Submitted:
04 September 2023
Posted:
06 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- minimalistic model containing only the elements directly involved in adsorption,
- reduction of the length scale of a problem,
- artificial speed-up of the system dynamics.
2. Experimental data on LDL Structure and the Modelling Details
3. Adsorption of the LDL at Normal Conditions
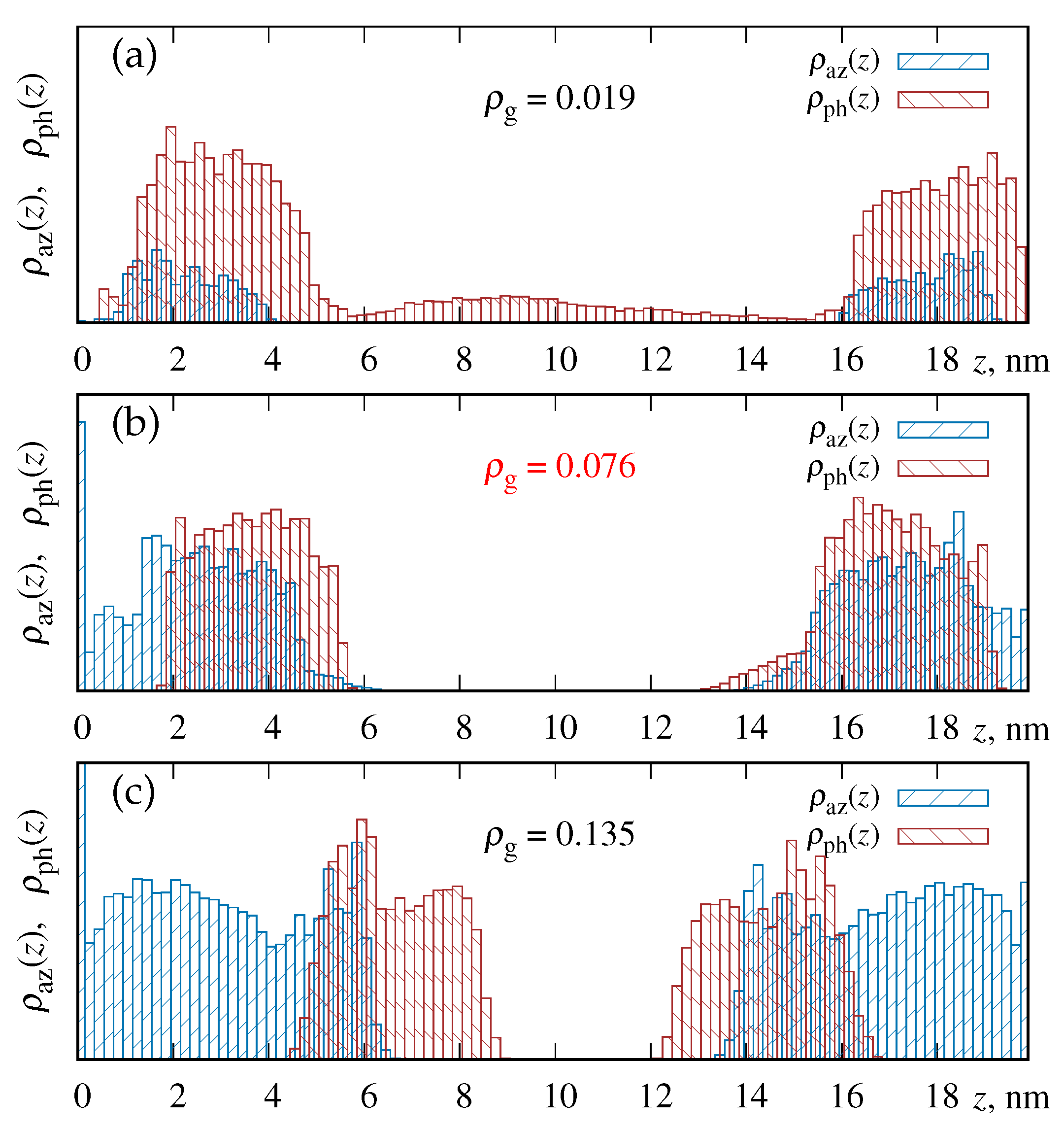
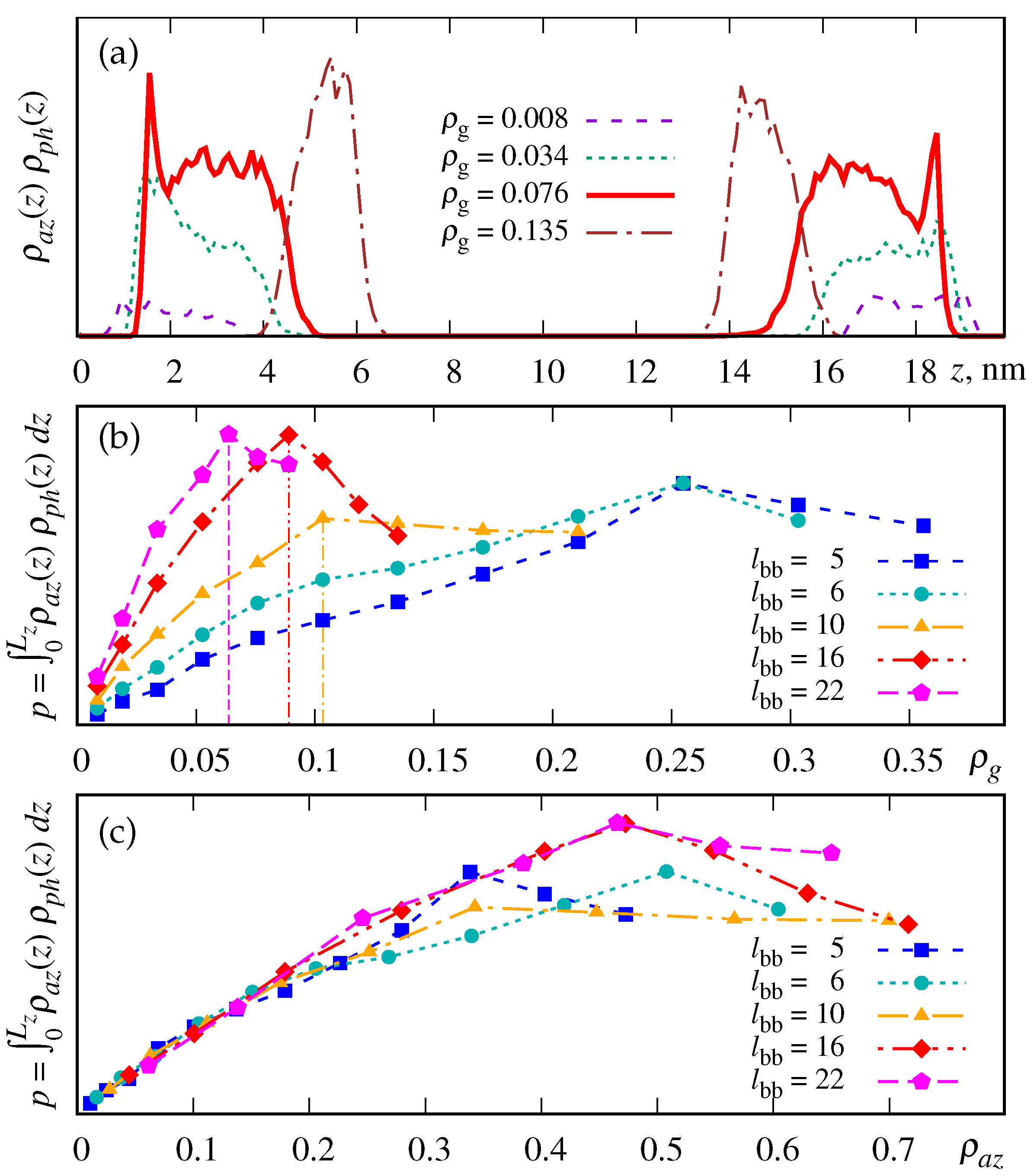
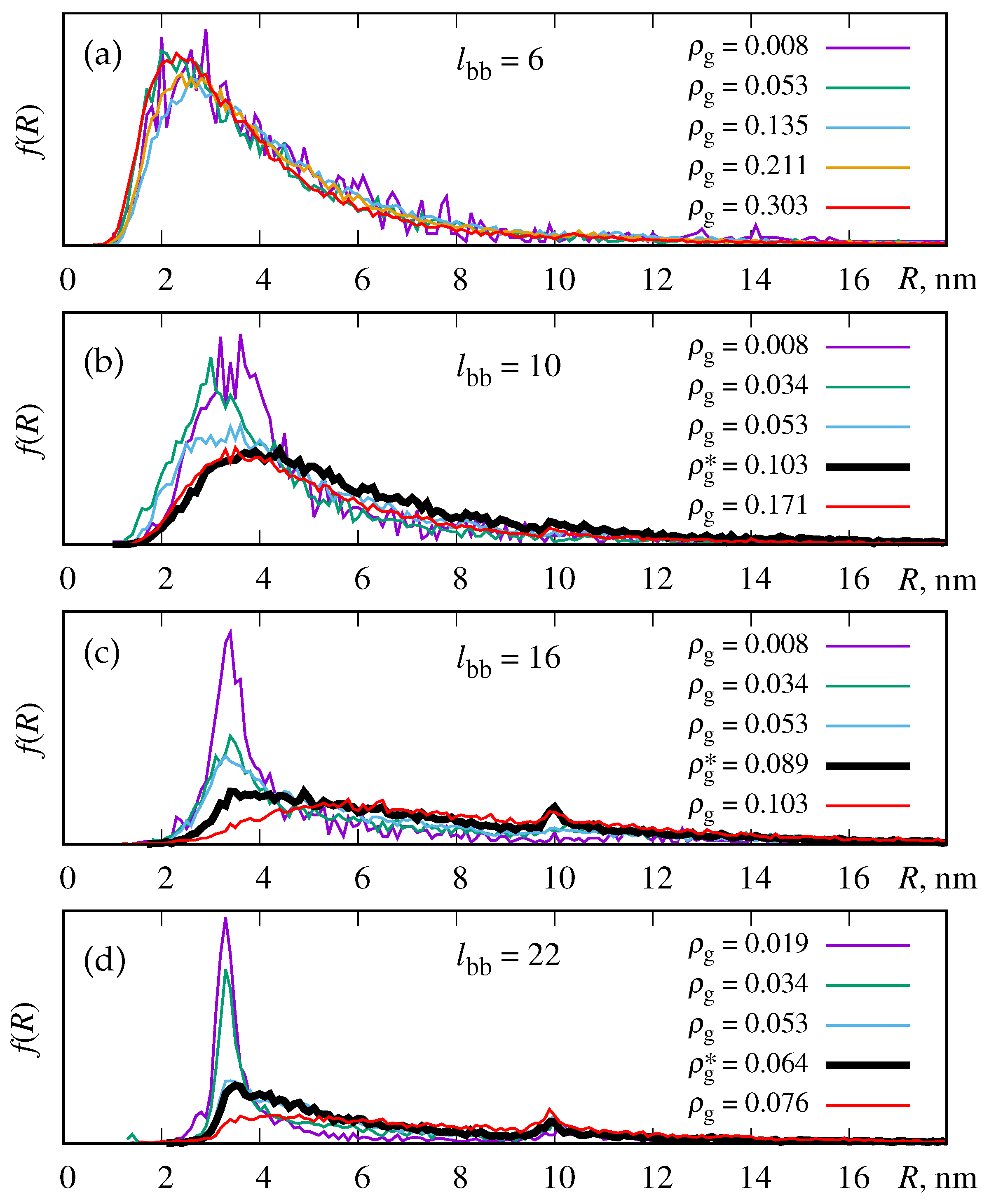
3.1. Density Profiles and the Probability for the Azobenzene-Phospholipid Contact
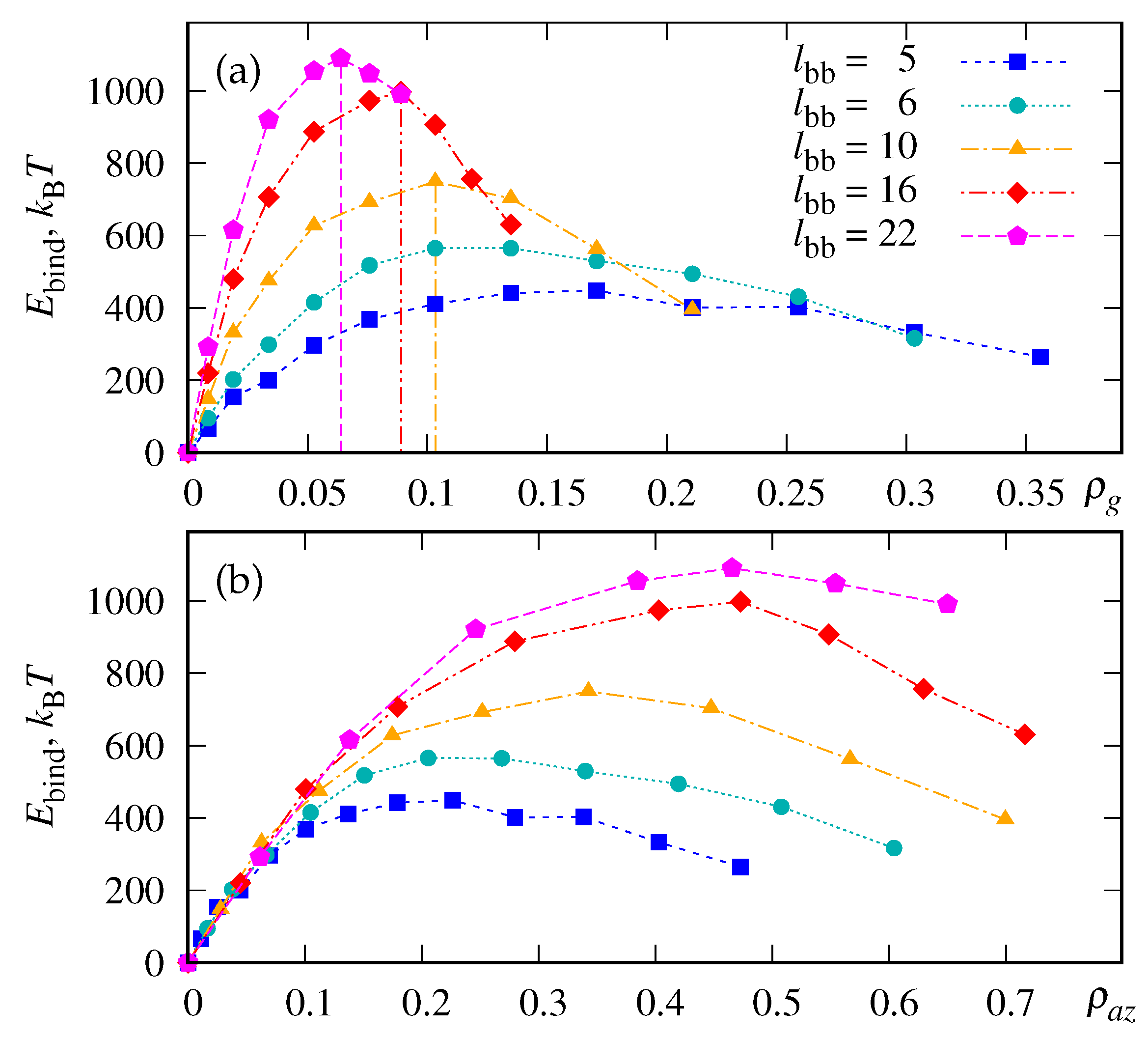
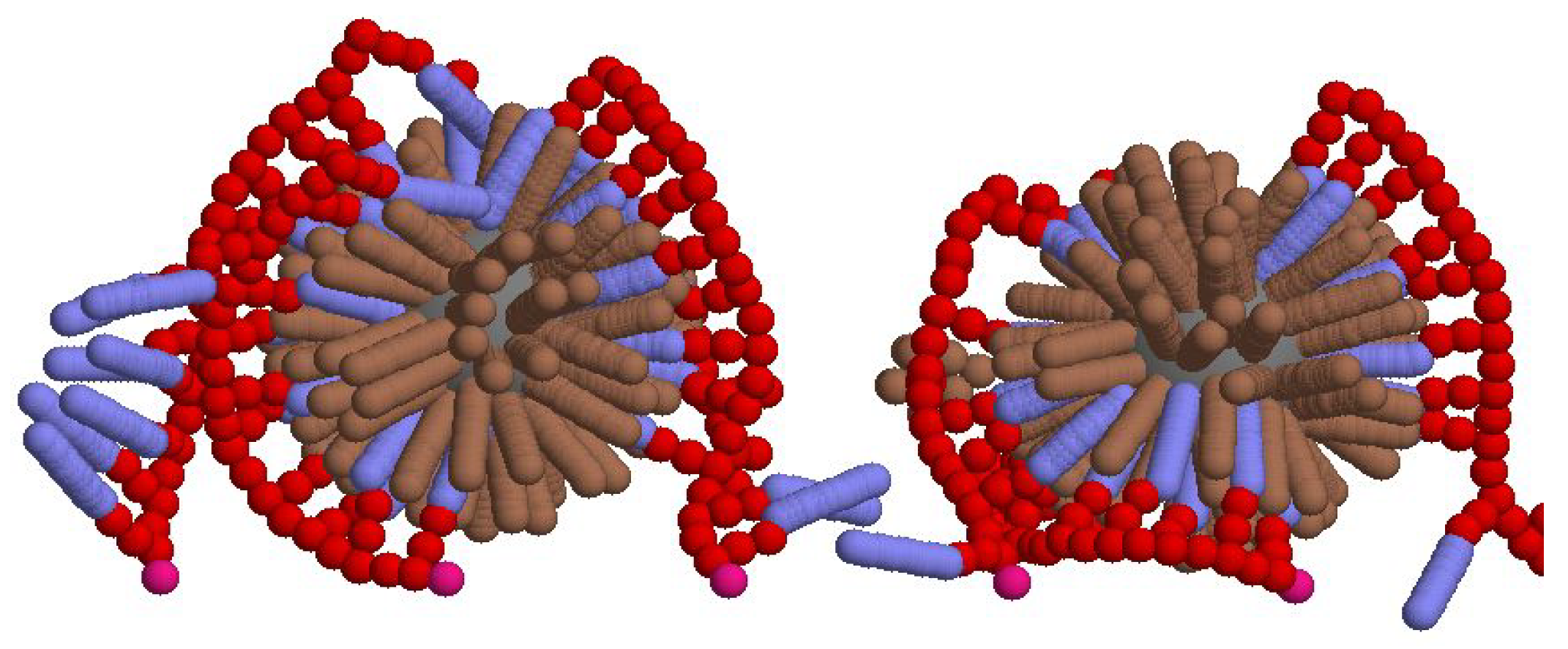
3.2. Binding Energy of LDLs
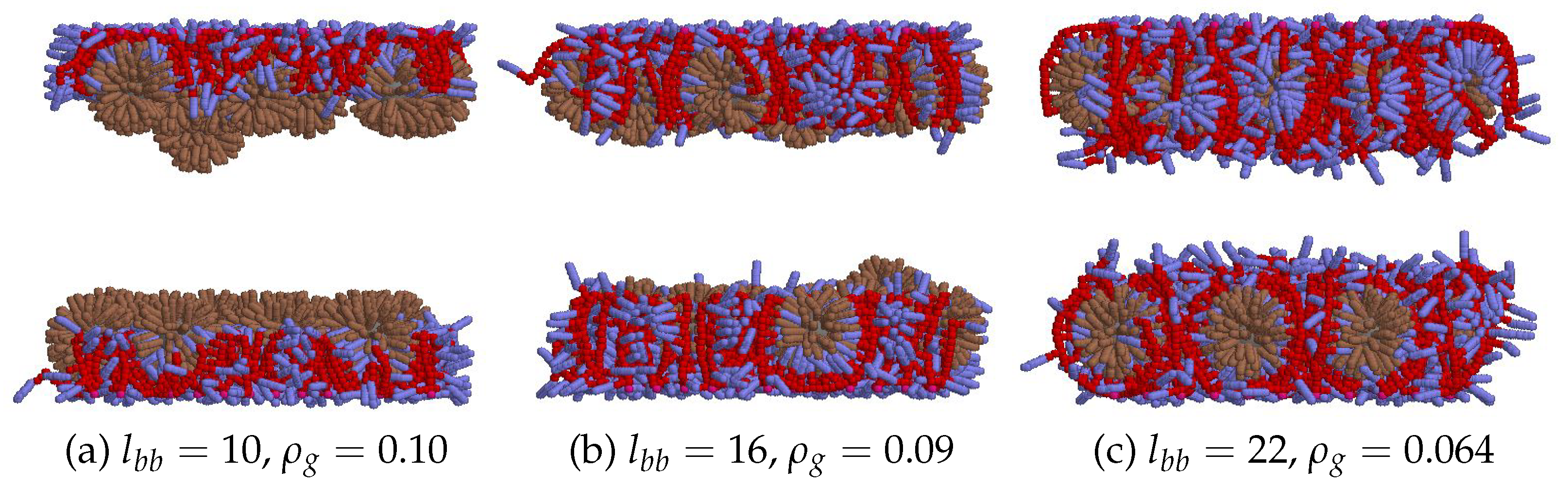
3.3. Polymer bending effect
3.4. Concludings Remarks on Adsorption Efficiency
4. Adsorber Regeneration under Ultraviolet Light
Conclusions
Acknowledgments
References
- Sacks, F.M.; Campos, H. Low-Density Lipoprotein Size and Cardiovascular Disease: A Reappraisal. The Journal of Clinical Endocrinology & Metabolism 2003, 88, 4525–4532. [Google Scholar] [CrossRef]
- Sikorski, J. Atherosclerosis/Lipoprotein/Cholesterol Metabolism. In Comprehensive Medicinal Chemistry II; Elsevier, 2007; pp. 459–494. [CrossRef]
- Mundi, S.; Massaro, M.; Scoditti, E.; Carluccio, M.A.; van Hinsbergh, V.W.M.; Iruela-Arispe, M.L.; Caterina, R.D. Endothelial permeability, LDL deposition, and cardiovascular risk factors—a review. Cardiovascular Research 2017, 114, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Kiberstis, P.A. Fatty liver—too much of a bad thing? Science 2019, 364, 1044.6–1045. [Google Scholar] [CrossRef]
- Thierer, J.H.; Ekker, S.C.; Farber, S.A. The LipoGlo reporter system for sensitive and specific monitoring of atherogenic lipoproteins. Nature Communications 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Dong, J.; Ma, B.; Jiang, X.; Guo, C.; Liu, Z.; Chai, Y.; Wang, L.; Sun, L.; Ou, L.; et al. Bio-inspired dual-functional phospholipid–poly(acrylic acid) brushes grafted porous poly(vinyl alcohol) beads for selective adsorption of low-density lipoprotein. Journal of Materials Chemistry B 2021, 9, 6364–6376. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, S.; Yu, Y.; Yuan, Y. In vitro, in vivo studies of a new amphiphilic adsorbent for the removal of low-density lipoprotein. Biomaterials 2003, 24, 2189–2194. [Google Scholar] [CrossRef]
