Submitted:
26 August 2023
Posted:
29 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Models and Methods
2.1. Problem Statement
2.2. Materials
2.3. Dataset Balancing
2.4. Data Preprocessing
2.5. Variable or Feature Selection
3. Experimental Results
3.1. Classification
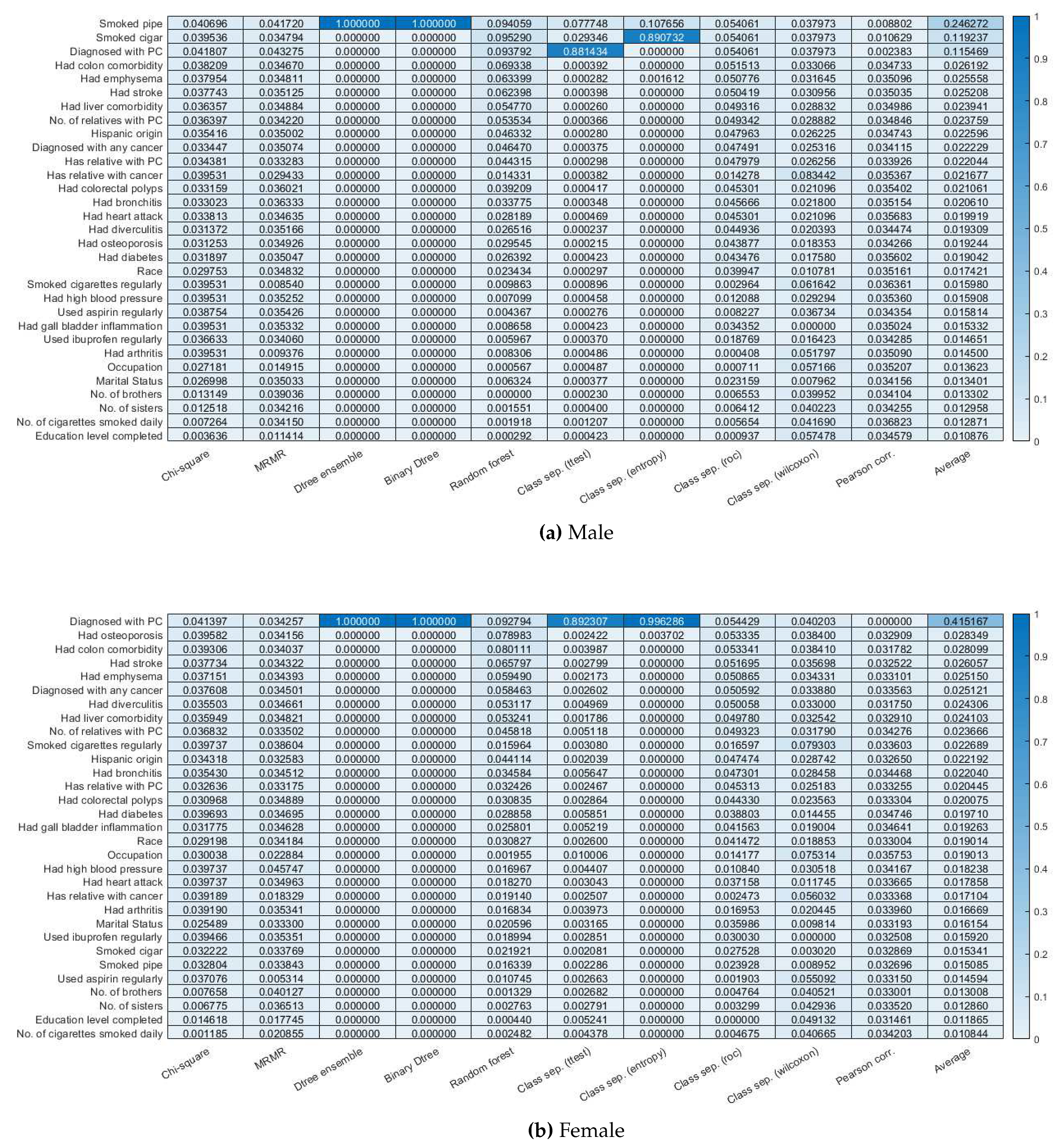
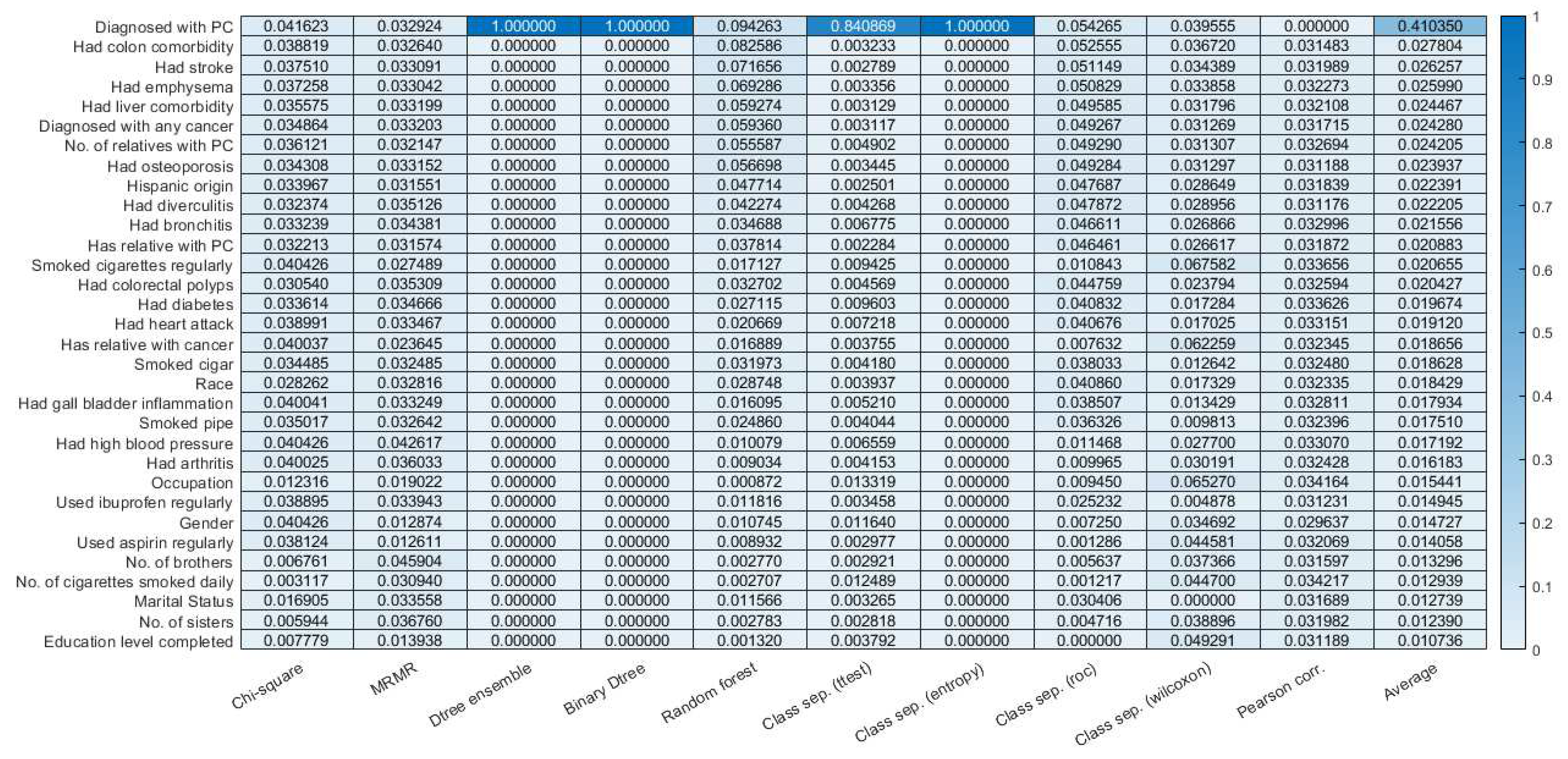
3.2. Feature selection.
3.3. Finding probability feature combination using a Bayesian Network
4. Discussions
- Hereditary breast and ovarian cancer syndrome, caused by mutations in the BRCA1 or BRCA2 genes,
- Hereditary breast cancer, caused by mutations in the PALB2 gene,
- Familial atypical multiple mole melanoma (FAMMM) syndrome, caused by mutations in the p16/CDKN2A gene and associated with skin and eye melanomas,
- Familial pancreatitis, usually caused by mutations in the PRSS1 gene,
- Lynch syndrome, also known as hereditary non-polyposis colorectal cancer (HNPCC), most often caused by a defect in the MLH1 or MSH2 genes,
- Peutz-Jeghers syndrome, caused by defects in the STK11 gene. This syndrome is also linked with polyps in the digestive tract and several other cancers.
