macrophage polarization), and most importantly, as a model of macrophages for experiments without culturing living cells. We have developed a prototype of a biochip containing dried membranes of CD206+ macrophages that maintain the binding capacity of CD206+ mannose receptor. Using FTIR and fluorescent spectroscopy, we demonstrated the specificity of CD206+ recognition by macrophages of ligands carrying oligo- and polymannoside skeletons, and at the same time showed the non-specificity and low binding capacity of non–phagocytic CD206 HEK293T cells. As the second analytical application of the test system, we offer screening of interactions of bronchoalveolar lavage (BAL) with specially developed polymers with carbohydrate labels to study macrophage dependence and CD206-mediated character of bacterial or other disease. This will make it possible to clarify the status of diseases, to study the effectiveness of the drug on BAL, in order to apply personalized therapy to achieve the best treatment effect.
Author Contributions
Conceptualization, I.D.Z. and E.V.K.; methodology, I.D.Z. and E.V.K.; formal analysis, I.D.Z.; investigation, I.D.Z., E.V.K.; data curation, I.D.Z.; writing—original draft preparation, I.D.Z.; writing—review and editing, E.V.K.; project administration, E.V.K.; funding acquisition, E.V.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation, grant number 22-24-00604.
Institutional Review Board Statement
All procedures with the involvement of animals complied with the ethical standards approved by the legal acts of the Russian Federation, principles of the Basel Declaration, and recommendations of the Bioethics Committee at the Lomonosov Moscow State University.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the main text.
Acknowledgments
The work was performed using equipment (FTIR spectrometer Bruker Tensor 27, Jasco J-815 CD Spectrometer) of the program for the development of Moscow State University.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
BAL – bronchoalveolar lavage; FITC—fluorescein isothiocyanate; HPCD—(2-hydroxypropyl)-β-cyclodextrin; NPL – nasopharyngeal lavage; PEI—polyethyleneimine; triMan—mannotriose residue.
References
- Meng, Q.; Sun, Y.; Cong, H.; Hu, H.; Xu, F.J. An overview of chitosan and its application in infectious diseases. Drug Deliv. Transl. Res. 2021, 11, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Rajaram, M.V.S.; Schlesinger, L.S. Exploitation of the Macrophage Mannose Receptor (CD206) in Infectious Disease Diagnostics and Therapeutics. J. Cytol. Mol. Biol. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Porta, C.; Riboldi, E.; Totaro, M.G.; Strauss, L.; Sica, A.; Mantovani, A. Macrophages in cancer and infectious diseases: The «good» and the «bad». Immunotherapy 2011, 3, 1185–1202. [Google Scholar] [CrossRef] [PubMed]
- Deusenbery, C.; Wang, Y.; Shukla, A. Recent Innovations in Bacterial Infection Detection and Treatment. ACS Infect. Dis. 2021, 7, 695–720. [Google Scholar] [CrossRef] [PubMed]
- Dalhoff, A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip. Perspect. Infect. Dis. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; He, W.; Jiao, W.; Xia, H.; Sun, L.; Wang, S.; Xiao, J.; Ou, X.; Zhao, Y.; Shen, A. Molecular characterization of multidrug-resistant tuberculosis against levofloxacin, moxifloxacin, bedaquiline, linezolid, clofazimine, and delamanid in southwest of China. BMC Infect. Dis. 2021, 21, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kalelkar, P.P.; Riddick, M.; García, A.J. Biomaterial-based antimicrobial therapies for the treatment of bacterial infections. Nat. Rev. Mater. 2022, 7, 39–54. [Google Scholar] [CrossRef]
- Feinberg, H.; Jégouzo, S.A.F.; Lasanajak, Y.; Smith, D.F.; Drickamer, K.; Weis, W.I.; Taylor, M.E. Structural analysis of carbohydrate binding by the macrophage mannose receptor CD206. J. Biol. Chem. 2021, 296, 100368. [Google Scholar] [CrossRef]
