1. Introduction
Zoonotic parasitic helminths pose a significant threat to the health of both humans and livestock on a global scale[
1]. Among these parasites,
Fasciola hepatica is particularly noteworthy as the causative agent of fascioliasis. This disease affects hundreds of millions of livestock, including bovines, ovines, equines, porcines, and camelids, as well as millions of humans worldwide who are parasitized by this zoonotic helminth [
2]. For many years, zoonotic fascioliasis has been effectively managed through the regular administration of anthelmintic drugs to humans, livestock, and various domestic animals. However, this preventative approach is facing challenges due to the emergence of anthelmintic resistance as a defensive mechanism employed by parasites against these drugs [
3]. The development of anthelmintic resistance in
F. hepatica is facilitated by the action of xenobiotic metabolizing enzymes (XMEs) [
4]. XMEs are present in most multicellular organisms and play a crucial role in protecting against the toxicity of natural chemicals [
5]. These enzymes have the ability to deactivate xenobiotic compounds by altering their hydrophilicity and speeding up their metabolism through a process called biotransformation [
6]. Among the XMEs found in parasitic helminths, cytochrome P450, monooxygenases, dehydrogenases, and carboxylesterases, among others, are involved [
7]. The expression and activity of XMEs regulate the susceptibility and persistence of various drugs in humans and animals [
3].
Carboxylesterase type B (CestB) plays a vital role within the xenobiotic metabolizing enzyme (XME) complex, exhibiting a wide array of functions in the metabolism and sequestration of various xenobiotics [
8]. One of CestB's primary functions is catalyzing hydrolytic reactions of carboxylic esters, phosphate esters, amides, thioesters, and other chemical compounds [
9]. Due to its large and flexible binding pocket, CestB displays a broad and overlapping substrate preference, allowing it to interact with numerous and structurally diverse substrates [
9]. However, the conformation of the binding pocket can be influenced by single nucleotide polymorphisms in the cestB gene sequence, leading to amino acid substitutions that can alter the enzyme's affinity for xenobiotics [
10]. Xenobiotic sequestration mediated by CestB has been observed in various organisms as a mechanism of resistance to pesticides. This phenomenon has been documented in examples such as the peach-potato aphid
Myzus persicae [
11], the hematophagous mosquito
Culex pipiens [
12], and the blowfly
Lucilia cuprina [
13]. In this type of pesticide resistance, there is a notable increase in CestB expression levels, and the pesticide exhibits a high affinity for the enzyme, even though hydrolysis of the pesticide may not necessarily occur. This mode of resistance, known as xenobiotic sequestration, is characterized by the presence of elevated levels of detectable CestB, which is capable of neutralizing xenobiotics by binding to an equivalent amount of toxic chemicals [
14].
Previous studies on
F. hepatica CestB have shown a significant increase in enzyme expression, which is inducible when the parasite is exposed to triclabendazole (TCBZ) [
15]. TCBZ is the most commonly used anthelmintic worldwide for the treatment of fascioliasis in both livestock and humans, and its use has led to the emergence of TCBZ-resistant strains of
F. hepatica [
16].
F. hepatica CestB is a 735 amino acid protein capable of hydrolyzing a wide range of synthetic chromogenic carboxylic esters, encompassing various chemical structures [
17]. Gene sequences of
F. hepatica CestB reported in the GenBank database have revealed single nucleotide polymorphisms (SNPs) that result in amino acid substitutions at the ligand-binding site. These substitutions, in turn, alter the affinity of the enzyme for the chromogenic substrate and impact its catalytic properties [
10].
The aim of this study was to investigate the impact of CestB polymorphisms on TCBZ-resistant parasites, specifically focusing on radical amino acid substitutions within the CestB amino acid sequence and their potential influence on the formation of a TCBZ-specific binding pocket.