| Symptom 1 | Symptom 2 | Probability |
|---|---|---|
| No of cigarettes smoked daily= 61-80 | No of relatives with PC=2+ | 0.156 |
| No of tubal/ectopic pregnancies=2+ | No of relatives with PC=2+ | 0.137 |
| Usually filtered or not filtered?=Both | No of relatives with PC=2+ | 0.115 |
| No of cigarettes smoked daily= 61-80 | No of relatives with PC=2+ | 0.095 |
| No of tubal/ectopic pregnancies=1 | No of relatives with PC=2+ | 0.084 |
| Heart attack history?=yes | No of relatives with PC=2+ | 0.08 |
| No of cigarettes smoked daily=21-30 | No of relatives with PC=2+ | 0.077 |
| No of relatives with PC=2+ | Race=Asian | 0.076 |
| No of relatives with PC=2+ | No of still births=1 | 0.074 |
| No of relatives with PC=2+ | Diabetes history?=Yes | 0.0737 |
| No of relatives with PC=2+ | Race=American Indian | 0.0737 |
| No of relatives with PC=2+ | Emphysema history?=Yes | 0.0737 |
| No of relatives with PC=2+ | No of cigarettes smoked daily=31-40 | 0.0708 |
| No of relatives with PC=2+ | Colorectal Polyps history?=Yes | 0.0708 |
| No of relatives with PC=2+ | Stroke history?=Yes | 0.0704 |
| No of relatives with PC=2+ | Age at hysterectomy=40-44 | 0.0686 |
| No of relatives with PC=2+ | No. of brothers=7+ | 0.0645 |
| No of relatives with PC=2+ | Bronchitis history?=2+ | 0.064 |
| No of relatives with PC=2+ | Liver comorbidities history?=Yes | 0.063 |
| No of relatives with PC=2+ | No of cigarettes smoked daily=11-20 | 0.063 |
| Male | |||
|---|---|---|---|
| Symptom 1 conditional probability | Symptom 2 conditional probability | Symptom 3 conditional probability | Total probability |
| No of cigarettes smoked daily is 61-80=0.005 | Age when told had enlarged prostate is 70+ =0.0175 | prior history of cancer is yes=0.05 | 0.00521 |
| No of cigarettes smoked daily is 61-80=0.005 | Age when told had enlarged prostate is 70+ =0.0175 | family history of PC=yes=0.533 | 0.002368 |
| No of cigarettes smoked daily is 61-80=0.005 | Age when told had enlarged prostate is 70+ =0.0175 | no. of relatives with PC is 1=0.04 | 0.004593 |
| prior history of cancer is yes=0.05 | Age when told had enlarged prostate is 70+ =0.0175 | family history of PC is yes=0.533 | 0.002153 |
| prior history of cancer is yes=0.05 | Age when told had enlarged prostate is 70+ =0.0175 | no. of relatives with PC is 1=0.04 | 0.004177 |
| Age when told had enlarged prostate is 70+ =0.0175 | family history of PC is yes=0.533 | no. of relatives with PC is 1=0.04 | 0.00189 |
| No of cigarettes smoked daily is 61-80=0.005 | prior history of cancer is yes=0.05 | family history of PC=yes=0.533 | 0.02742 |
| No of cigarettes smoked daily is 61-80=0.005 | prior history of cancer is yes=0.05 | no. of relatives with PC is 1=0.04 | 0.05197055 |
| No of cigarettes smoked daily is 61-80=0.005 | family history of PC is yes=0.533 | no. of relatives with PC is 1=0.04 | 0.024218 |
| prior history of cancer is yes=0.05 | family history of PC is yes=0.533 | no. of relatives with PC is 1=0.04 | 0.02206 |
| Female | |||
| No of tubal/ectopic pregnancies is 1=0.003 | No. of relatives with PC is 2+=0.011 | no. of cigarettes smoked is 61-80=0.007 | 0.3578 |
5. Conclusion
6. Appendix
6.1. Data Visualization Methods
6.2. Data Balancing Methods
6.3. Feature Selection Methods
- Rank the magnitudes (absolute values) of the deviations of the observed values from the hypothesized median, adjusting for ties if they exist.
- Assign to each rank the sign (+ or - ) of the deviation (thus, the name “signed rank”).
- Compute the sum of positive ranks,T(+) , or negative ranks,T(-) , the choice depending on which is easier to calculate. The sum of T(+) and T(-) is n(n+1)/2, so either can be calculated from the other.
- Choose the smaller of T(+) and T(-), and call this T.
- Since the test statistic is the minimum of T(+) and T(-), the critical region consists of the left tail of the distribution, containing a probability of at most . If n is large, the sampling distribution of T is approximately normal withwhich can be used to compute a z-statistic for the hypothesis test.
- Initialize set F to the whole set of p features. S is an empty set.
- For all features compute J(f) coefficient.
- Find feature f that maximizes J(f) and move it to ,
- Repeat until the cardinal of S is p
- As is actually true for any statistical inference, the data are derived from a random, or at least representative, sample. If the data are not representative of the population of interest, one cannot draw meaningful conclusions about that population.
- Both variables are continuous, jointly normally distributed, random variables. They follow a bivariate normal distribution in the population from which they were sampled. The bivariate normal distribution is beyond the scope of this tutorial but need not be fully understood to use a Pearson coefficient. The equation for correlation coefficent is represented as follows [77]:
6.4. Classification Methods
6.5. Evaluation Matrices
References
- Wikipedia contributors. Wikipedia, The Free Encyclopedia. Available online: https://en.wikipedia.org/ (accessed on 25 August 2019).
- Ik-Gyu, J. Method of providing information for the diagnosis of pancreatic cancer using bayesian network based on artificial intelligence, computer program, and computer-readable recording media using the same, 2019. US Patent App. 15/833,828.
