Submitted:
29 August 2023
Posted:
01 September 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Material & Methods
Developing the framework
Testing the Index for Adaptive Responses
Application examples for the Index for Adaptive Responses
Results
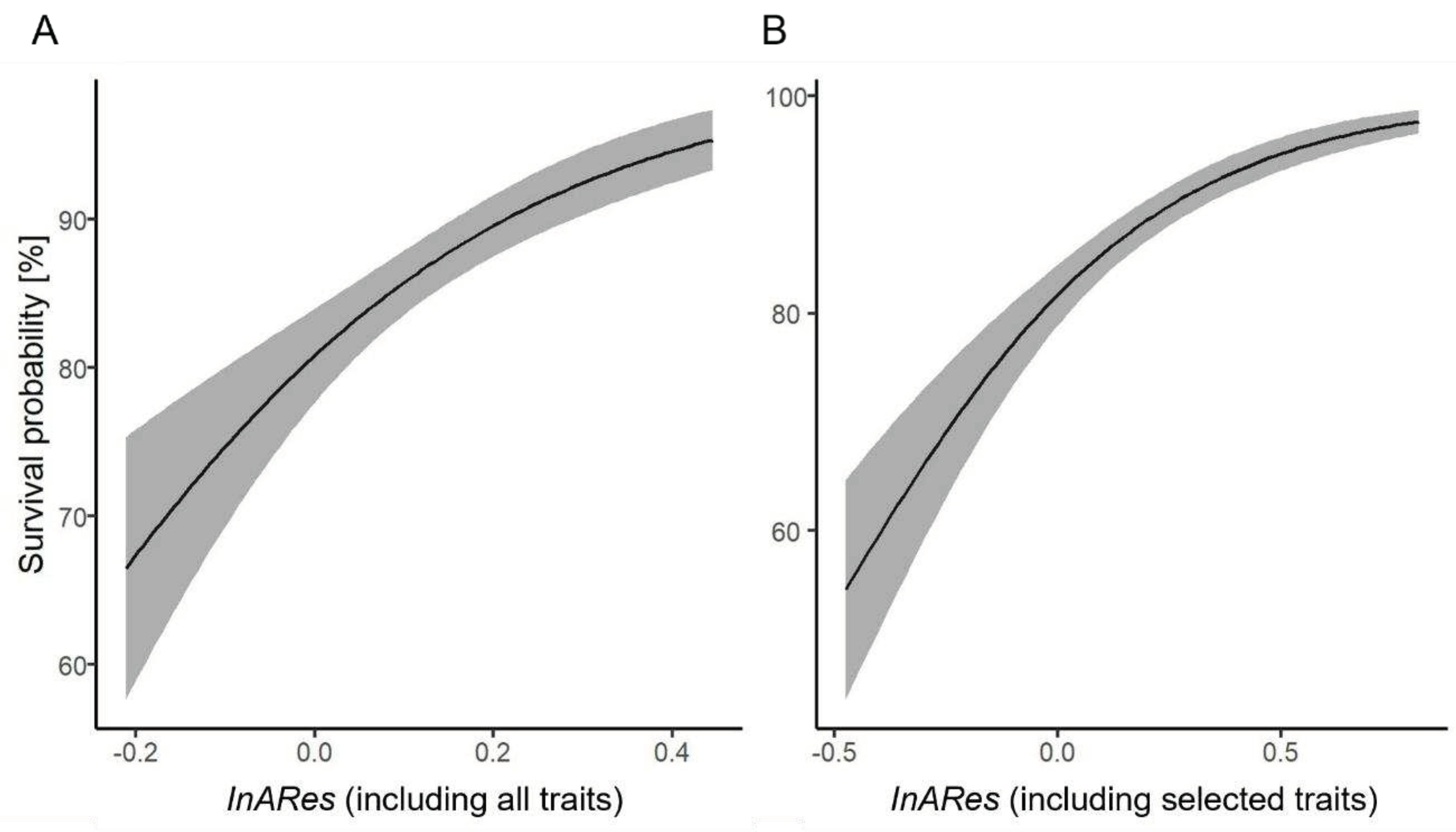
Re-evaluation of the case study
The transferability of the relation between the Index for Adaptive Responses value and defence effectiveness.