2. Materials and Methods
2.1. Animals and parasite strains
The animals used in this study were cared for in accordance with the ethical guidelines of our research institutions, as outlined in the Mexican norm NOM-062-ZOO-1999 and its technical specifications for the production, care, and use of animals. The
F. hepatica reference strains were maintained for anthelmintic bioassays at the Centro Nacional de Investigación Disciplinaria en Salud Animal (CENIDSAI/INIFAP, Mexico). These strains have been registered in the NCBI BioSamples database, where they can be accessed with the identifiers SAMN16822856 for anthelmintic-susceptible
F. hepatica and SAMN16822858 for TCBZ-resistant
F. hepatica [
18]. The complete transcriptome sequences for these BioSamples have been previously uploaded to GenBank and are available at
https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR13076124&display=metadata. Adult parasites were isolated from the livers of parasitized sheep for carboxylesterase-specific enzymatic activity assays, following previously described methods [
10].
2.2. Enzyme analysis.
For protein extraction, five parasites from each strain were washed in phosphate-buffered saline (PBS) with a pH of 7.2. The parasites were then frozen at -196 °C and finely powdered following a previously published procedure [
2]. The macerated samples were subsequently homogenized with PBS (pH 7.2) at 1:1 ratio. After homogenization, the samples were centrifuged at 5,000 × g for 5 minutes. The resulting supernatant was carefully collected and subjected to further centrifugation at 20,000 × g for 30 minutes. The supernatant obtained from this centrifugation step was considered the soluble cytosol fraction, which was used for determination of enzymatic specific activity. All protein fractions obtained were collected for protein content determination using the Lowry method, with bovine serum albumin serving as the standard [
19]. The protein extracts were stored at -80 °C until they were ready to be assayed. To determine the carboxylesterase enzymatic activity in
F. hepatica, the protein extract samples were used. This was done using α-naphthyl acetate (ANA) coupled to the diazonium salt Fast Gardner. This method has been previously reported [
20]. A total of 100 μg of protein extract from each sample was assayed with four replications. The amount of α-naphthol-Azo dye released from the enzymatic reaction was measured spectrophotometrically at a wavelength of 524 nm. The absorbance values were then converted to micromoles of hydrolyzed substrate per minute per milligram of protein, following a method that had been previously described [
2].
2.3. Protein sequences and 3D models
The amino acid sequences A0A8A1L7B4 and A0A4E0S0J7 from UniProt, along with their respective Alphafold 3D models in PDB files, were downloaded from the websites
www.uniprot.org [
21] and
https://alphafold.com [
22,
23]. In addition, the
F. hepatica CestB complementary amino acid sequences MT843326, MW655750, OP537815, and THD28967.1 were obtained from the website
www.ncbi.nlm.nih.gov. These sequences were then converted to 3D models using Fasta amino acid sequences submitted to the website
https://robetta.bakerlab.org [
24].
2.4. Protein-ligand docking 3D modeling
The oxidized anthelmintic ligand TCBZSOX and the chromogenic carboxylic ester ANA were obtained as 3D models from
https://pubchem.ncbi.nlm.nih.gov. To assess ligand docking and analyze the ligand-binding site in 3D, we utilized the CB-Dock2 online algorithm available at
https://cadd.labshare.cn/cb-dock2/php/blinddock.php [
25,
26]. The resulting protein-ligand complex 3D models were downloaded as pdb files. Further modeling, visualization, and recording of the protein-ligand complexes were performed using the Mol* online algorithm, accessible at
https://molstar.org [
27].