- Huxley, R.; Ansary-Moghaddam, A.; De González, A.B.; Barzi, F.; Woodward, M. Type-II diabetes and pancreatic cancer: a meta-analysis of 36 studies. British journal of cancer 2005, 92, 2076. [Google Scholar] [CrossRef] [PubMed]
- Everhart, J.; Wright, D. Diabetes mellitus as a risk factor for pancreatic cancer: a meta-analysis. Jama 1995, 273, 1605–1609. [Google Scholar] [CrossRef] [PubMed]
- Ben, Q.; Xu, M.; Ning, X.; Liu, J.; Hong, S.; Huang, W.; Zhang, H.; Li, Z. Diabetes mellitus and risk of pancreatic cancer: a meta-analysis of cohort studies. European journal of cancer 2011, 47, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Hruban, R.H.; Kamiyama, M.; Borges, M.; Zhang, X.; Parsons, D.W.; Lin, J.C.H.; Palmisano, E.; Brune, K.; Jaffee, E.M.; others. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009, 324, 217–217. [Google Scholar] [CrossRef]
- Barton, C.; Staddon, S.; Hughes, C.; Hall, P.; O’sullivan, C.; Klöppel, G.; Theis, B.; Russell, R.; Neoptolemos, J.; Williamson, R.; others. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. British journal of cancer 1991, 64, 1076. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; others. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. Journal of clinical oncology 2009, 27, 1806. [Google Scholar] [CrossRef]
- Das, A.; Nguyen, C.C.; Li, F.; Li, B. Digital image analysis of EUS images accurately differentiates pancreatic cancer from chronic pancreatitis and normal tissue. Gastrointestinal endoscopy 2008, 67, 861–867. [Google Scholar] [CrossRef]
- Ge, G.; Wong, G.W. Classification of premalignant pancreatic cancer mass-spectrometry data using decision tree ensembles. BMC bioinformatics 2008, 9, 275. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Gheonea, D.I.; Gorunescu, M.; Ciurea, T.; Popescu, G.L.; Iordache, A.; Hassan, H.; Iordache, S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointestinal endoscopy 2008, 68, 1086–1094. [Google Scholar] [CrossRef]
- Zhang, M.M.; Yang, H.; Jin, Z.D.; Yu, J.G.; Cai, Z.Y.; Li, Z.S. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointestinal endoscopy 2010, 72, 978–985. [Google Scholar] [CrossRef]
- Baruah, M.; Banerjee, B. Modality selection for classification on time-series data. MileTS 2020, 20, 6th. [Google Scholar]
- Biometry.nci.nih.gov. (2019). Pancreas-Datasets-PLCO-The Cancer Data Access System. Available online: https://biometry.nci.nih.gov/cdas/datasets/plco/10/ (accessed on 25 August 2019).
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence). IEEE, 2008, pp. 1322–1328.
- Maaten, L.v.d.; Hinton, G. Visualizing data using t-SNE. Journal of machine learning research 2008, 9, 2579–2605. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Khalilia, M.; Chakraborty, S.; Popescu, M. Predicting disease risks from highly imbalanced data using random forest. BMC medical informatics and decision making 2011, 11, 1–13. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. Journal of machine learning research 2003, 3, 1157–1182. [Google Scholar]
- Tang, J.; Alelyani, S.; Liu, H. Feature selection for classification: A review. Data classification: Algorithms and applications 2014, 37. [Google Scholar]
- Roffo, G.; Melzi, S. Ranking to Learn. International Workshop on New Frontiers in Mining Complex Patterns. Springer, 2016, pp. 19–35.
- Mangasarian, O.; Bradley, P. Feature Selection via Concave Minimization and Support Vector Machines. Technical report, 1998.
- He, X.; Cai, D.; Niyogi, P. Laplacian score for feature selection. Advances in neural information processing systems, 2006, pp. 507–514.
- Yang, Y.; Shen, H.T.; Ma, Z.; Huang, Z.; Zhou, X. L2, 1-Norm Regularized Discriminative Feature Selection for Unsupervised. Twenty-Second International Joint Conference on Artificial Intelligence, 2011.
- Du, L.; Shen, Y.D. Unsupervised feature selection with adaptive structure learning. Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining. ACM, 2015, pp. 209–218.
- Hall, M.A. Correlation-based feature selection for machine learning 1999.
- Fonti, V.; Belitser, E. Feature selection using lasso. VU Amsterdam Research Paper in Business Analytics 2017. [Google Scholar]
- Guo, J.; Zhu, W. Dependence guided unsupervised feature selection. Thirty-Second AAAI Conference on Artificial Intelligence, 2018.
- Roffo, G. Ranking to learn and learning to rank: On the role of ranking in pattern recognition applications. arXiv 2017, arXiv:1706.05933 2017. [Google Scholar]
- Roffo, G.; Melzi, S.; Castellani, U.; Vinciarelli, A. Infinite latent feature selection: A probabilistic latent graph-based ranking approach. Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 1398–1406.
- Baine, M.; Sahak, F.; Lin, C.; Chakraborty, S.; Lyden, E.; Batra, S.K. Marital status and survival in pancreatic cancer patients: a SEER based analysis. PloS one 2011, 6, e21052. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Liu, X.Y. On multi-class cost-sensitive learning. Computational Intelligence 2010, 26, 232–257. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and regression trees. Belmont, CA: Wadsworth. International Group 1984, 432, 151–166. [Google Scholar]
- Zadrozny, B.; Langford, J.; Abe, N. Cost-Sensitive Learning by Cost-Proportionate Example Weighting. ICDM, 2003, Vol. 3, p. 435.
- Russell, S.J.; Norvig, P. Artificial intelligence: a modern approach; Malaysia; Pearson Education Limited, 2016.
- ”What are risk factors for Pancreatic Cancer?” The Sol Goldman Pancreatic Cancer Research Center, JHU. Available online: https://pathology.jhu.edu/pancreas/BasicRisk.php?area=ba.
- Pancreatic Cancer: Risk Factors Approved by the Cancer.Net Editorial Board, 05/2018. Available online: https://www.cancer.net/cancer-types/pancreatic-cancer/risk-factors.