- Yu, Y.; Ma, B.; Jiang, X.; Guo, C.; Liu, Z.; Li, N.; Chai, Y.; Wang, L.; Du, Y.; Wang, B.; et al. Amphiphilic shell nanomagnetic adsorbents for selective and highly efficient capture of low-density lipoprotein from hyperlipidaemia serum. Journal of Materials Chemistry B 2022, 10, 4856–4866. [Google Scholar] [CrossRef]
- Lanzalaco, S.; Armelin, E. Poly(N-isopropylacrylamide) and Copolymers: A Review on Recent Progresses in Biomedical Applications. Gels 2017, 3, 36. [Google Scholar] [CrossRef]
- Sudre, G.; Siband, E.; Gallas, B.; Cousin, F.; Hourdet, D.; Tran, Y. Responsive Adsorption of N-Isopropylacrylamide Based Copolymers on Polymer Brushes. Polymers 2020, 12, 153. [Google Scholar] [CrossRef]
- Yaremchuk, D.; Kalyuzhnyi, O.; Ilnytskyi, J. Modelling thermoresponsive polymer brush by mesoscale computer simulations. Condensed Matter Physics 2023, 26, 33302. [Google Scholar] [CrossRef]
- Tian, L.; Dou, H.; Shao, Y.; Yi, Y.; Fu, X.; Zhao, J.; Fan, Y.; Ming, W.; Ren, L. Magnetically controlled super-wetting surface switching between ultra-low and ultra-high droplet adhesion. Chemical Engineering Journal 2023, 456, 141093. [Google Scholar] [CrossRef]
- Malm, J.; Sahramo, E.; Karppinen, M.; Ras, R.H.A. Photo-Controlled Wettability Switching by Conformal Coating of Nanoscale Topographies with Ultrathin Oxide Films. Chemistry of Materials 2010, 22, 3349–3352. [Google Scholar] [CrossRef]
- Bahl, S.; Nagar, H.; Singh, I.; Sehgal, S. Smart materials types, properties and applications: A review. Materials Today: Proceedings 2020, 28, 1302–1306. [Google Scholar] [CrossRef]
- Zhao, Y. Smart light-responsive materials; Wiley-Blackwell: Hoboken, NJ, 2009. [Google Scholar]
- Roling, O.; Stricker, L.; Voskuhl, J.; Lamping, S.; Ravoo, B.J. Supramolecular surface adhesion mediated by azobenzene polymer brushes. Chemical Communications 2016, 52, 1964–1966. [Google Scholar] [CrossRef] [PubMed]
- Santer, S. Remote control of soft nano-objects by light using azobenzene containing surfactants. Journal of Physics D: Applied Physics 2017, 51, 013002. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Jiang, X.; Xie, T.; Li, H.; Zhang, G.; Jiang, J. Photoswitchable de/adsorption of an azobenzene-derived surfactant on a silica surface. Physical Chemistry Chemical Physics 2019, 21, 21030–21037. [Google Scholar] [CrossRef] [PubMed]