3. Results
3.1. Carboxylesterase enzymatic specific activity.
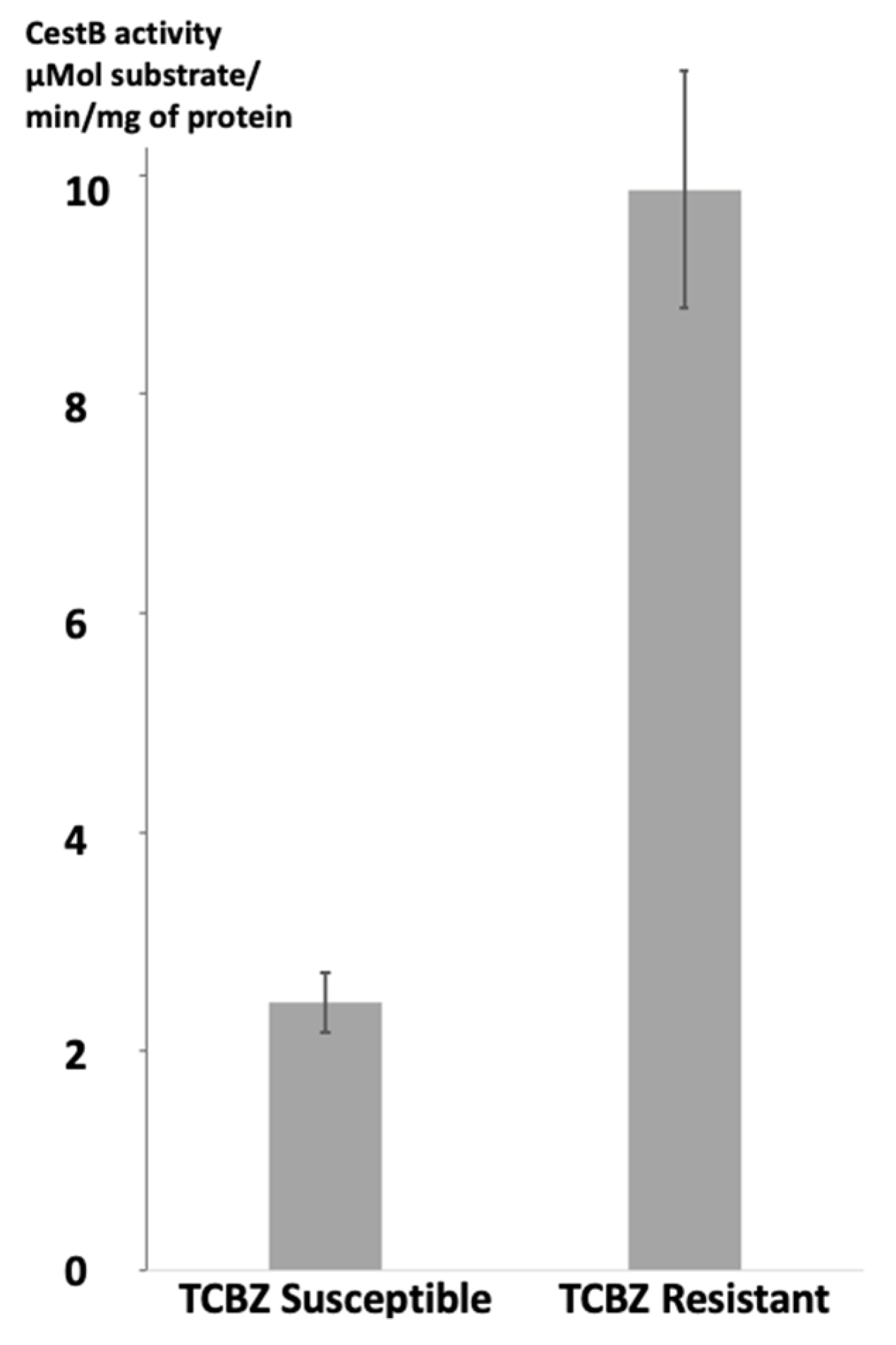
The enzymatic specific activity of
F. hepatica CestB on the parasite's protein extracts was determined for each strain of parasites. Statistical analysis using a Student's unpaired t test revealed significant differences, with a two-tailed P value of less than 0.0001. The susceptible strain exhibited a mean enzymatic specific activity of 2.325 µMol substrate/mg of protein/min, while the resistant strain showed a higher activity of 9.675 µMol substrate hydrolyzed for mg of protein per minute. These results are summarized and presented in
Figure 1.
3.2. Protein 3D modeling and protein-ligand docking analysis.
The amino acid sequences of
F. hepatica carboxylesterase B from both susceptible and resistant strains were retrieved from the GenBank and UniProt databases. These sequences were then converted to Alphafold and Rosettafold 3D models. The models were subsequently utilized for protein-ligand docking studies with the metabolically oxidized anthelmintic TCBZSOX and the synthetic chromogenic carboxylesterase substrate ANA. The findings of these experiments are summarized in
Table 1. Additionally, detailed 3D models resulting from the docking process are visually presented in
Figure 2 and
Figure 3.