- Pancreatic Cancer Risk Factors. 2019. Available online: https://www.cancer.org/cancer/pancreatic-cancer/causes-risks-prevention/risk-factors.html.
- Li, D.; Xie, K.; Wolff, R.; Abbruzzese, J.L. Pancreatic cancer. The Lancet 2004, 363, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.M.; Vrieling, A.; Lubin, J.H.; Kraft, P.; Mendelsohn, J.B.; Hartge, P.; Canzian, F.; Steplowski, E.; Arslan, A.A.; Gross, M.; others. Cigarette smoking and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. American journal of epidemiology 2009, 170, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Louizos, C.; Welling, M.; Kingma, D.P. Learning Sparse Neural Networks through L_0 Regularization. arXiv 2017, arXiv:1712.01312 2017. [Google Scholar]
- Muscat, J.E.; Stellman, S.D.; Hoffmann, D.; Wynder, E.L. Smoking and pancreatic cancer in men and women. Cancer Epidemiology and Prevention Biomarkers 1997, 6, 15–19. [Google Scholar]
- Yadav, D.; Lowenfels, A.B. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013, 144, 1252–1261. [Google Scholar] [CrossRef]
- Raimondi, S.; Maisonneuve, P.; Lowenfels, A.B. Epidemiology of pancreatic cancer: an overview. Nature reviews Gastroenterology & hepatology 2009, 6, 699. [Google Scholar]
- Silverman, D.; Schiffman, M.; Everhart, J.; Goldstein, A.; Lillemoe, K.; Swanson, G.; Schwartz, A.; Brown, L.; Greenberg, R.; Schoenberg, J.; others. Diabetes mellitus, other medical conditions and familial history of cancer as risk factors for pancreatic cancer. British journal of cancer 1999, 80, 1830. [Google Scholar] [CrossRef]
- Liao, K.F.; Lai, S.W.; Li, C.I.; Chen, W.C. Diabetes mellitus correlates with increased risk of pancreatic cancer: a population-based cohort study in Taiwan. Journal of gastroenterology and hepatology 2012, 27, 709–713. [Google Scholar] [CrossRef]
- Lo, A.C.; Soliman, A.S.; El-Ghawalby, N.; Abdel-Wahab, M.; Fathy, O.; Khaled, H.M.; Omar, S.; Hamilton, S.R.; Greenson, J.K.; Abbruzzese, J.L. Lifestyle, occupational, and reproductive factors in relation to pancreatic cancer risk. Pancreas 2007, 35, 120–129. [Google Scholar] [CrossRef]
- Kreiger, N.; Lacroix, J.; Sloan, M. Hormonal factors and pancreatic cancer in women. Annals of epidemiology 2001, 11, 563–567. [Google Scholar] [CrossRef]
- Aizer, A.A.; Chen, M.H.; McCarthy, E.P.; Mendu, M.L.; Koo, S.; Wilhite, T.J.; Graham, P.L.; Choueiri, T.K.; Hoffman, K.E.; Martin, N.E.; others. Marital status and survival in patients with cancer. Journal of clinical oncology 2013, 31, 3869. [Google Scholar] [CrossRef] [PubMed]
- Logan, W. Cancer mortality by occupation and social class 1851-1971 1982.
- Ghadirian, P.; Simard, A.; Baillargeon, J. Cancer of the pancreas in two brothers and one sister. International journal of pancreatology 1987, 2, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.L.; Lombardo, K.M.R.; Bamlet, W.R.; Oberg, A.L.; Robinson, D.P.; Anderson, K.E.; Petersen, G.M. Aspirin, nonsteroidal anti-inflammatory drugs, acetaminophen, and pancreatic cancer risk: a clinic-based case–control study. Cancer prevention research 2011, 4, 1835–1841. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Giovannucci, E.; Bergkvist, L.; Wolk, A. Aspirin and nonsteroidal anti-inflammatory drug use and risk of pancreatic cancer: a meta-analysis. Cancer Epidemiology and Prevention Biomarkers 2006, 15, 2561–2564. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.E.; Beebe-Donk, J.; Doss, H.; Doss, D.B. Aspirin, ibuprofen, and other non-steroidal anti-inflammatory drugs in cancer prevention: a critical review of non-selective COX-2 blockade. Oncology reports 2005, 13, 559–583. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, L.; Palmer, J.R.; Zauber, A.G.; Warshauer, M.E.; Strom, B.L.; Harlap, S.; Shapiro, S. The relation of vasectomy to the risk of cancer. American journal of epidemiology 1994, 140, 431–438. [Google Scholar] [CrossRef]
- Andersson, G.; Borgquist, S.; Jirstrom, K. Hormonal factors and pancreatic cancer risk in women: The Malmo Diet and Cancer Study. International Journal of Cancer 2018, 143, 52–62. [Google Scholar] [CrossRef]
- Stolzenberg-Solomon, R.Z.; Pietinen, P.; Taylor, P.R.; Virtamo, J.; Albanes, D. A prospective study of medical conditions, anthropometry, physical activity, and pancreatic cancer in male smokers (Finland). Cancer causes and control 13.