- Hisham, S.; Sarih, N.M.; Tajuddin, H.A.; Abidin, Z.H.Z.; Abdullah, Z. Unraveling the surface properties of PMMA/azobenzene blends as coating films with photoreversible surface polarity. RSC Advances 2021, 11, 15428–15437. [Google Scholar] [CrossRef]
- Mukai, K.; Imai, K.; Hara, M.; Nagano, S.; Seki, T. A High-Density Azobenzene Side Chain Polymer Brush for Azimuthal and Zenithal Orientational Photoswitching of a Nematic Liquid Crystal. ChemPhotoChem 2019, 3, 495–500. [Google Scholar] [CrossRef]
- Guo, C.; Yu, Y.; Jiang, X.; Ma, B.; Liu, Z.; Chai, Y.; Wang, L.; Wang, B.; Du, Y.; Li, N.; et al. Photorenewable Azobenzene Polymer Brush-Modified Nanoadsorbent for Selective Adsorption of LDL in Serum. ACS Applied Materials & Interfaces 2022, 14, 34388–34399. [Google Scholar] [CrossRef]
- Antwerpen, R.V.; Gilkey, J.C. Cryo-electron microscopy reveals human low density lipoprotein substructure. Journal of Lipid Research 1994, 35, 2223–2231. [Google Scholar] [CrossRef]
- Orlova, E.V.; Sherman, M.B.; Chiu, W.; Mowri, H.; Smith, L.C.; Gotto, A.M. Three-dimensional structure of low density lipoproteins by electron cryomicroscopy. Proceedings of the National Academy of Sciences 1999, 96, 8420–8425. [Google Scholar] [CrossRef] [PubMed]
- Hevonoja, T.; Pentikäinen, M.O.; Hyvönen, M.T.; Kovanen, P.T.; Ala-Korpela, M. Structure of low density lipoprotein (LDL) particles: Basis for understanding molecular changes in modified LDL. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2000, 1488, 189–210. [Google Scholar] [CrossRef]
- Segrest, J.P.; Jones, M.K.; Loof, H.D.; Dashti, N. Structure of apolipoprotein B-100 in low density lipoproteins. Journal of Lipid Research 2001, 42, 1346–1367. [Google Scholar] [CrossRef] [PubMed]
- Teerlink, T.; Scheffer, P.G.; Bakker, S.J.; Heine, R.J. Combined data from LDL composition and size measurement are compatible with a discoid particle shape. Journal of Lipid Research 2004, 45, 954–966. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Rudenko, G.; Ludtke, S.J.; Deisenhofer, J.; Chiu, W.; Pownall, H.J. Model of human low-density lipoprotein and bound receptor based on CryoEM. Proceedings of the National Academy of Sciences 2009, 107, 1059–1064. [Google Scholar] [CrossRef]