The CB-Dock2 algorithm was used to identify the amino acids present at the TCBZSOX-binding site in the susceptible CestB enzyme isolated from the susceptible strain. The identified amino acids at this binding site were F146, R147, A199, S204, C206, G208, C225, T227, H262, R263, G265, L266, P267, P269, H637, and Y639. Interestingly, the CestB amino acid sequence from the resistant strain revealed a different set of amino acids at the TCBZSOX-binding pocket, which included A281, I282, and S283 in addition to those found in the susceptible CestB amino acid polymorphism. These results, along with the corresponding TCBZSOX-protein 3D models, are summarized in
Table 1 and depicted in
Figure 2.
In the analysis of docking modeling using the esterase chromogenic substrate ANA, the CB-Dock2 algorithm identified specific amino acids at the susceptible CestB catalytic site. These amino acids were K210, N211, G214, E215, L216, V217, G256, L259, Y284, S336, T339, and I340. However, in the resistant CestB, amino acid N735 replaced I340, which was detected in the susceptible polymorphism. These findings are summarized in
Table 1, and corresponding 3D models of the ANA-protein complex are depicted in
Figure 3.
4. Discussion
Anthelmintic resistance of parasites is a complex phenomenon involving genetic mechanisms that often require the collaboration of one or multiple genes to result in noticeable levels of resistance to anthelmintic treatment [
6]. These genetic mechanisms can confer resistance through different phenotypes, including modifications in the target site of the anthelmintic [
28] and/or increased enzymatic detoxification [
7].
According to our experimental findings, there was a significant difference in CestB enzymatic specific activity between TCBZ-susceptible and TCBZ-resistant parasites protein extracts. This observation is consistent with previous reports indicating that CestB exhibits increased enzymatic specific activity in anthelmintic-resistant parasites [
2,
15].
To further investigate the underlying mechanisms, we employed a combined experimental and bioinformatics approach to develop a 3D model that could explain the observed enzymatic differences between the susceptible and resistant parasite strains. Our study revealed that the amino acid sequences of CestB exhibited polymorphisms between strains displaying different levels of susceptibility to TCBZ. This suggests that these differences may be responsible for the development of anthelmintic resistance in the liver fluke. Our findings align with the hypothesis of enzymatic detoxification, wherein the CestB amino acid polymorphisms may bind to, sequester, or enzymatically neutralize the anthelmintic compound, thus rendering it less effective.
Our assessment was conducted using TCBZ-CestB 3D docking modeling, which involves the use of the metabolically oxidized form of the anthelmintic triclabendazole, known as triclabendazole sulfoxide (TCBZSOX). This biotransformation occurs at the host's liver level and enhances the toxic effect of TCBZ on the parasites [
16]. Therefore, the 3D modeling analysis employed TCBZSOX for ligand-docking evaluation, specifically looking at how amino acid polymorphisms found in CestB impact the TCBZSOX-binding pocket.