- Navi, B.B.; Reiner, A.S.; Kamel, H.; Ladecola, C.; Okin, P.M.; Tagawa, S.T.; Panageas, K.S.; DeAngelis, L.M. Arterial thromboembolic events preceding the diagnosis of cancer in older persons. Clinical trials and observations 2019, 133, 781–789. [Google Scholar] [CrossRef]
- Bertero, E.; Canepa, M.; Maack, C.; Ameri, P. Linking heart failure to cancer. Circulations 2018, 138, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; White, D.L.; Hoogeveen, R.; Chen, L.; Whitsel, E.A.; Richardson, P.A.; Virani, S.S.; Garcia, J.M.; El-Serag, H.B.; Jiao, L. Anti-Hypertensive Medication Use, Soluble Receptor for Glycation End Products and Risk of Pancreatic Cancer in the Women’s Health Initiative Study. Journal of clinical medicine 197. [CrossRef] [PubMed]
- Zhou, H.; Wang, F.; Tao, P. t-Distributed stochastic neighbor embedding method with the least information loss for macromolecular simulations. Journal of chemical theory and computation 2018, 14, 5499–5510. [Google Scholar] [CrossRef] [PubMed]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The global k-means clustering algorithm. Pattern recognition 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Mathworks.com. Train models to classify data using supervised machine learning - MATLAB. Available online: https://www.mathworks.com/help/stats/classificationlearner-app.html (accessed on 25 August 2019).
- Manning, C.; Raghavan, P.; Schütze, H. Introduction to Information Retrieval; Vol. 39, Cambridge University Press, 2008.
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC bioinformatics 2017, 18, 1–14. [Google Scholar] [CrossRef]
- Ding, C.; Peng, H. Minimum redundancy feature selection from microarray gene expression data. Journal of bioinformatics and computational biology 2005, 3, 185–205. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Machine learning 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and regression trees 2017.
- Coppersmith, D.; Hong, S.J.; Hosking, J.R. Partitioning nominal attributes in decision trees. Data Mining and Knowledge Discovery 1999, 3, 197–217. [Google Scholar] [CrossRef]
- Rugeles, D. Study of feature ranking using Bhattacharyya distance 2012. 1, 1.
- King, A.P.; Eckersley, R. Statistics for biomedical engineers and scientists: How to visualize and analyze data; Academic Press, 2019.
- Biesiada, J.; Duch, W.; Kachel, A.; Maczka, K.; Palucha, S. Feature ranking methods based on information entropy with parzen windows 2005. 1, 1.
- Liu, H.; Motoda, H. Feature selection for knowledge discovery and data mining 2012. 454.
- Wikipedia contributors. Wikipedia, The Free Encyclopedia. Available online: https://en.wikipedia.org/Wilcoxon_signed-rank_test (accessed on 5 June 2022).
- Loh, W.; Shih, Y. Split selection methods for classification trees. Statistica sinica.
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation coefficients: appropriate use and interpretation. Anesthesia & Analgesia 2018, 126, 1763–1768. [Google Scholar]
- Wikipedia contributors. Wikipedia, The Free Encyclopedia. Available online: https://en.wikipedia.org/Pearson_correlation_coefficient (accessed on 26 June 2022).
- Roffo, G.; Melzi, S.; Cristani, M. Infinite feature selection. Proceedings of the IEEE International Conference on Computer Vision, 2015, pp. 4202–4210.
- Kira, K.; Rendell, L.A. ; others. The feature selection problem: Traditional methods and a new algorithm. 1992, 2, 129–134. [Google Scholar]
- Kira, K.; Rendell, L.A. A practical approach to feature selection 1992. pp. 249–256.
- Wikipedia contributors. Wikipedia, The Free Encyclopedia. 2022. Available online: https://en.wikipedia.org/Relief_(feature_selection) (accessed on 5 June 2022).
- Wikipedia contributors. Wikipedia, The Free Encyclopedia 2022. [Online; accessed 5-Jun-2022].
- Seiffert, C.; Khoshgoftaar, T.M.; Van Hulse, J.; Napolitano, A. RUSBoost: A hybrid approach to alleviating class imbalance. IEEE Transactions on Systems, Man, and Cybernetics-Part A: Systems and Humans 2009, 40, 185–197. [Google Scholar] [CrossRef]
- Wikipedia contributors. Wikipedia, The Free Encyclopedia. Available online: https://en.wikipedia.org/Precision_and_recall) (accessed on 5 June 2022).