- Kumar, V.; Butcher, S.J.; Öörni, K.; Engelhardt, P.; Heikkonen, J.; Kaski, K.; Ala-Korpela, M.; Kovanen, P.T. Three-Dimensional cryoEM Reconstruction of Native LDL Particles to 16Å Resolution at Physiological Body Temperature. PLoS ONE 2011, 6, e18841. [Google Scholar] [CrossRef]
- Prassl, R.; Laggner, P. Lipoprotein Structure and Dynamics: Low Density Lipoprotein Viewed as a Highly Dynamic and Flexible Nanoparticle. In Lipoproteins - Role in Health and Diseases; InTech, 2012. [CrossRef]
- Heikelä, M.; Vattulainen, I.; Hyvönen, M.T. Atomistic Simulation Studies of Cholesteryl Oleates: Model for the Core of Lipoprotein Particles. Biophysical Journal 2006, 90, 2247–2257. [Google Scholar] [CrossRef]
- Vattulainen, I.; Rog, T. Lipid Simulations: A Perspective on Lipids in Action. Cold Spring Harbor Perspectives in Biology 2011, 3, a004655–a004655. [Google Scholar] [CrossRef]
- Murtola, T.; Vuorela, T.A.; Hyvönen, M.T.; Marrink, S.J.; Karttunen, M.; Vattulainen, I. Low density lipoprotein: structure, dynamics, and interactions of apoB-100 with lipids. Soft Matter 2011, 7, 8135. [Google Scholar] [CrossRef]
- Koivuniemi, A.; Vuorela, T.; Kovanen, P.T.; Vattulainen, I.; Hyvönen, M.T. Lipid Exchange Mechanism of the Cholesteryl Ester Transfer Protein Clarified by Atomistic and Coarse-grained Simulations. PLoS Computational Biology 2012, 8, e1002299. [Google Scholar] [CrossRef]
- Lewis, D.R.; Kholodovych, V.; Tomasini, M.D.; Abdelhamid, D.; Petersen, L.K.; Welsh, W.J.; Uhrich, K.E.; Moghe, P.V. In silico design of anti-atherogenic biomaterials. Biomaterials 2013, 34, 7950–7959. [Google Scholar] [CrossRef] [PubMed]
- Antonijevic, T.; Lancaster, J.L.; Starobin, J.M. Modeling order-disorder transition in Low-Density Lipoprotein. In Proceedings of the 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2014. [Google Scholar] [CrossRef]
- Cilpa-Karhu, G.; Jauhiainen, M.; Riekkola, M.L. Atomistic MD simulation reveals the mechanism by which CETP penetrates into HDL enabling lipid transfer from HDL to CETP. Journal of Lipid Research 2015, 56, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Angarica, V.E.; Orozco, M.; Sancho, J. Exploring the complete mutational space of the LDL receptor LA5 domain using molecular dynamics: linking SNPs with disease phenotypes in familial