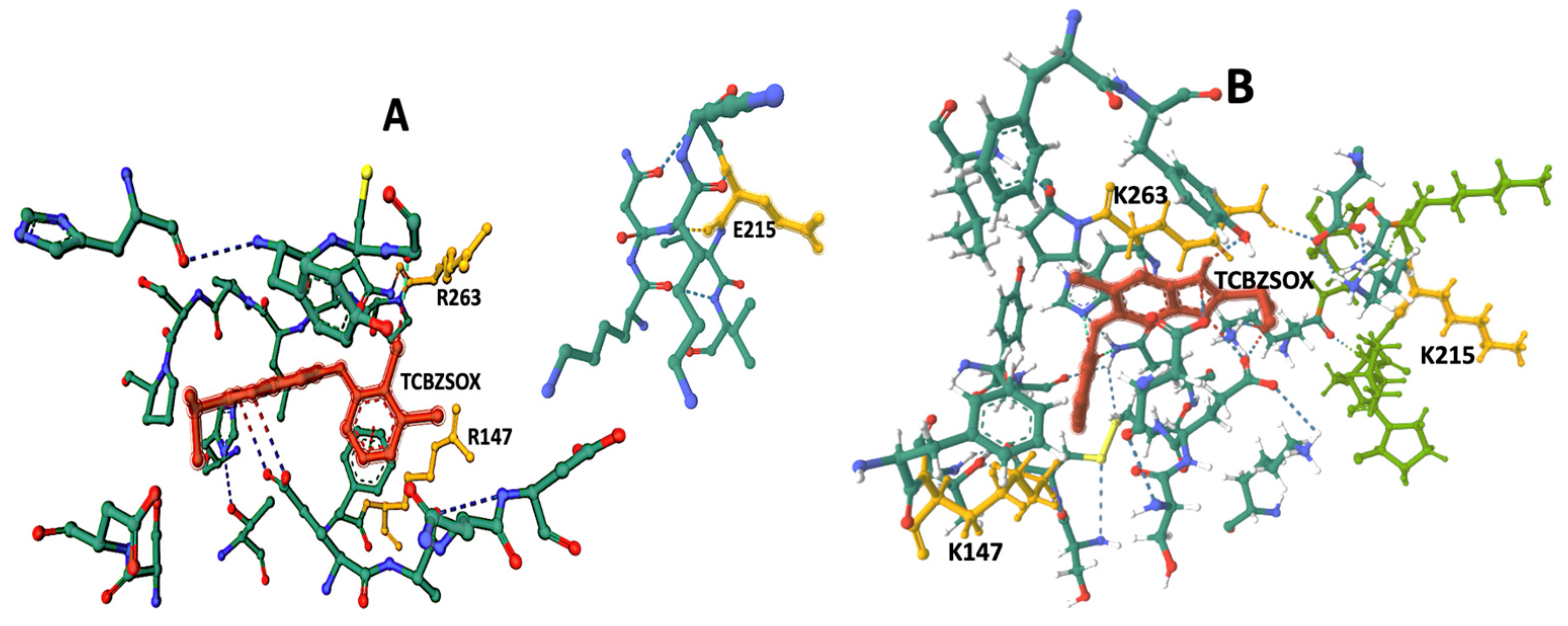
The docking results indicate the presence of TCBZSOX-binding pockets in the enzyme of both strains, although there are noticeable differences. In the susceptible CestB strain, amino acid substitutions R147, E215, and R263 are located in separate, unconnected domains within the 3D model shown in
Figure 3A. On the other hand, in the resistant strain, amino acid substitutions K147, K215, and K263 alter the conformation of the binding pocket to form a compact domain surrounding TCBZSOX, as depicted in
Figure 3B. Additionally, in the resistant strain, amino acid substitutions K147 and K263 directly interact with the anthelmintic, while T639 forms an ionic bond with TCBZSOX. None of these interactions were observed in the TCBZ-susceptible CestB model, as shown in
Table 1.
Another significant difference observed was the presence of a considerable number of hydrogen bonds between the amino acids in the TCBZSOX-binding pocket of the resistant strain and the anthelmintic. Notably, amino acids such as D202, K210, H262, and P264 formed hydrogen bonds in the resistant enzyme, as depicted in
Table 1 and
Figure 2B. However, these specific interactions were not observed in the susceptible enzyme.
The results of the ligand docking modeling with ANA revealed that the synthetic chromogenic substrate consistently positioned itself in close proximity to S336. This specific amino acid is known to be the catalytic serine responsible for the hydrolysis of ANA into acetic acid and naphthol [
10]. Moreover, the positioning of ANA in the docking model provided insights into the location of the enzyme's catalytic site and highlighted the distinct conformations of constitutive amino acids between parasite strains. In the resistant strains, S336 exhibited hydrogen bond interactions with G256, Y258, L259, T339, I340, and N735, as shown in
Table 1 and Figure 4. In contrast, the catalytic serine in the susceptible polymorphism formed hydrogen bonds with G256, Y258, L259, T339, and I340. These differences in the active site of the enzyme are consistent with previous reports and may help explain the observed increase in enzymatic specific activity in the resistant strain compared to the susceptible strain [
10].