| Risk factor categories | Risk factors(values) | Male risk factors (total 47, removed 15) | Female risk factors (total 52, removed 20) |
|---|---|---|---|
| Cancer history | 59. Has relative with cancer (yes, no) | ||
| 60. Has relative with PC (yes, no) | |||
| 61. No. of relatives with PC (0, 1, 2, 3, ...) | |||
| 62. Diagnosed with any cancer (yes, no) | |||
| 63. Diagnosed with PC (yes, no) | |||
| Demo- graphics | 64. Gender (male, female) | ||
| 38. Race (White, Black, Asian, Pacific Islander, American Indian/Alaskan Native) | |||
| 39. Hispanic origin (yes, no) | |||
| 1. Education level completed (<8 yrs, 8-11 yrs, 12 yrs, 12 yrs + some college, college grad, post grad) | |||
| 2. Marital status (married, widowed, divorced, separated, never married) | |||
| 3. Occupation (homemaker, working, unemployed, retired, extended sick leave, disabled, other) | |||
| 6. No. of sisters (0, 1, 2, 3, 4, 5, 6, ≥7) | |||
| 7. No. of brothers (0, 1, 2, 3, 4, 5, 6, ≥7) | |||
| Medi- cation usage | 8. Used aspirin regularly (yes, no) | ||
| 9. Used ibuprofen regularly (yes, no) | |||
| 52. Taken birth control pills (yes, no) | (removed) | ||
| 20. Age started taking birth control pills (<30 yrs, 30-39 yrs, 40-49 yrs, 50-59 yrs, ≥60 yrs) | (removed) | ||
| 21. Currently taking female hormones (yes, no) | (removed) | ||
| 22. No. of years taking female hormones (≤1, 2-3, 4-5, 6-9, ≥10) | (removed) | ||
| 53. Taken female hormones (yes, no, don’t know) | (removed) | ||
| Health history | 27. Had high blood pressure (yes, no) | ||
| 28. Had heart attack (yes, no) | |||
| 29. Had stroke (yes, no) | |||
| 30. Had emphysema (yes, no) | |||
| 31. Had bronchitis (yes, no) | |||
| 32. Had diabetes (yes, no) | |||
| 33. Had colorectal polyps (yes, no) | |||
| 34. Had arthritis (yes, no) | |||
| 35. Had osteoporosis (yes, no) | |||
| 36. Had diverculitis (yes, no) | |||
| 37. Had gall bladder inflammation (yes, no) | |||
| 57. Had colon comorbidity (yes, no) | |||
| 58. Had liver comorbidity (yes, no) | |||
| 40. Had biopsy of prostrate (yes, no) | (removed) | ||
| 41. Had transurethral resection of prostate (yes, no) | (removed) | ||
| 42. Had prostatetomy of benign disease (yes, no) | (removed) | ||
| 43. Had prostate surgery (yes, no) | (removed) | ||
| 47. Had enlarged prostate (yes, no) | (removed) | ||
| 48. Had inflamed prostate (yes, no) | (removed) | ||
| 49. Had prostate problem (yes, no) | (removed) | ||
| 50. No. of times wakes up to urinate at night (0, 1, 2, 3, >3) | (removed) | ||
| 23. Age started to urinate more than once at night (<30 yrs, 30-39 yrs, 40-49 yrs, 50-59 yrs, 60-69 yrs, ≥70 yrs) | (removed) | ||
| 24. Age when told had enlarged prostate (<30 yrs, 30-39 yrs, 40-49 yrs, 50-59 yrs, 60-69 yrs, ≥70 yrs) | (removed) | ||
| 25. Age when told had inflammed prostate (<30 yrs, 30-39 yrs, 40-49 yrs, 50-59 yrs, 60-69 yrs, ≥70 yrs) | (removed) | ||
| 26. Age at vasectomy (<25 yrs, 25-34 yrs, 35-44 yrs, ≥45 yrs) | (removed) | ||
| 51. Had vasectomy (yes, no) | (removed) | ||
| 44. Been pregnant (yes, no, don’t know) | (removed) | ||
| 45. Had hysterectomy (yes, no) | (removed) | ||
| 46. Had ovaries removed (yes, no) | (removed) | ||
| 10. No. of tubal pregnancies (0, 1, ≥2) | (removed) | ||
| 11. Had tubal ligation (yes, no, don’t know) | (removed) | ||
| 12. Had benign ovarian tumor (yes, no) | (removed) | ||
| 13. Had benign breast disease (yes, no) | (removed) | ||
| 14. Had endometriosis (yes, no) | (removed) | ||
| 15. Had uterine fibroid tumors (yes, no) | (removed) | ||
| 16. Tried to become pregnant without success (yes, no) | (removed) | ||
| 17. No. of pregnancies (0, 1, 2, 3, 4-9, ≥10) | (removed) | ||
| 18. No. of stillbirth pregnancies (0, 1, ≥2) | (removed) | ||
| 19. Age at hysterectomy (<40 yrs, 40-44 yrs, 45-49 yrs, 50-54 yrs, ≥55 yrs) | (removed) | ||
| Smoking habits | 4. Smoked pipe (never, currently, formerly) | ||
| 5. Smoked cigar (never, currently, formerly) | |||
| 54. Smoked cigarettes regularly (yes, no) | |||
| 55. Smoke regularly now (yes, no) | (removed) | (removed) | |
| 56. Usually filtered or not filtered (filter more often, non-filter more often, both about equally) | (removed) | (removed) | |
| 65. No. of cigarettes smoked daily (0, 1-10, 11-20, 21-30, 31-40, 41-60, 61-80, >80) |
| Symptoms | Results | Conclusion |
|---|---|---|
| Occupation | All subjects in HPT were retired(category 4) and in LPT, they were extended sick leave(category 5) | Older people have a greater risk of PC |
| Smoked pipe | Subjects in HPT were in ratio 0.22(never smoked):0.5(current smoker):0.28(past smoker) whereas subjects in LPT were in ratio 0.37(never smoked):0.27(current smoker):0.36(past smoker) | Subjects who never smoked have a lesser risk than past smokers and risk for current smokers was doubled |
