Key Points
Question: What were the associations between genetic sequence variations of the Helicobacter pylori (H. pylori) ureC gene and the pathogenesis of peptic ulcer diseases?
Findings: The study found distinct genetic variations in the H. pylori ureC gene among Taiwanese samples. Gastric ulcer samples tended to cluster together, while gastric cancer samples were distributed among other groups. UreC sequences from China formed a separate and unique group.
Meaning: These findings suggest a potential correlation between genetic sequence variations of the ureC gene and the development of peptic ulcer diseases. The clustering patterns observed and the distinct group from China indicate the presence of different genotypes or strains of H. pylori. Further research is needed to explore the implications of these genetic variations and their role in the pathogenesis of peptic ulcer diseases.
1. Introduction
Helicobacter pylori (
H. pylori) plays an important mediator role for development of peptic ulcer diseases, including gastric ulcer (GU) and duodenal ulcer (DU), and gastric cancer [
1]. Numerous investigations reported that the prevalence of
H. pylori infection worldwide was decreasing because of the improvement of living-style and sanitary environment, as well as availability of antibiotics and proton pump inhibitor for the treatment of
H. pylori [
2,
3]. Nevertheless, the incidence of
H. pylori infection-mediated gastric cancer remains high in Taiwan [
4], and it is unclear what factors predispose the final development of gastric cancer in
H. pylori-infected patients.
Our previous study demonstrated that
ureC genotypes was associated with patients with GU and gastric malignancy, by use of a polymerase chain reaction (PCR)-based restriction fragment length polymorphism (RFLP) method [
5]. However, the associations between the genetic information (DNA sequences) of
ureC gene and pathogenesis of peptic ulcer diseases remain elusive. In the present study,
ureC sequences derived from
H. pylori-infected gastric tissues of different pathologies were amplified and subjected to DNA sequencing. Based on these sequences, phylogenetic analysis was conducted.
2. Materials and Methods
2.1. Patients and histopathological analysis
H. pylori-positive gastric specimens of different pathologies, including gastritis, gastric ulcer (GU), duodenal ulcer (DU) and gastric cancer, were obtained from Chang Gung Memorial Hospital. All tissues were retrieved from our tissue bank. The research had been approved by IRB committee (IRB: 202200046A3). Clinical parameters of patients in this study were shown in
Table 1, including gender, age and pathology.
2.2. DNA extraction and polymerase chain reaction (PCR)
Genomic DNA extraction from frozen tissues was performed as previously described [
5]. The nested PCR procedure was used: 95 °C for 10 min, 30 cycles of 95 °C for 30 s and 55 °C for 1 min, and 72 °C for 1 min using primers ureC-1 forward primer: 5’-TTTGGGACTGATGGCGTGAGGGGTAA-3’ and ureC-1 reverse primer: 5’-GGACATTCAAATTCACCAGGTTTTGAGG-3’ for the first round PCR and ureC-2 forward primer: 5’-TGGGACTGATGGCGTGAGGG-3’ and ureC-2 reverse primer: 5’-AAGGGCGTTTTTAGATTTTT-3’ for the second round PCR. The PCR amplicon was monitored by 1.2% agarose gel electrophoresis with DNA view (TOOLs). Subsequently, the
ureC sequences in these samples were isolated and sent for DNA sequencing.
2.3. Data and statistical analysis
The
ureC sequences in our samples and previous characterized sequences obtained from GenBank were aligned using CLUSTAL W mode. Moreover, phylogenetic tree analysis was conducted by MEGA X software (version 10.2.4) with Maximum-Likeihood method and bootstrap analysis (1000 replicates)[
6].
3. Results
3.1. Identification and determination of ureC sequences in Taiwanese patients
To determine whether
H. pylori infection based on
ureC sequences was correlated the pathologies of gastric mucosal diseases, including gastritis, gastric ulcer (GU), duodenal ulcer (DU) and gastric cancer, 33 Taiwanese patients having received esophagogastroduodenoscopy examination and positive for urease test were enrolled for investigation (
Table 1).