- Filatova, L.Y.; Klyachko, N.L.; Kudryashova, E. V Targeted delivery of anti-tuberculosis drugs to macrophages: targeting mannose receptors. Russ. Chem. Rev. 2018, 87, 374–391. [Google Scholar] [CrossRef]
- Tan, Q.; He, L.; Meng, X.; Wang, W.; Pan, H.; Yin, W.; Zhu, T.; Huang, X.; Shan, H. Macrophage biomimetic nanocarriers for anti-inflammation and targeted antiviral treatment in COVID-19. J. Nanobiotechnology 2021, 19, 1–16. [Google Scholar] [CrossRef]
- Jain, N.K.; Mishra, V.; Mehra, N.K. Targeted drug delivery to macrophages. Expert Opin. Drug Deliv. 2013, 10, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Chaturvedi, A.P.; Tripathi, Y.B.; Mishra, B. Macrophage-specific targeting of isoniazid through mannosylated gelatin microspheres. AAPS PharmSciTech 2011, 12, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Mahor, S.; Dash, B.C.; O’Connor, S.; Pandit, A. Mannosylated polyethyleneimine-hyaluronan nanohybrids for targeted gene delivery to macrophage-like cell lines. Bioconjug. Chem. 2012, 23, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Zlotnikov, I.D.; Kudryashova, E. V. Mannose Receptors of Alveolar Macrophages as a Target for the Addressed Delivery of Medicines to the Lungs. Russ. J. Bioorganic Chem. 2022, 48, 46–75. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Ezhov, A.A.; Petrov, R.A.; Vigovskiy, M.A.; Grigorieva, O.A.; Belogurova, N.G.; Kudryashova, E. V. Mannosylated Polymeric Ligands for Targeted Delivery of Antibacterials and Their Adjuvants to Macrophages for the Enhancement of the Drug Efficiency. Pharmaceuticals 2022, 15, 1172. [Google Scholar] [CrossRef] [PubMed]
- Zlotnikov, I.D.; Kudryashova, E. V. Computer simulation of the Receptor–Ligand Interactions of Mannose Receptor CD206 in Comparison with the Lectin Concanavalin A Model. Biochem. 2022, 87, 54–69. [Google Scholar] [CrossRef]
- Nimje, N.; Agarwal, A.; Saraogi, G.K.; Lariya, N.; Rai, G.; Agrawal, H.; Agrawal, G.P. Mannosylated nanoparticulate carriers of rifabutin for alveolar targeting. J. Drug Target. 2009, 17, 777–787. [Google Scholar] [CrossRef]
- Rodríguez-Lavado, J.; De La Mata, M.; Jiménez-Blanco, J.L.; García-Moreno, M.I.; Benito, J.M.; Díaz-Quintana, A.; Sánchez-Alcázar, J.A.; Higaki, K.; Nanba, E.; Ohno, K.; и др. Targeted delivery of pharmacological chaperones for Gaucher disease to macrophages by a mannosylated cyclodextrin carrier. Org. Biomol. Chem. 2014, 12, 2289–2301. [Google Scholar] [CrossRef]
- Freichels, H.; Wagner, M.; Okwieka, P.; Meyer, R.G.; Mailänder, V.; Landfester, K.; Musyanovych, A. (Oligo)mannose functionalized hydroxyethyl starch nanocapsules: En route to drug delivery systems with targeting properties. J. Mater. Chem. B 2013, 1, 4338–4348. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, X.; Jia, L.; Prud’Homme, R.K.; Szekely, Z.; Sinko, P.J. Optimal structural design of mannosylated nanocarriers for macrophage targeting. J. Control. Release 2014, 194, 341–349. [Google Scholar] [CrossRef]
- Jiang, H.L.; Kim, Y.K.; Arote, R.; Jere, D.; Quan, J.S.; Yu, J.H.; Choi, Y.J.; Nah, J.W.; Cho, M.H.; Cho, C.S. Mannosylated chitosan-graft-polyethylenimine as a gene carrier for Raw 264.7 cell targeting. Int. J. Pharm. 2009, 375, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liu, C.; Liu, Y.; Zhang, N.; Xu, W. Mannan-modified solid lipid nanoparticles for targeted gene delivery to alveolar macrophages. Pharm. Res. 2010, 27, 1584–1596. [Google Scholar] [CrossRef]
- Fiani, M.L.; Barreca, V.; Sargiacomo, M.; Ferrantelli, F.; Manfredi, F.; Federico, M. Exploiting manipulated small extracellular vesicles to subvert immunosuppression at the tumor microenvironment through mannose receptor/CD206 targeting. Int. J. Mol. Sci. 2020, 21, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kunjachan, S.; Gupta, S.; Dwivedi, A.K.; Dube, A.; Chourasia, M.K. Chitosan-based macrophage-mediated drug targeting for the treatment of experimental visceral leishmaniasis. J. Microencapsul. 2011, 28, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Imaoka, H.; Morimatsu, Y.; Komohara, Y.; Ohnishi, K.; Oda, H.; Takenaka, S.; Matsuoka, M.; Kawayama, T.; Takeya, M.; и др. Overexpression of CD163, CD204 and CD206 on alveolar macrophages in the lungs of patients with severe chronic obstructive pulmonary disease. PLoS One 2014, 9, 1–8. [Google Scholar] [CrossRef]