| Heart Attack | Subjects in HPT were in ratio 0.23(never had heart attack):0.77(had heart attack) whereas subjects in LPT were in ratio 0.8(never had heart attack):0.2(had heart attack) | Subjects who had heart attack at least once have a greater risk for PC |
| Hypertension | Subjects in HPT were in ratio 0.36 (not diagnosed with hypertension):0.63(diagnosed with hypertension) whereas subjects in LPT were in ratio 0.68 (not diagnosed with hypertension):0.32(diagnosed with hypertension) | Stress(or hypertension) is directly proportional to risk for PC |
| Taken female hormones | Subjects in HPT were in ratio 0.7(never taken):0.3(taken) whereas subjects in LPT were in ratio 0.23(never taken):0.77(taken) | Somehow female hormones reduces risk of PC |
| Race | Subjects in HPT were mostly Asian(0.38) and only 0.3 were Pacific Islander whereas subjects in LPT were mostly American Indian(0.85) | Clearly shows that Asians are at a higher risk of PC while Pacific Islander and American Indian were at lower risk. |
| Diabetes | Subjects in HPT were in ratio 0.17(never had diabetes):0.83(had diabetes) whereas subjects in LPT were in ratio 0.75(never had diabetes):0.25(had diabetes) | Diabetes is a clear risk factor for PC |
| Bronchitis | Subjects in HPT were in ratio 0.27(never had):0.73(had) whereas subjects in LPT were in ratio 0.68(never had ):0.32(had) | Bronchitis is a risk factor for PC |
| Liver comorbidities | Subjects in HPT were in ratio 0.39(never had):0.61(had) whereas subjects in LPT were in ratio 0.62(never had ):0.38(had) | Liver comorbidities is a risk factor for PC |
| Colorectal Polyps | Subjects in HPT were in ratio 0.36(never had):0.64(had) whereas subjects in LPT were in ratio 0.62(never had ):0.38(had) | Colorectal Polyps is a risk factor for PC |
| Gender | Subjects in HPT were in ratio 0.53(male):0.47(female) whereas subjects in LPT were in ratio 0.35(male):0.65(female) | Male were at higher risk of PC than female |
| No of relatives with pancreatic cancer | Subjects in HPT were in ratio 0.02(no relative):0.1(1 relative):0.88(2 relatives) whereas subjects in LPT were in ratio 0.71(no relative:0.29(1 relative) | Risk of PC increases as incidence of PC on family members increases. |
| Ever take birth control pills? | Subjects in HPT were in ratio 0.76(no history):0.24(has history) whereas subjects in LPT were in ratio 0.17(no history):0.83(has history) | birth control pills may lower risk of PC |
| Smoke regularly now? | Subjects in HPT were in ratio 0.12(no history):0.88(has history) whereas subjects in LPT were in ratio 0.95(no history):0.05(has history) | Current smokers have higher risk of PC |
| Ever smoke regularly more than 6 months? | Subjects in HPT were in ratio 0.22(no history):0.78(has history) whereas subjects in LPT were in ratio 0.85(no history):0.15(has history) | Smoking in excess of 6 months also poses higher risk of PC |
| Symptom 1 | Symptom 2 | Probability |
|---|---|---|
| Age when told had inflamed prostate= 70+ | No of cigarettes smoked daily=80+ | 0.032 |
| Age when told had inflamed prostate= 70+ | Prior history of any cancer?= Yes | 0.03 |
| Prior history of any cancer?= Yes | No of cigarettes smoked daily=80+ | 0.026 |
| Age when told had inflamed prostate= 70+ | Age when told had enlarged prostate= 70+ | 0.026 |
| Age when told had inflamed prostate= 70+ | Family history of PC?=Yes | 0.026 |
| Age when told had inflamed prostate= 70+ | No of relatives with PC=1 | 0.026 |
| Age when told had enlarged prostate= 70+ | No of cigarettes smoked daily=80+ | 0.024 |
| Family history of PC=Yes | No of cigarettes smoked daily=80+ | 0.024 |
| No of relatives with PC=1 | No of cigarettes smoked daily=80+ | 0.024 |
| Age when told had inflamed prostate= 70+ | Bronchitis history?=Yes | 0.022 |
| Age when told had enlarged prostate= 70+ | Prior history of any cancer?=Yes | 0.022 |
| Prior history of any cance?r=Yes | Family history of PC=Yes | 0.022 |
| Prior history of any cancer?=Yes | No of relatives with PC=1 | 0.021 |
| Age when told had inflamed prostate= 70+ | Gall bladder stone or inflammation=Yes | 0.021 |
| Age when told had inflamed prostate= 70+ | Smoke regularly now?=Yes | 0.021 |
| No of cigarettes smoked daily=80+ | Bronchitis history?=Yes | 0.021 |
| Age when told had inflamed prostate= 70+ | During past year, how many times wake up in the night to urinate?=Thrice | 0.021 |
| Age when told had inflamed prostate= 70+ | Smoked pipe=current smoker | 0.021 |
| Age when told had inflamed prostate= 70+ | Diabetes history=yes | 0.02 |
| Age when told had inflamed prostate= 70+ | No. of brother=7+ | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