hypercholesterolemia. Human Molecular Genetics 2016, 25, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Lei, D.; Rames, M.; Zhang, X.; Zhang, L.; Zhang, S.; Ren, G. Insights into the Tunnel Mechanism of Cholesteryl Ester Transfer Protein through All-atom Molecular Dynamics Simulations. Journal of Biological Chemistry 2016, 291, 14034–14044. [Google Scholar] [CrossRef]
- Lauer, M.E.; Graff-Meyer, A.; Rufer, A.C.; Maugeais, C.; von der Mark, E.; Matile, H.; D’Arcy, B.; Magg, C.; Ringler, P.; Müller, S.A.; et al. Cholesteryl ester transfer between lipoproteins does not require a ternary tunnel complex with CETP. Journal of Structural Biology 2016, 194, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Ilnytskyi, J.; Saphiannikova, M.; Neher, D. Photo-induced deformations in azobenzene-containing side-chain polymers: molecular dynamics study. Condensed Matter Physics 2006, 9, 87. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Neher, D.; Saphiannikova, M. Opposite photo-induced deformations in azobenzene-containing polymers with different molecular architecture: Molecular dynamics study. The Journal of Chemical Physics 2011, 135, 044901. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Saphiannikova, M. Reorientation Dynamics of Chromophores in Photosensitive Polymers by Means of Coarse-Grained Modeling. ChemPhysChem 2015, 16, 3180–3189. [Google Scholar] [CrossRef]
- Ilnytskyi, J.; Slyusarchuk, A.; Saphiannikova, M. Photo-controllable percolation of decorated nanoparticles in a nanopore: molecular dynamics simulation study. Mathematical Modeling and Computing 2016, 3, 33–42. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Slyusarchuk, A.; Saphiannikova, M. Photocontrollable Self-Assembly of Azobenzene-Decorated Nanoparticles in Bulk: Computer Simulation Study. Macromolecules 2016, 49, 9272–9282. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Toshchevikov, V.; Saphiannikova, M. Modeling of the photo-induced stress in azobenzene polymers by combining theory and computer simulations. Soft Matter 2019, 15, 9894–9908. [Google Scholar] [CrossRef] [PubMed]
- Slyusarchuk, A.Y. ; Yaremchuk, D.L.; Ilnytskyi, J.M.; and. Adsorption of decorated nanoparticles on a liquid crystalline polymer brush: molecular dynamics study. Mathematical Modeling and Computing 2020, 7, 207–218. [Google Scholar] [CrossRef]