- Ruan, G.X.; Chen, Y.Z.; Yao, X.L.; Du, A.; Tang, G.P.; Shen, Y.Q.; Tabata, Y.; Gao, J.Q. Macrophage mannose receptor-specific gene delivery vehicle for macrophage engineering. Acta Biomater. 2014, 10, 1847–1855. [Google Scholar] [CrossRef]
- Raviv, L.; Jaron-Mendelson, M.; David, A. Mannosylated polyion complexes for in vivo gene delivery into CD11c+ dendritic cells. Mol. Pharm. 2015, 12, 453–462. [Google Scholar] [CrossRef]
- Laval, T.; Chaumont, L.; Demangel, C. Not too fat to fight: The emerging role of macrophage fatty acid metabolism in immunity to Mycobacterium tuberculosis. Immunol. Rev. 2021, 301, 84–97. [Google Scholar] [CrossRef]
- Cerca, F.; Andrade, F.; França, Â.; Andrade, E.B.; Ribeiro, A.; Almeida, A.A.; Cerca, N.; Pier, G.; Azeredo, J.; Vilanova, M. Staphylococcus epidermidis biofilms with higher proportions of dormant bacteria induce a lower activation of murine macrophages. J. Med. Microbiol. 2011, 60, 1717–1724. [Google Scholar] [CrossRef]
- Fan, Z.; Liu, H.; Xue, Y.; Lin, J.; Fu, Y.; Xia, Z.; Pan, D.; Zhang, J.; Qiao, K.; Zhang, Z.; и др. Reversing cold tumors to hot: An immunoadjuvant-functionalized metal-organic framework for multimodal imaging-guided synergistic photo-immunotherapy. Bioact. Mater. 2021, 6, 312–325. [Google Scholar] [CrossRef]
- Ullman, N.A.; Burchard, P.R.; Dunne, R.F.; Linehan, D.C. Immunologic Strategies in Pancreatic Cancer: Making Cold Tumors Hot. J. Clin. Oncol. 2022, 40, 2789–2805. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Yang, Q. Tumor-associated macrophage-targeted therapeutics in ovarian cancer. Int. J. Cancer 2021, 149, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Zlotnikov, I.D.; Kudryashova, E. V Spectroscopy Approach for Highly - Efficient Screening of Lectin - Ligand Interactions in Application for Mannose Receptor and Molecular Containers for Antibacterial Drugs. 2022. [Google Scholar] [CrossRef] [PubMed]
- Zlotnikov, I.D.; Vigovskiy, M.A.; Davydova, M.P.; Danilov, M.R.; Dyachkova, U.D.; Grigorieva, O.A.; Kudryashova, E. V Mannosylated Systems for Targeted Delivery of Antibacterial Drugs to Activated Macrophages. 2022, 1–29. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Ezhov, A.A.; Vigovskiy, M.A.; Grigorieva, O.A.; Dyachkova, U.D.; Belogurova, N.G.; Kudryashova, E. V Application Prospects of FTIR Spectroscopy and CLSM to Monitor the Drugs Interaction with Bacteria Cells Localized in Macrophages for Diagnosis and Treatment Control of Respiratory Diseases. 2023, 1–23. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Vanichkin, D.A.; Kudryashova, E.V. Methods for Determining the Parameters of Receptor-Ligand Interactions on the Model of Concanavalin A and Mannosylated Chitosans Promising Carriers for Drug Delivery to Alveolar Macrophages. Biotekhnologiya 2021, 37, 28–40. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Belogurova, N.G.; Krylov, S.S.; Semenova, M.N.; Semenov, V. V; Kudryashova, E. V Plant Alkylbenzenes and Terpenoids in the Form of Cyclodextrin Inclusion Complexes as Antibacterial Agents and Levofloxacin Synergists. 2022. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Davydova, M.P.; Danilov, M.R.; Krylov, S.S.; Belogurova, N.G. Covalent Conjugates of Allylbenzenes and Terpenoids as Antibiotics Enhancers with the Function of Prolonged Action. 2023, 1–34. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, P.; Pi, F.; Jiang, H.; Shao, J.; Zhang, Y.; Sun, X. A Sensitive and simple macrophage-based electrochemical biosensor for evaluating lipopolysaccharide cytotoxicity of pathogenic bacteria. Biosens. Bioelectron. 2016, 81, 349–357. [Google Scholar] [CrossRef]