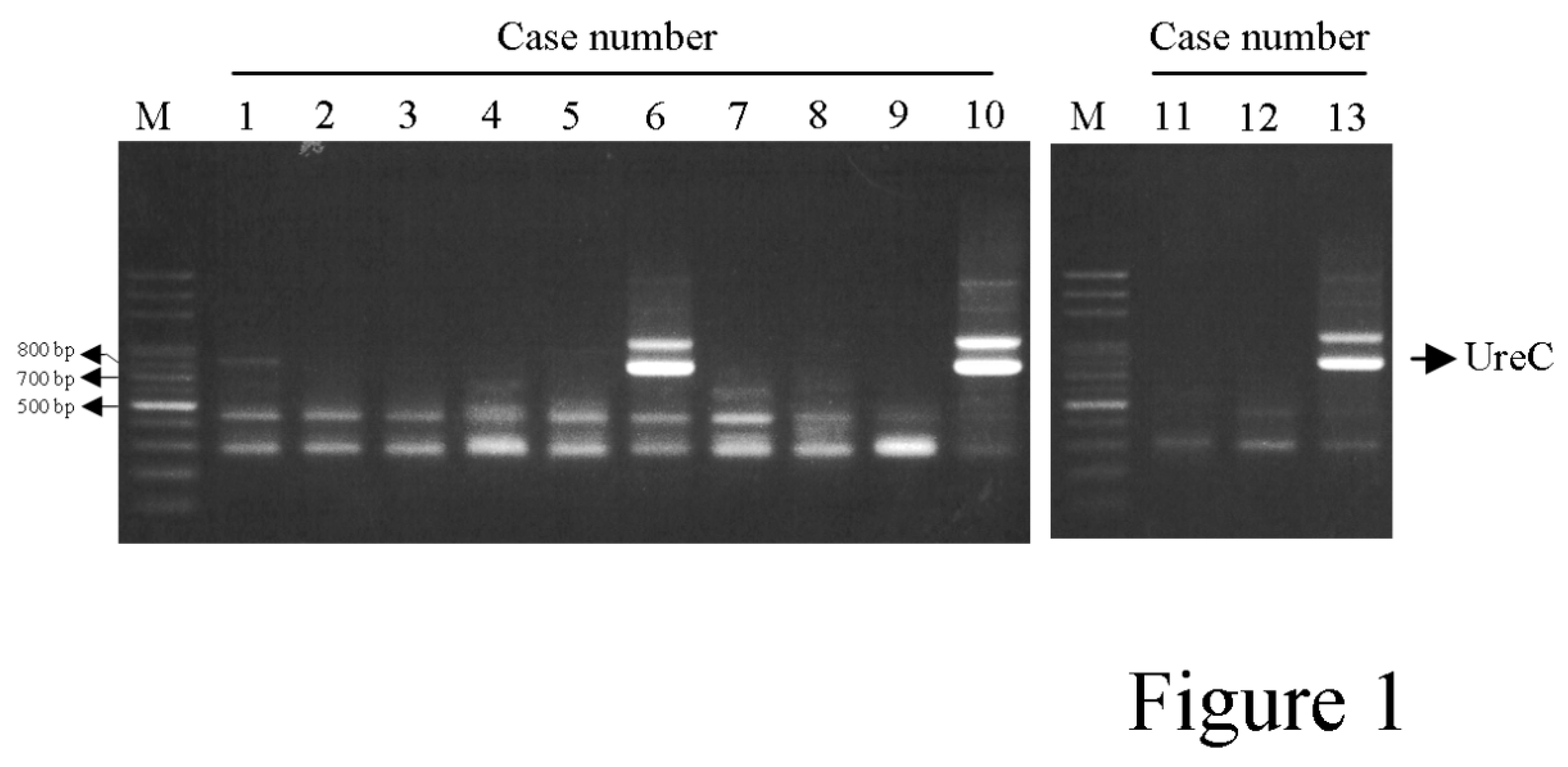
UreC sequences in these samples were amplified by PCR (
Figure 1) and subjected to DNA sequencing. Totally, 32
ureC PCR signals were detectable. Subsequently, a phylogenetic tree based on these
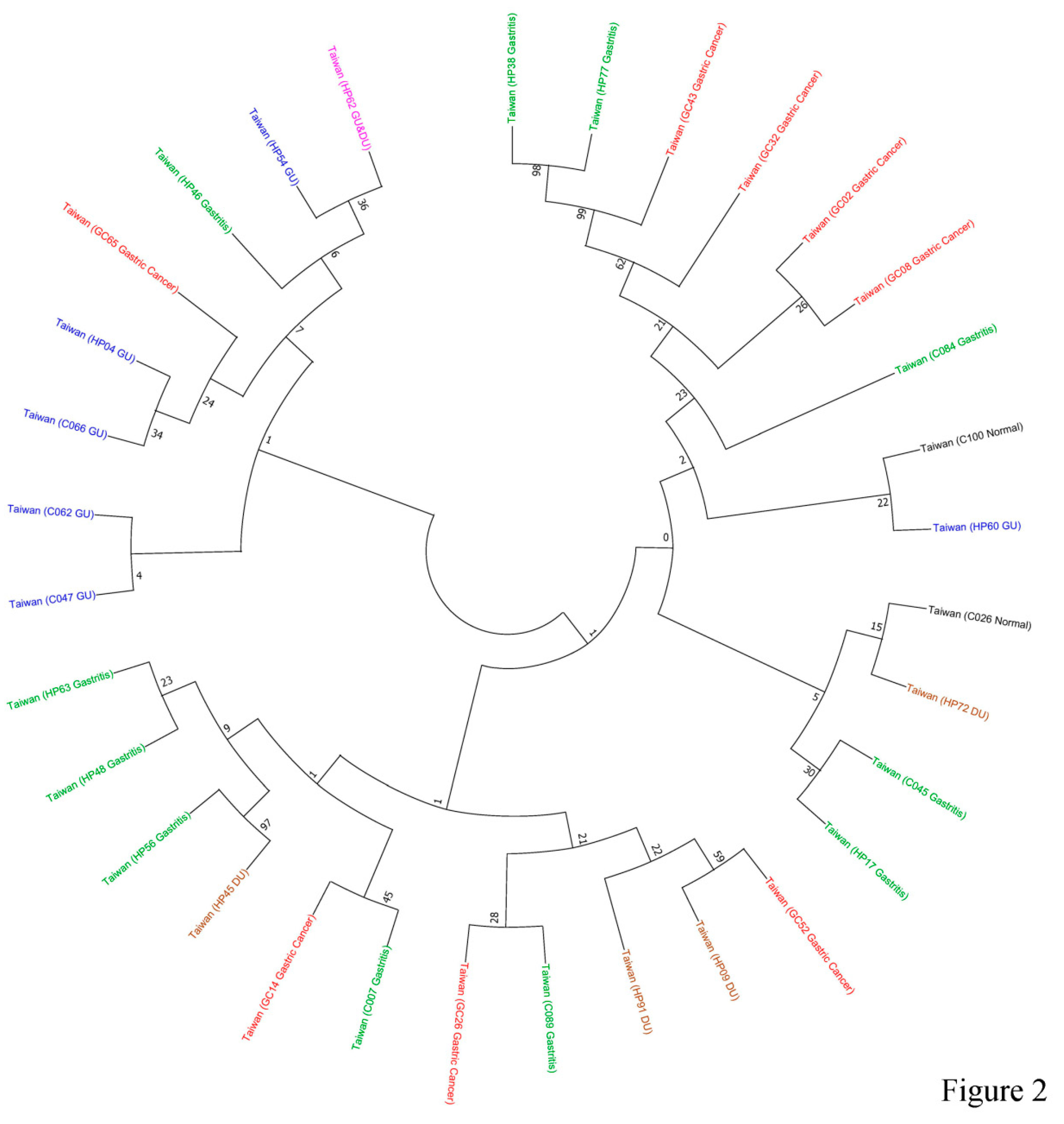
ureC sequences was constructed by use of MEGA X software. The results indicated that
ureC sequences in 6 of 7 GU samples (except for patient-HP60) were clustered together (
Figure 2), implying that the
ureC sequences in GU patients likely possessing common features. Interestingly,
ureC sequences in only 1 of the 8 gastric cancer samples (GC65) was grouped to the GU-related group, whereas GC43, GC32, GC02 and GC08 were grouped in a separated cluster (
Figure 2). The other sequences were randomly distributed in the phylogenetic tree (
Figure 2).