- Yaremchuk, D.; Patsahan, T.; Ilnytskyi, J. Photo-switchable liquid crystalline brush as an aligning surface for liquid crystals: modelling via mesoscopic computer simulations. Condensed Matter Physics 2022, 25, 33601. [Google Scholar] [CrossRef]
- Austin, M.; Krauss, R. GENETIC CONTROL OF LOW-DENSITY-LIPOPROTEIN SUBCLASSES. The Lancet 1986, 328, 592–595. [Google Scholar] [CrossRef]
- Campos, H.; Genest, J.J.; Blijlevens, E.; McNamara, J.R.; Jenner, J.L.; Ordovas, J.M.; Wilson, P.W.; Schaefer, E.J. Low density lipoprotein particle size and coronary artery disease. Arteriosclerosis and Thrombosis: A Journal of Vascular Biology 1992, 12, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.A. Low-Density Lipoprotein Subclass Patterns and Risk of Myocardial Infarction. JAMA: The Journal of the American Medical Association 1988, 260, 1917. [Google Scholar] [CrossRef]
- Stampfer, M.J. A Prospective Study of Triglyceride Level, Low-Density Lipoprotein Particle Diameter, and Risk of Myocardial Infarction. JAMA: The Journal of the American Medical Association 1996, 276, 882. [Google Scholar] [CrossRef]
- Lamarche, B.; Tchernof, A.; Moorjani, S.; Cantin, B.; Dagenais, G.R.; Lupien, P.J.; Despre´s, J.P. Small, Dense Low-Density Lipoprotein Particles as a Predictor of the Risk of Ischemic Heart Disease in Men. Circulation 1997, 95, 69–75. [Google Scholar] [CrossRef]
- Scheffer, P.G.; Bakker, S.J.L.; Heine, R.J.; Teerlink, T. Measurement of low-density lipoprotein particle size by high-performance gel-filtration chromatography. Clinical Chemistry 1997, 43, 1904–1912. [Google Scholar] [CrossRef]
- Ilnytskyi. ; Lintuvuori.; Wilson. Simulation of bulk phases formed by polyphilic liquid crystal dendrimers. Condensed Matter Physics 2010, 13, 33001. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Slyusarchuk, A.; Sokołowski, S. Gelation of patchy ligand shell nanoparticles decorated by liquid-crystalline ligands: computer simulation study. Soft Matter 2018, 14, 3799–3810. [Google Scholar] [CrossRef] [PubMed]
- Hughes, Z.E.; Wilson, M.R.; Stimson, L.M. Coarse-grained simulation studies of a liquid crystal dendrimer: towards computational predictions of nanoscale structure through microphase separation. Soft Matter 2005, 1, 436. [Google Scholar] [CrossRef] [PubMed]