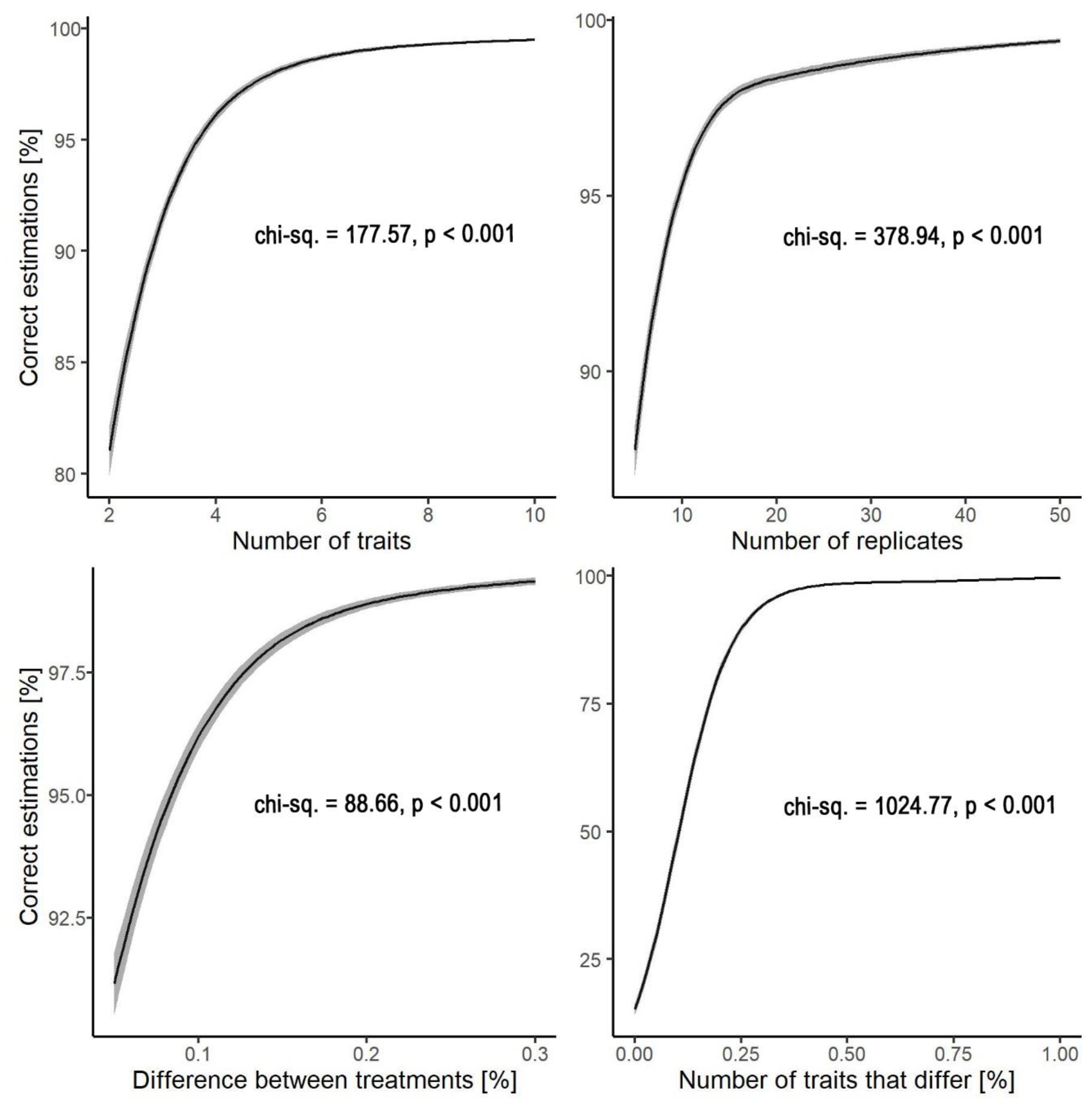
Performance of the prediction of the adaptiveness of traits
Discussion
Conclusions
Supplementary Materials
Author Contributions
Data Availability Statement
Acknowledgements
Conflict of Interest
References
- Belluau, M. , & Shipley, B. (2017). Predicting habitat affinities of herbaceous dicots to soil wetness based on physiological traits of drought tolerance. ( 119(6), 1073–1084. [CrossRef] [PubMed]
- Blumenthal, D. M. , Mueller, K. E., Kray, J. A., Ocheltree, T. W., Augustine, D. J., & Wilcox, K. R. (2020). Traits link drought resistance with herbivore defence and plant economics in semi-arid grasslands: The central roles of phenology and leaf dry matter content. Journal of Ecology, 2351. [Google Scholar] [CrossRef]
- Borden, K. A. , Anglaaere, L. C. N., Owusu, S., Martin, A. R., Buchanan, S. W., Addo-Danso, S. D., & Isaac, M. E. (2020). Soil texture moderates root functional traits in agroforestry systems across a climatic gradient. Agriculture, Ecosystems & Environment. [CrossRef]
- Candeias, M. , & Fraterrigo, J. (2020). Trait coordination and environmental filters shape functional trait distributions of forest understory herbs. ( 10(24), 14098–14112. [CrossRef] [PubMed]
- Christjani, M. , Fink, P., & von Elert, E. (2016). Phenotypic plasticity in three Daphnia genotypes in response to predator kairomone: evidence for an involvement of chitin deacetylases. The Journal of Experimental Biology, 1697. [Google Scholar] [CrossRef]
- Colpaert, J. v. , Muller, L. A. H., Lambaerts, M., Adriaensen, K., & Vangronsveld, J. (2004). Evolutionary adaptation to Zn toxicity in populations of Suilloid fungi. ( 162(2), 549–559. [CrossRef]
- Core Development Team, R. (2020). A Language and Environment for Statistical Computing. R Foundation for Statistical Computing.