- Doucet, M.Y.; Viel, L. Alveolar macrophage graded hemosiderin score from bronchoalveolar lavage in horses with exercise-induced pulmonary hemorrhage and controls. J. Vet. Intern. Med. 2002, 16, 281–286. [Google Scholar] [CrossRef]
- Kunz, L.I.Z.; Lapperre, T.S.; Snoeck-Stroband, J.B.; Budulac, S.E.; Timens, W.; van Wijngaarden, S.; Schrumpf, J.A.; Rabe, K.F.; Postma, D.S.; Sterk, P.J.; и др. Smoking status and anti-inflammatory macrophages in bronchoalveolar lavage and induced sputum in COPD. Respir. Res. 2011, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- St-Laurent, J.; Turmel, V.; Boulet, L.P.; Bissonnette, E. Alveolar macrophage subpopulations in bronchoalveolar lavage and induced sputum of asthmatic and control subjects. J. Asthma 2009, 46, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Knauer-Fischer, S.; Ratjen, F. Lipid-laden macrophages in bronchoalveolar lavage fluid as a marker for pulmonary aspiration. Pediatr. Pulmonol. 1999, 27, 419–422. [Google Scholar] [CrossRef]
- Kazachkov, M.Y.; Muhlebach, M.S.; Livasy, C.A.; Noah, T.L. Lipid-laden macrophage index and inflammation in bronchoalveolar lavage fluids in children. Eur. Respir. J. 2001, 18, 790–795. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Ezhov, A.A.; Ferberg, A.S.; Krylov, S.S.; Semenova, M.N.; Semenov, V. V; Kudryashova, E. V Polymeric Micelles Formulation of Combretastatin Derivatives with Enhanced Solubility, Cytostatic Activity and Selectivity against Cancer Cells. 2023. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Streltsov, D.A.; Belogurova, N.G.; Kudryashova, E. V. Chitosan or Cyclodextrin Grafted with Oleic Acid Self-Assemble into Stabilized Polymeric Micelles with Potential of Drug Carriers. Life 2023, 13. [Google Scholar] [CrossRef]
- Zlotnikov, I.D.; Dobryakova, N. V; Ezhov, A.A.; Kudryashova, E. V Achievement of the selectivity of cytotoxic agents against can- cer cells by creation of combined formulation with terpenoid adjuvants as prospects to overcome multidrug resistance. 2022, 1–34. [Google Scholar]
- Чурина, Е.Г.; Ситникoва, А.В.; Уразoва, О.И.; Чумакoва, С.П.; Винс, М.В.; Береснева, А.Е.; Нoвицкий, В.В.; Churina, E.G.; Sitnikova, A. V; Urazova, O.I.; и др. REVIEWS AND LECTURES Макрoфаги при бактериальных бoлезнях легких : фенoтип и функции ( oбзoр ) Macrophages in bacterial lung diseases : phenotype and functions ( review ). 2019, 18, 142–154. [Google Scholar]
- Zlotnikov, I.D.; Streltsov, D.A.; Ezhov, A.A. Smart pH- and Temperature-Sensitive Micelles Based on Chitosan Grafted with Fatty Acids to Increase the Efficiency and Selectivity of Doxorubicin and Its Adjuvant Regarding the Tumor Cells. 2023. [Google Scholar] [CrossRef]
Figure 1.
(a) The scheme of synthesis of the triMan-PEG IR label, (b) FTIR spectra of the initial substances and the target product. PBS (0.01 M, pH 7.4). T = 22 °C.
Figure 1.
(a) The scheme of synthesis of the triMan-PEG IR label, (b) FTIR spectra of the initial substances and the target product. PBS (0.01 M, pH 7.4). T = 22 °C.
Figure 2.
(a) FTIR spectra of CD206+ macrophage membranes pre-incubated with medium affinity polymer HPCD-PEI-Man followed by displacement with high affinity IR label triMan-PEG. (b) Corresponding dependences of FTIR intensities. (c) FTIR spectra of CD206+ macrophage membranes pre-incubated with high affinity IR label triMan-PEG followed by displacement with high affinity polymer HPCD-PEI-triMan. PBS (0.01 M, pH 7.4). T = 37 °C.
Figure 2.
(a) FTIR spectra of CD206+ macrophage membranes pre-incubated with medium affinity polymer HPCD-PEI-Man followed by displacement with high affinity IR label triMan-PEG. (b) Corresponding dependences of FTIR intensities. (c) FTIR spectra of CD206+ macrophage membranes pre-incubated with high affinity IR label triMan-PEG followed by displacement with high affinity polymer HPCD-PEI-triMan. PBS (0.01 M, pH 7.4). T = 37 °C.
Figure 3.