Partial ureC DNA sequences were amplified by PCR and subject to DNA sequencing. The arrowheads indicated ureC amplicon after sequence verification.
A phylogenetic tree was constructed based on 32 partial ureC sequences by Mega X software with Maximum-Likelihood method and bootstrap analysis (1000 replicates). The status of gastritis, GU, DU and gastric cancer was displayed by green, blue, brown and red color.
3.2. Comparison of ureC sequences from Taiwan and other countries
Previous studies reported that the geographical factor was associated with diversity of
H. pylori strains [
7,
8]. To understand the sequence variations between
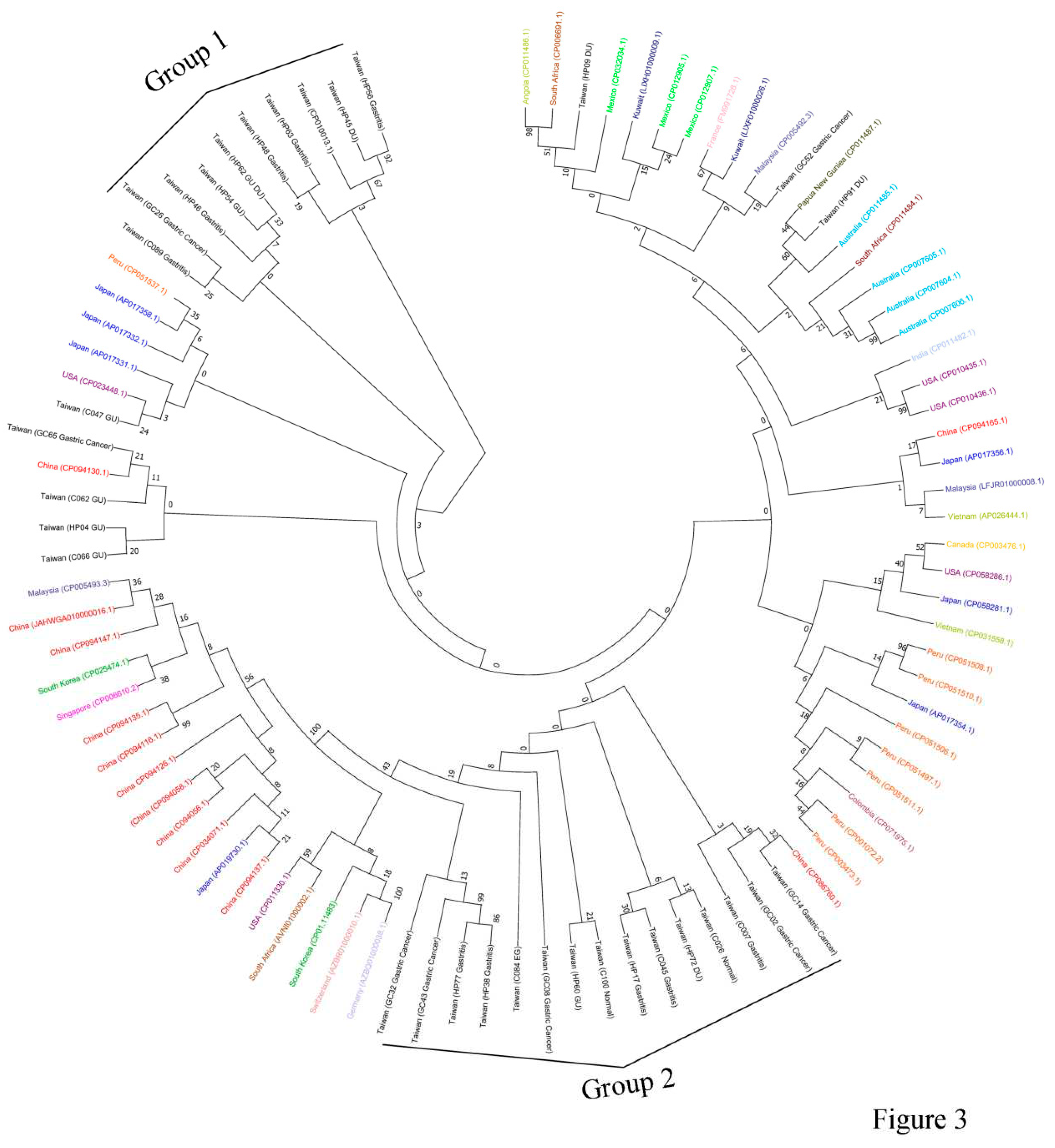
ureC sequences from Taiwan (our cohort) and those from 20 different countries (Kuwait, South Africa, Mexico, France, Angola, Malaysia, Australia, Papua New Guniea, India, USA, Vietnam, Japan, Canada, China, Peru, Colombia, Germany, Switerland, South Korea and Singapore) downloadable from GenBank, a phylogenetical tree analysis was performed. The results indicated that two groups of Taiwanese
ureC sequences (group 1: HP56, HP45, HP63, HP48, HP62, HP54, HP46, GC26 and C089; group 2: GC32, GC43, HP77, HP38, C084, GC08, HP60, C100, HP17, C045, HP72, C026, C007, GC02 and GC14) were separately clustered, but in low bootstrap values (
Figure 3). Notably, the 5 of 8
ureC sequences from gastric cancer were clustered in group 2.
The 32 partial ureC sequences obtained in the current study were indicated by blue line. UreC sequences from various geographic regions were retrieved from GenBank and were shown in different color. These included sequences from Kuwait, South Africa, Mexico, France, Angola, Malaysia, Australia, Papua New Guniea, India, USA, Vietnam, Japan, Canada, China, Peru, Colombia, Germany, Switerland, South Korea, Singapore and Taiwan. The Genbank accession number was shown in brackets. Phylogenetic tree of ureC DNA sequence was analyzed using the Maximum-Likelihood method with General Time Reversible model and bootstrap analysis (1000 replicates).
When examining sequences from different geographical regions, it was found that ureC sequences from China formed a distinct group with a very high bootstrap value (bootstrap value = 100). Only two Chinese ureC sequences, with Genbank accession number CP094130.1 and CP086760.1 were grouped with Taiwanese group 1 and group 2 (gastric cancer group) sequences, respectively, whereas all other 9 Chinese ureC sequences clustered to a separated group. Sequences from Peru also formed a separated group, but with low bootstrap values.