- Dewitt, T. , & Scheiner, S. (2004). Future research directions. Phenotypic Plasticity: Functional and Conceptual Approaches, Oxford University Press, 2004. [Google Scholar]
- Diel, P. , Rabus, M., & Laforsch, C. (2021). Pricklier with the proper predator? Predator-induced small-scale changes of spinescence in Daphnia. Ecology and Evolution. [CrossRef]
- Dodson, S. I. (1974). Adaptive change in plankton morphology in response to size-selective predation: A new hypothesis of cyclomorphosis. Limnology and Oceanography. [CrossRef]
- Eshun-Wilson, F. , Wolf, R., Andersen, T., Hessen, D. O., & Sperfeld, E. (2020). UV radiation affects antipredatory defense traits in Daphnia pulex. Ecology and Evolution. [CrossRef]
- Forsman, A. (2015). Rethinking phenotypic plasticity and its consequences for individuals, populations and species. Heredity. [CrossRef]
- Fox, J. , & Weisberg, S. (2019). An R Companion to Applied Regression.
- Galarowicz, T. L. , & Wahl, D. H. (2005). Foraging by a young-of-the-year piscivore: The role of predator size, prey type, and density. Canadian Journal of Fisheries and Aquatic Sciences, 2342. [Google Scholar] [CrossRef]
- Gilbert, J. J. (2011). Temperature, kairomones, and phenotypic plasticity in the rotifer Keratella tropica (Apstein, 1907). Hydrobiologia. [CrossRef]
- Herzog, Q. , Tittgen, C., & Laforsch, C. (2016). Predator-specific reversibility of morphological defenses in Daphnia barbata. Journal of Plankton Research. [CrossRef]
- Hofmann, P. , Chatzinotas, A., Harpole, W. S., & Dunker, S. (2019). Temperature and stoichiometric dependence of phytoplankton traits. Ecology. [CrossRef]
- Hu, Y. , Zuo, X., Yue, P., Zhao, S., Guo, X., Li, X., & Medina-Roldán, E. (2020). Increased Precipitation Shapes Relationship between Biochemical and Functional Traits of Stipa glareosa in Grass-Dominated Rather than Shrub-Dominated Community in a Desert Steppe. Plants 2020, Vol. 9, Page 1463. [CrossRef]
- Karpestam, E. , Wennersten, L., & Forsman, A. (2012). Matching habitat choice by experimentally mismatched phenotypes. ( 26(4), 893–907. [CrossRef]
- Kruppert, S. , Horstmann, M., Weiss, L. C., Schaber, C. F., Gorb, S. N., & Tollrian, R. (2016). Push or Pull? The light-weight architecture of the Daphnia pulex carapace is adapted to withstand tension, not compression. Journal of Morphology. [CrossRef]
- Laforsch, C. , & Tollrian, R. (2004). Inducible defenses in multipredator environments: Cyclomorphosis in Daphnia cucullata. Ecology, 2311. [Google Scholar] [CrossRef]
- Laforsch, C. , & Tollrian, R. (2009). Cyclomorphosis and Phenotypic Changes. In Encyclopedia of Inland Waters (pp. 643–650). Elsevier. [CrossRef]
- Leech, D. M. , & Williamson, C. E. (2000). Is tolerance to UV radiation in zooplankton related to body size, taxon, or lake transparency? Ecological Applications, 1540. [Google Scholar] [CrossRef]
- Li, H. , Xu, W., Wu, L., Dong, B., Jin, J., Han, D., Zhu, X., Yang, Y., Liu, H., & Xie, S. (2020). Distinct dietary cadmium toxic effects and defense strategies in two strains of gibel carp (Carassius gibelio) revealed by a comprehensive perspective. ( 261, 127597. [CrossRef] [PubMed]
- Martin-Creuzburg, D. , Wacker, A., Ziese, C., & Kainz, M. J. (2012). Dietary lipid quality affects temperature-mediated reaction norms of a freshwater key herbivore. Oecologia. [CrossRef]
- Murray, G. P. D. , Stillman, R. A., & Britton, J. R. (2016). Habitat complexity and food item size modify the foraging behaviour of a freshwater fish. R. ( 766(1), 321–332. [CrossRef]
- Piersma, T. , & Gils, J. van. (2011). The flexible phenotype: a body-centred integration of ecology, physiology, and behaviour.