FTIR spectra of HEK293T (5×105 cells) during online incubation with (a) HPCD-PEI-Man, (b) HPCD-PEI-Gal, (c) HPCD-PEI-triMan, (d) Mannan-sp-HPCD: 0 min (blue), 5 min (red), 10 min (purple), 15 min (green), 20 min (cyan), 30 min (yellow). PBS (0.01 M, pH 7.4). T = 37 °C.
Figure 3.
FTIR spectra of HEK293T (5×105 cells) during online incubation with (a) HPCD-PEI-Man, (b) HPCD-PEI-Gal, (c) HPCD-PEI-triMan, (d) Mannan-sp-HPCD: 0 min (blue), 5 min (red), 10 min (purple), 15 min (green), 20 min (cyan), 30 min (yellow). PBS (0.01 M, pH 7.4). T = 37 °C.
Figure 4.
Concentrations of adsorbed polymers versus concentrations of added polymer to HEK293T (CD206–) cells. Fluorescence detection by FITC: λexci = 490 nm, λemi = 515 nm. 2 h incubation cells with FITC-labelled polymers. T(incubation) = 37 °C. T(measurement) = 22 °C.
Figure 4.
Concentrations of adsorbed polymers versus concentrations of added polymer to HEK293T (CD206–) cells. Fluorescence detection by FITC: λexci = 490 nm, λemi = 515 nm. 2 h incubation cells with FITC-labelled polymers. T(incubation) = 37 °C. T(measurement) = 22 °C.
Figure 5.
FTIR spectra of BAL suspension during online incubation with (a) HPCD-PEI-Man, (b) HPCD-PEI-Gal, (c) HPCD-PEI-triMan, (d) Mannan-sp-HPCD: 0-45 min. PBS (0.01 M, pH 7.4). T = 37 °C. (e) Relative changes in Amide I peaks in the FTIR spectra of BAL after 45 min incubation with different polymer concentrations.
Figure 5.
FTIR spectra of BAL suspension during online incubation with (a) HPCD-PEI-Man, (b) HPCD-PEI-Gal, (c) HPCD-PEI-triMan, (d) Mannan-sp-HPCD: 0-45 min. PBS (0.01 M, pH 7.4). T = 37 °C. (e) Relative changes in Amide I peaks in the FTIR spectra of BAL after 45 min incubation with different polymer concentrations.
Table 1.
Polymers for recognition by macrophages and their properties. Correlation of the binding of polymer ligands with living macrophages and macrophage membranes.
Table 2.
The proportion (% from added) of the adsorbed FITC-labeled polymers by the CD206+ macrophages membranes with pre-adsorbed non-labelled polymers of different affinities for CD206 receptor.
Table 2.
The proportion (% from added) of the adsorbed FITC-labeled polymers by the CD206+ macrophages membranes with pre-adsorbed non-labelled polymers of different affinities for CD206 receptor.
| |
|
Membranes of CD206+ macrophages with pre-adsorbed polymer |
| Displacing polymer |
Concentration of displacing polymer |
Mannan-sp-HPCD |
HPCD-PEI-Gal |
HPCD-PEI-Man |
HPCD-PEItriMan |
| HPCD-PEI-triMan-FITC, High affinity |
5 µg/mL |
14±2 |
60±4 |
36±5 |
21±3 |
| 100 µg/mL |
4±1 |
29±3 |
15±2 |
9±1 |
HPCD-PEI-Gal-FITC,
Low affinity |
5 µg/mL |
8±1 |
43±5 |
18±1 |
10±2 |
| 100 µg/mL |
3±1 |
17±3 |
6±1 |
2±0.5 |
Table 3.
Binding of FITC-labeled ligands to bronchoalveolar and nasopharyngeal lavage and inhibition of this interaction by mannan to elucidate the CD206-dependent mechanism.
Table 3.
Binding of FITC-labeled ligands to bronchoalveolar and nasopharyngeal lavage and inhibition of this interaction by mannan to elucidate the CD206-dependent mechanism.
| Polymer, 0.5 mg/mL |
Bronchoalveolar lavage |
Nasopharyngeal lavage |
| free |
+ mannan |
free |
+ mannan |
| HPCD-PEI-Man-FITC |
10±1 |
4±1 |
12±1 |
46±3 |
| HPCD-PEI-Gal-FITC |
7±1 |
9±1 |
4±1 |
54±4 |
| HPCD-PEI-triMan-FITC |
27±3 |
18±2 |
70±4 |
58±4 |
| Mannan-sp-HPCD-FITC |
16±2 |
9±1 |
63±3 |
59±5 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).