- Steinhauser, M.O. Computational Multiscale Modeling of Fluids and Solids; Springer Berlin Heidelberg, 2008. [CrossRef]
- Karplus, M. Development of Multiscale Models for Complex Chemical Systems: From 745 HHsub2/subto Biomolecules (Nobel Lecture). Angewandte Chemie International Edition 2014, 53, 9992–10005. [Google Scholar] [CrossRef] [PubMed]
- Groot, R.D.; Warren, P.B. Dissipative particle dynamics: Bridging the gap between atomistic and mesoscopic simulation. The Journal of Chemical Physics 1997, 107, 4423–4435. [Google Scholar] [CrossRef]
- Wilson, M.R.; Ilnytskyi, J.M.; Stimson, L.M. Computer simulations of a liquid crystalline dendrimer in liquid crystalline solvents. The Journal of Chemical Physics 2003, 119, 3509–3515. [Google Scholar] [CrossRef]
- Lintuvuori, J.S.; Wilson, M.R. A new anisotropic soft-core model for the simulation of liquid crystal mesophases. J Chem Phys 2008, 128, 044906. [Google Scholar] [CrossRef]
- Akiyama, H.; Tamada, K.; Nagasawa, J.; Abe, K.; Tamaki, T. Photoreactivity in Self-Assembled Monolayers Formed from Asymmetric Disulfides Having para-Substituted Azobenzenes. The Journal of Physical Chemistry B 2002, 107, 130–135. [Google Scholar] [CrossRef]
- Wilson, M.R. Molecular dynamics simulations of flexible liquid crystal molecules using a Gay-Berne/Lennard-Jones model. The Journal of Chemical Physics 1997, 107, 8654–8663. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, S.; Zhang, D.; Zhang, W.; Zhang, H.; Zou, J.; Mao, Z.; Yuan, Y.; Gao, C.; Liu, R. Impact of Antifouling PEG Layer on the Performance of Functional Peptides in Regulating Cell Behaviors. Journal of the American Chemical Society 2019, 141, 16772–16780. [Google Scholar] [CrossRef]
- Ilnytskyi, J.; Wilson, M.R. A domain decomposition molecular dynamics program for the simulation of flexible molecules with an arbitrary topology of Lennard–Jones and/or Gay–Berne sites. Computer Physics Communications 2001, 134, 23–32. [Google Scholar] [CrossRef]
- Ilnytskyi, J.M.; Wilson, M.R. A domain decomposition molecular dynamics program for the simulation of flexible molecules of spherically-symmetrical and nonspherical sites. II. Extension to NVT and NPT ensembles. Computer Physics Communications 2002, 148, 43–58. [Google Scholar] [CrossRef]