- Pigliucci, M. , & Preston, K. (2004). Phenotypic integration: studying the ecology and evolution of complex phenotypes.
- Polat, B. , Suleyman, H., & Alp, H. H. (2010). Adaptation of rat gastric tissue against indomethacin toxicity. Chemico-Biological Interactions. [CrossRef]
- Rabus, M. , Söllradl, T., Clausen-Schaumann, H., & Laforsch, C. (2013). Uncovering Ultrastructural Defences in Daphnia magna - An Interdisciplinary Approach to Assess the Predator-Induced Fortification of the Carapace. PLoS ONE. [CrossRef]
- Ritschar, S. , Bangalore Narayana, V. K., Rabus, M., & Laforsch, C. (2020). Uncovering the chemistry behind inducible morphological defences in the crustacean Daphnia magna via micro-Raman spectroscopy. Scientific Reports. [CrossRef]
- Rose, N. L. , Yang, H., Turner, S. D., & Simpson, G. L. (2012). An assessment of the mechanisms for the transfer of lead and mercury from atmospherically contaminated organic soils to lake sediments with particular reference to Scotland, UK. L. ( 82, 113–135. [CrossRef]
- Sarrazin, J. , & Sperfeld, E. (2022). Food quality mediates responses of Daphnia magna life history traits and heat tolerance to elevated temperature. Freshwater Biology. [CrossRef]
- Shudo, E. , & Iwasa, Y. (2001). Inducible defense against pathogens and parasites: optimal choice among multiple options. Journal of Theoretical Biology, 0022. [Google Scholar]
- Stoks, R. , Govaert, L., Pauwels, K., Jansen, B., & de Meester, L. (2016). Resurrecting complexity: the interplay of plasticity and rapid evolution in the multiple trait response to strong changes in predation pressure in the water flea Daphnia magna. Ecology Letters. [CrossRef]
- Turschwell, M. P. , Connolly, S. R., Schäfer, R. B., de Laender, F., Campbell, M. D., Mantyka-Pringle, C., Jackson, M. C., Kattwinkel, M., Sievers, M., Ashauer, R., Côté, I. M., Connolly, R. M., van den Brink, P. J., Brown, C. J., & Byers, J. (2022). Interactive effects of multiple stressors vary with consumer interactions, stressor dynamics and magnitude. Ecology Letters. [CrossRef]
- Valladares, F. , Gianoli, E., & Gómez, J. (2007). Ecological limits to plant phenotypic plasticity. ( 176(4), 749–763. [CrossRef]
- Valladares, F. , Sanchez-Gomez, D., & Zavala, M. A. (2006). Quantitative estimation of phenotypic plasticity: bridging the gap between the evolutionary concept and its ecological applications. Journal of Ecology, 1116. [Google Scholar] [CrossRef]
- Whitman, D. W. , & Agrawal, A. A. (2009). What is Phenotypic Plasticity and Why is it Important? Phenotypic Plasticity of Insects: Mechanisms and Consequences.
- Wickham, H. (2016). ggplot2 Elegant Graphics for Data Analysis (Use R!). Springer.
- Wigley, B. J. , Fritz, H., & Coetsee, C. (2018). Defence strategies in African savanna trees. ( 187(3), 797–809. [CrossRef]
- Wood, S. , Scheipl, F., Stat, M. W.-A., & 2017, undefined. (2020). Package “gamm4.” 152.19.134.44, 9. ftp://152.19.134.44/CRAN/web/packages/gamm4/gamm4.
- Wright, I. J. , Falster, D. S., Pickup, M., & Westoby, M. (2006). Cross-species patterns in the coordination between leaf and stem traits, and their implications for plant hydraulics. Physiologia Plantarum. [CrossRef]
- Zhang, H. , He, Y., Liang He, |, Zhao, | Kangshun, Molinos, J. G., Hansson, L.-A., & Xu, J. (2022). Plasticity in rotifer morphology induced by conflicting threats from multiple predators. Wiley Online Library. [CrossRef]
| Model | Deviance |
|---|---|
| Full trait-based model from the corresponding study, including 19 parameters | 630.5 |
| Selected version of the full-trait-based model | 648.0 |
| Trait-based model, including only the two majorly affecting terms of the corresponding study | 560.5 |
| Model, based on the InARes, including all 10 traits | 568.4 |
| Model, based on the InARes, including all traits except the two collinear traits | 570.2 |
| Model, based on the InARes, including terms selected by prior contribution analyses (as part of the framework) | 563.6 |
| Model, based on the InARes, including only the two majorly affecting terms of the corresponding study | 560.4 |
| Model, based on the InARes, including the two major terms and the minor affecting terms (in specific orientation) of the corresponding study | 564.2 |
| Model, based on the principal component analysis including components 1--3 | 564.3 |
| Model, based on the principal component analysis including just component 1 | 570.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