4. Discussion
H. pylori urease C gene encodes an open reading frame close to those of the urease AB genes, from which a phosphoglucosamine mutase is derived [
9]. This gene is an important regulator to modulate bacterial growth. Comparison of sequences between H. pylori-encoded urease C and Escherichia coli-encoded GlmM, a similarity of 43% is found. Biochemically, GlmM was responsible for converting glucosamine-6-phosphate to glucosamine-1-phosphate, which is an important component of cell wall as well as a building unit of lipopolysaccharides. A meta-analysis reported that the prevalence of
H. pylori infection in Taiwan was decreasing in the past decades [
10]. Concordantly, the incidence of gastric cancer in Taiwan also reduced overtime [
11]. It is generally believed that
H. pylori infection is a risk factor for gastric cancer [
12]. However, the cellular and/or molecular mechanisms for gastric cancer oncogenesis have not been completely illustrated. In
H. pylori positive patients, the associations between urease C (or
ureC sequences) and pathogenesis of gastric cancer have never been raised. In the present study, by performing phylogenetic analysis, it was found that Taiwanese
ureC sequences could be separated into two genotypes, with one of them associated with gastric cancer.
On the other hand, it was found that the ureC sequences from China formed a robust group with high bootstrap value, separating them from the ureC sequences derived from other parts of the world. It implies that Chinese H. pylori, based on the ureC sequence analysis, have a unique evolutional path, comparing to H. pylori from other countries.
Previous study reported that the prevalence of
H. pylori infection in China ranged from 41.5% to 72.3% [
13]. According to our analysis, only two Chinese
ureC sequences, with Genbank accession number CP094130.1 and CP086760.1 were grouped with Taiwanese group 1 and group 2 (gastric cancer group) sequences, respectively, while all other Chinese
ureC sequences formed a separated group. This finding is consistent with the long-standing separation between Taiwanese and Chinese people, albeit geologically close between the two areas.