- Jerca, F.A.; Jerca, V.V.; Hoogenboom, R. Advances and opportunities in the exciting world of azobenzenes. Nature Reviews Chemistry 2021, 6, 51–69. [Google Scholar] [CrossRef]
- Zannoni, C. Liquid Crystals and Their Computer Simulations; Cambridge University Press, 2022. [CrossRef]
- Jekel, C.F. Digital Image Correlation on Steel Ball; 2016; chapter Appendix A, pp. 83–87.
- Paige, C.C.; Saunders, M.A. LSQR: An Algorithm for Sparse Linear Equations and Sparse Least Squares. ACM Transactions on Mathematical Software 1982, 8, 43–71. [Google Scholar] [CrossRef]
- Paige, C.C.; Saunders, M.A. Algorithm 583: LSQR: Sparse Linear Equations and Least Squares Problems. ACM Transactions on Mathematical Software 1982, 8, 195–209. [Google Scholar] [CrossRef]
- Saunders, M.A. Solution of sparse rectangular systems using LSQR and CRAIG. BIT Numerical Mathematics 1995, 35, 588–604. [Google Scholar] [CrossRef]
- LSQR. https://github.com/jacobwilliams/LSQR. Accessed: 2023-07-23.
- Todorov, T.; Nikolova, L.; Tomova, N. Polarization holography 1: A new high-efficiency organic material with reversible photoinduced birefringence. Applied Optics 1984, 23, 4309. [Google Scholar] [CrossRef]
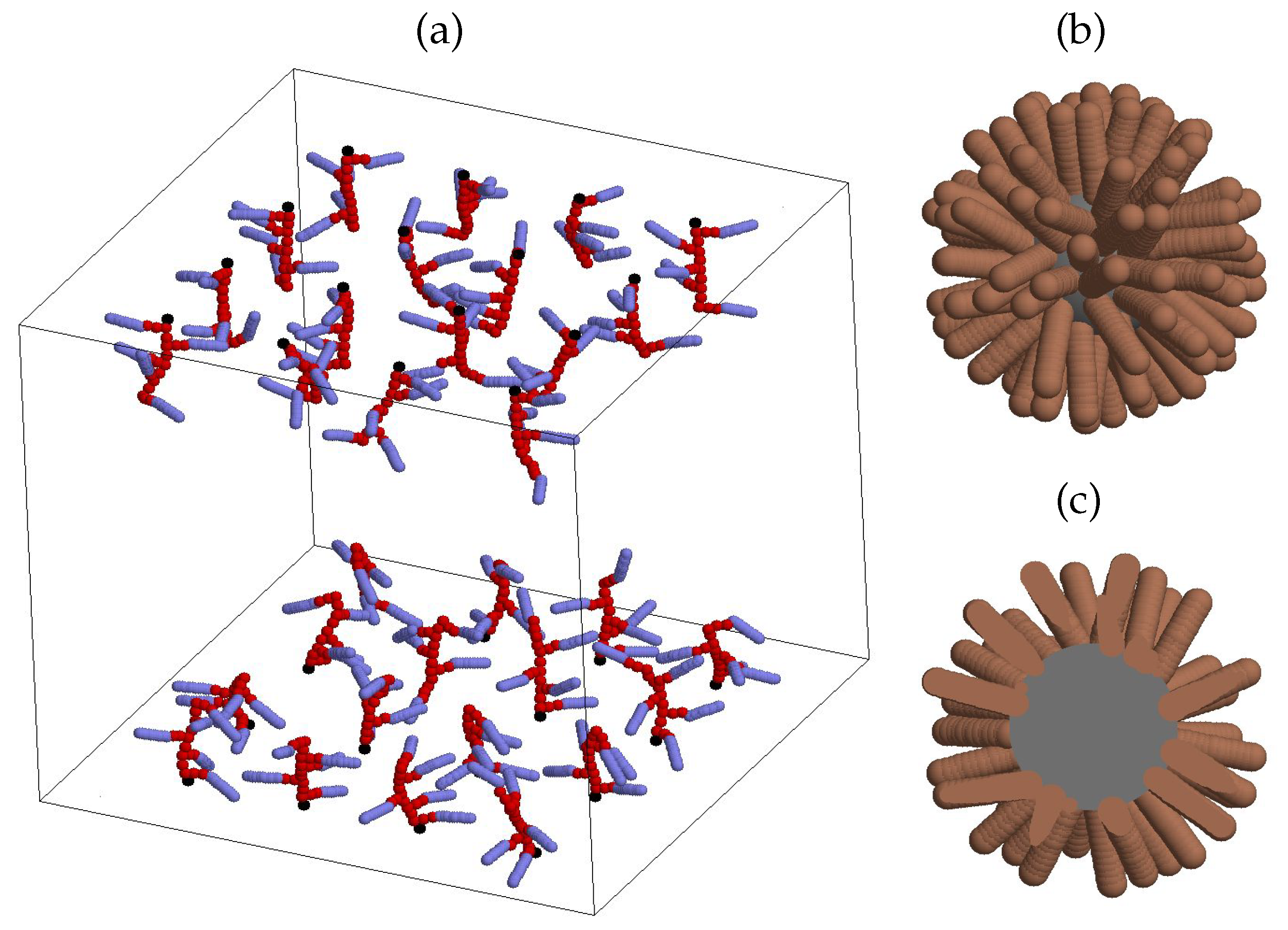
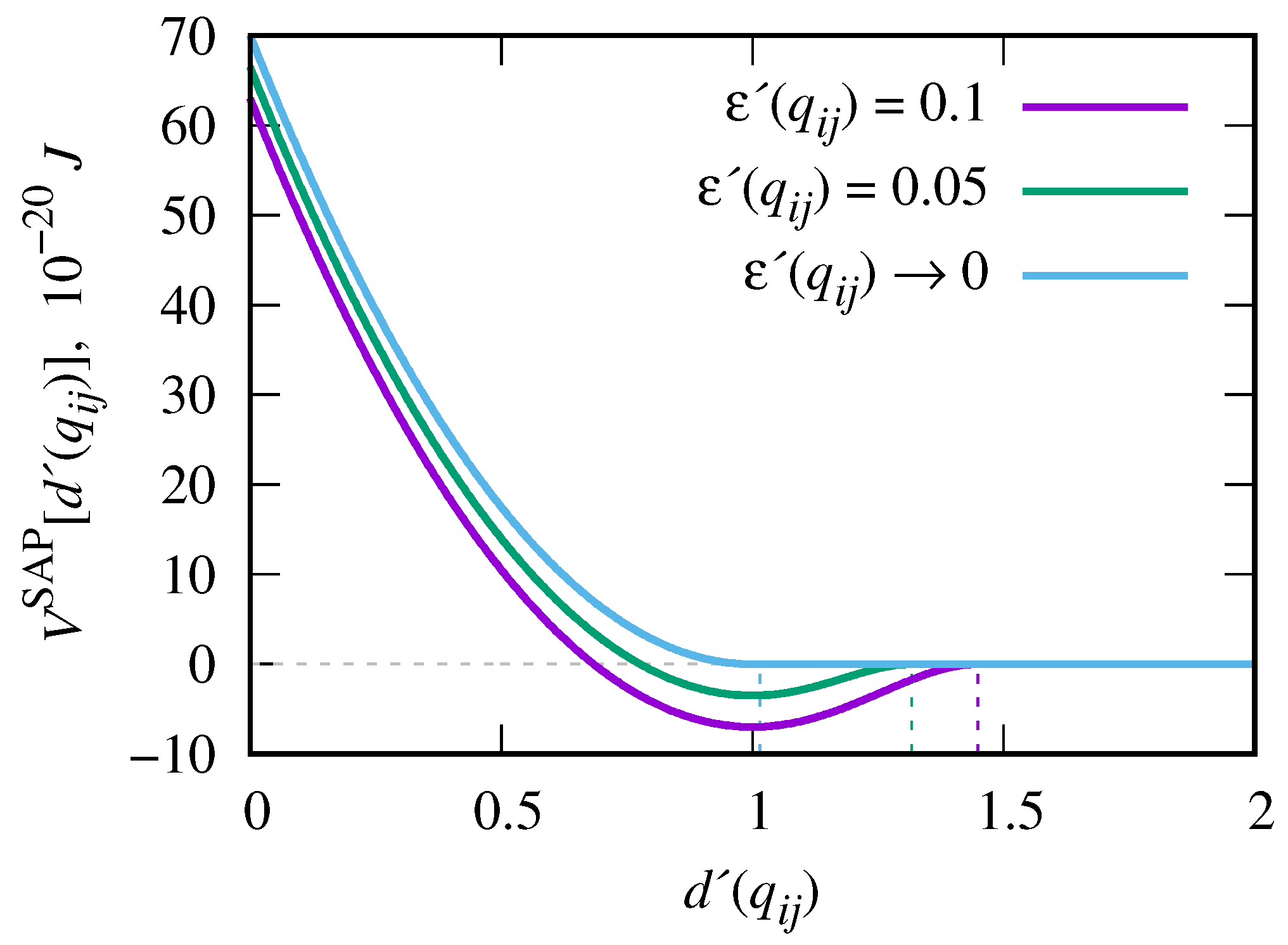

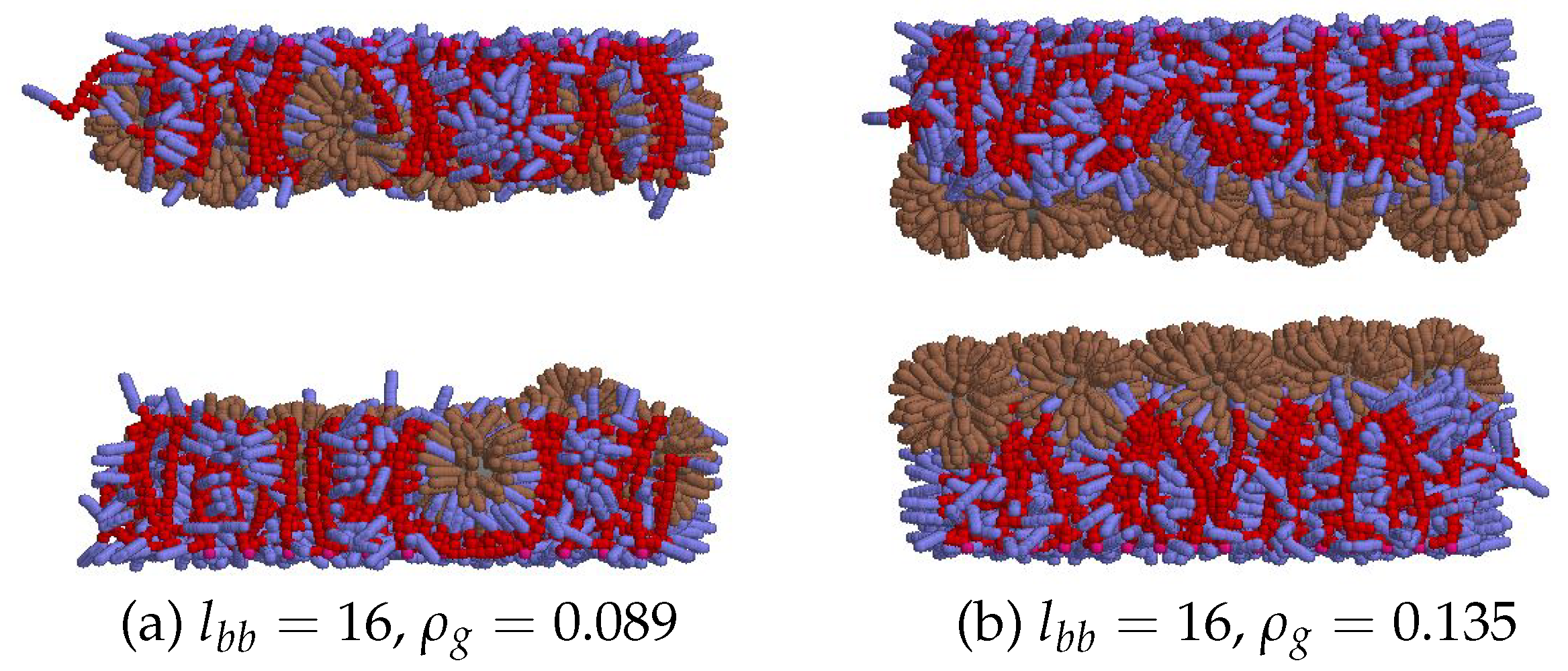
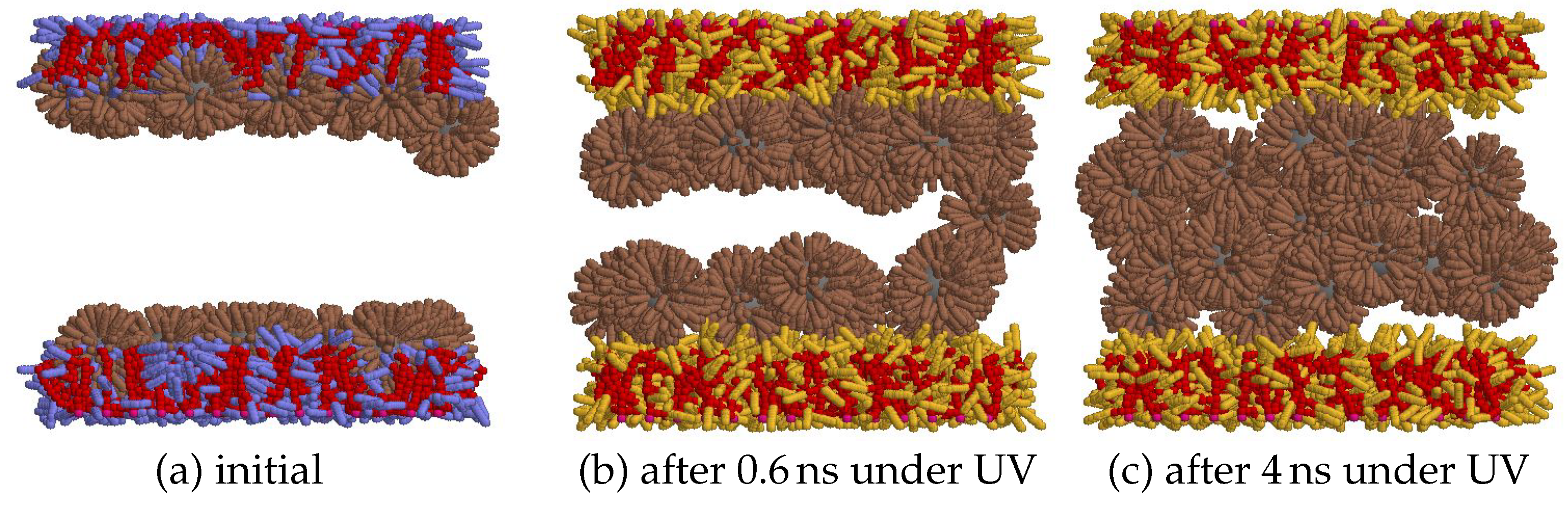
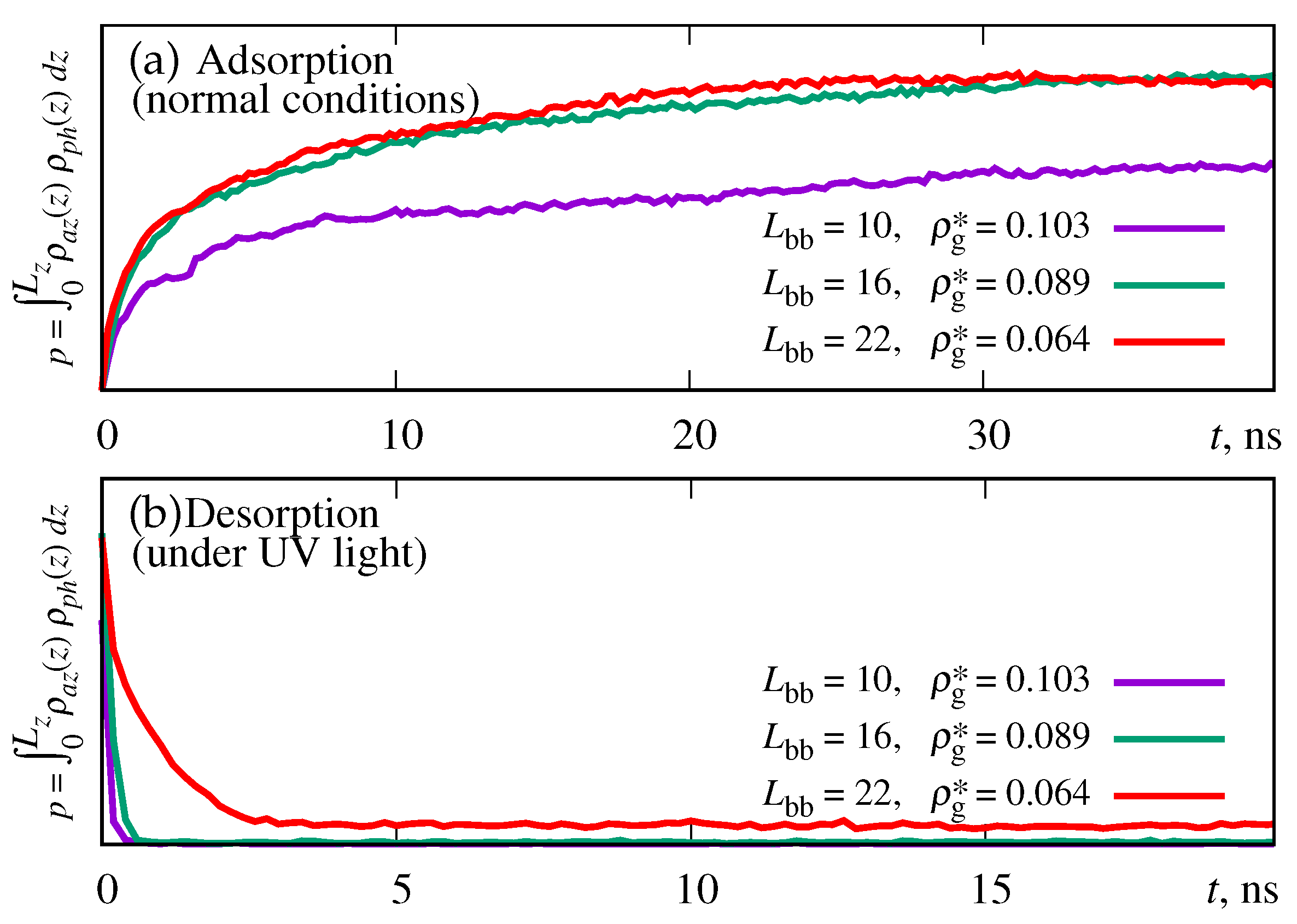
| parameter | description | value |
|---|---|---|
| core diameter | ||
| monomer and solvent bead diameter | ||
| D | azobenzene and phospholipid cap diameter | |
| azobenzene and phospholipid aspect ratio | 3 | |
| U | energy factor for regular repulsion | |
| energy factor for strong repulsion | ||
| attractive energy parameter | ||
| attractive energy parameter | ||
| attractive energy parameter | ||
| core mass | ||
| m | monomer and solvent bead mass | |
| azobenzene and phospholipid mass | ||
| azobenzene and phospholipid moment of inertia | ||
| bond length: core – phospholipid | ||
| l | bond length: monomer-monomer | |
| bond length: last spacer bead – azobenzene | ||
| bond spring constant | ||
| pseudo-valent angle at branching points | ||
| pseudo-valent angle spring constant | ||
| terminal angle for azobenzene and phospholipid | 0 | |
| terminal angle for azobenzene and phospholipid spring constant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