The limitations in this study are: (1) This is a retrospective study. With the availability of anti-H. pylori treatment and increasing contact between people over the world, the genetic differences may change. (2) The sample size is small, owing to the decreased number of H. pylori-positive gastric samples in Taiwan. (3) Only ureC sequences, but not the whole genomes, were used for analysis. (4) The functional association between urease C and gastric cancer remains undetermined.
In conclusion, by analyzing H. pylori ureC sequences obtained from Taiwan and the world, we discovered a subgroup of ureC, which was associated with gastric cancer. The Chinese ureC sequences formed a separated genetic group, distinguishable from sequences from Taiwan and other parts of the world.
Funding
This work was supported by grants from Chang Gung Memorial Hospital, Taoyuan, Taiwan (CMRPG3L1811 to THC).
Conflicts of Interest
The authors have no conflicts to disclose.
List of Abbreviations
H. pylori, Helicobacter pylori; GU, gastric ulcer; DU, duodenal ulcer; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism.
References
- Yang L, Kartsonaki C, Yao P, de Martel C, Plummer M, Chapman D, et al. The relative and attributable risks of cardia and non-cardia gastric cancer associated with Helicobacter pylori infection in China: a case-cohort study. Lancet Public Health. 2021, 6, e888–e96. [Google Scholar] [CrossRef] [PubMed]
- Habbash F, Alalwan TA, Perna S, Ahmed N, Sharif O, Al Sayyad A, et al. Association between Dietary Habits and Helicobacter pylori Infection among Bahraini Adults. Nutrients. 2022, 14. [Google Scholar]
- 3. Malfertheiner P, Megraud F, Rokkas T, Gisbert JP, Liou JM, Schulz C, et al. Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report. Gut.
- Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, et al. Screening and eradication of Helicobacter pylori for gastric cancer prevention: the Taipei global consensus. Gut. 2020, 69, 2093–112. [Google Scholar] [CrossRef] [PubMed]
- Chen TH, Cheng HT, Yeh CT. Epidemiology changes in peptic ulcer diseases 18 years apart explored from the genetic aspects of Helicobacter pylori. Transl Res. 2021, 232, 115–20. [Google Scholar] [CrossRef] [PubMed]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018, 35, 1547–9. [Google Scholar] [CrossRef] [PubMed]
- Aftab H, Miftahussurur M, Subsomwong P, Ahmed F, Khan AKA, Matsumoto T, et al. Two populations of less-virulent Helicobacter pylori genotypes in Bangladesh. PLoS One. 2017, 12, e0182947. [Google Scholar]
- Vazirzadeh J, Karbasizadeh V, Falahi J, Moghim S, Narimani T, Rafiei R. Genetic Diversity of Helicobacter pylori Isolates from Patients with Gastric Diseases in Isfahan. Adv Biomed Res. 2022, 11, 4. [Google Scholar]
- De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 1997, 179, 3488–93. [Google Scholar] [CrossRef] [PubMed]
- Chen MJ, Bair MJ, Chen PY, Lee JY, Yang TH, Fang YJ, et al. Declining trends of prevalence of Helicobacter pylori infection and incidence of gastric cancer in Taiwan: An updated cross-sectional survey and meta-analysis. Helicobacter. 2022, 27, e12914. [Google Scholar] [CrossRef] [PubMed]
- Lin YT, Chiang CJ, Yang YW, Huang SP, You SL. Secular decreasing trends in gastric cancer incidence in Taiwan: A population-based cancer registry study. World J Gastroenterol. 2021, 27, 5764–74. [Google Scholar] [CrossRef] [PubMed]
- Yan L, Chen Y, Chen F, Tao T, Hu Z, Wang J, et al. Effect of Helicobacter pylori Eradication on Gastric Cancer Prevention: Updated Report From a Randomized Controlled Trial With 26.5 Years of Follow-up. Gastroenterology. 2022, 163, 154–62. [Google Scholar] [CrossRef] [PubMed]
- Xie C, Lu NH. Review: clinical management of Helicobacter pylori infection in China. Helicobacter. 2015, 20, 1–10. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).