The docking model further reveals that the TCBZ-binding pocket is located far away from the enzyme's catalytic site, suggesting that there is no theoretical possibility of enzymatic hydrolysis of TCBZSOX. However, the high level of expression observed is consistent with the hypothesis of anthelmintic sequestration. This hypothesis suggests that excessive expression of the enzyme would result in a sufficient number of free enzyme molecules capable of neutralizing the toxic effects of the anthelmintic, even in the absence of hydrolytic activity on the anthelmintic itself.
Previous studies have indicated that sequestration of TCBZ by exosomes could be an important mechanism of anthelmintic resistance in
F. hepatica [
16]. Our data support this observation and may provide a complementary explanation. Exosomes are membranous vesicles that act as carriers for biomolecules, including enzymes [
29]. As CestB is a membrane-bound enzyme [
2], it is possible that it plays a role in the transportation of anthelmintics between different cellular compartments, and eventually secretion into exosomes.
Our study provides evidence suggesting that a protein in TCBZ-resistant F. hepatica is capable of binding and potentially sequestering TCBZ, which may be a significant factor in explaining TCBZ resistance. Furthermore, our data suggest that radical amino acid substitutions at positions 147, 215, and 263, along with the single nucleotide polymorphisms that led to these amino acid changes in the CestB nucleic acid sequences, could serve as valuable molecular markers for anthelmintic resistance in F. hepatica.
5. Conclusions
Our findings provide support for a hypothesis in which anthelmintic resistance in the zoonotic parasitic trematode F. hepatica arises from amino acid substitutions at the TCBZ-binding site of CestB, without involving enzymatic hydrolytic degradation of the drug. Through our study, we determined that there is a significant level of polymorphism in F. hepatica CestB, and radical amino acid substitutions K147, K215, and K243 are exclusively found in TCBZ-resistant parasites. These substitutions alter the TCBZ-binding pocket, enabling direct binding of the anthelmintic.
Additionally, our investigation has revealed that these amino acid substitutions also lead to a reconfiguration of the enzyme's catalytic site, resulting in changes in enzymatic specific activity against chromogenic carboxylic esters. This difference in enzymatic activity was found to be statistically significant when comparing TCBZ-susceptible parasites to those exhibiting anthelmintic resistance.
In conclusion, our findings have implications for the design of a molecular test to detect anthelmintic resistance in F. hepatica. This could involve the use of qPCR, SNTPs analysis, and measurement of Carboxylesterase specific activity using protein extracts from F hepatica.
Author Contributions
EMM: Experimental execution, Conceptualization, Methodology, Writing-original draft preparation. RCB: Experimental Execution, Analysis, Investigation, Writing-Original draft preparation. LTC: Samples Processing, Analysis and Investigation. HAD: Writing-Reviewing and Analysis.
Funding
This research received funding from INIFAP SIGI:1172635360.
Institutional Review Board Statement
Animal management was performed according to the ethical guidelines of our institutions. Animal care and use were performed according to the Mexican norm NOM-062-ZOO-1999, and its technical specifications for the production, care and use of laboratory animals and supervised by the laboratory animal handling ethics committee.
Data Availability Statement
Access to RNAseq data Transcriptome from TCBZ-resistant and TCBZ-susceptible F. hepatica, and Supplemental information of the Transcriptome is provided as links within the manuscript in the form of raw sequencing readings, and tables that include differential expression for each transcript in FPKM, GO and KEGG numbers as well as GENBANK accession numbers for each sequence, details of raw read generated, assembly and annotation information, overall transcriptomic annotation information such as mapping rate, number of known and unknown transcripts identified, splicing events and long noncoding RNA transcripts as well as the annotated gene ontology divided in number of genes found as cellular components or fulfilling a biological process or molecular function.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships, that have, or could be perceived to have, influenced the work reported in this article.
References
- Zumaquero-Ríos, J.L.; Sarracent-Pérez, J.; Rojas-García, R.; Rojas-Rivero, L.; Martínez-Tovilla, Y.; Valero, M.A.; Mas-Coma, S. Fascioliasis and Intestinal Parasitoses Affecting Schoolchildren in Atlixco, Puebla State, Mexico: Epidemiology and Treatment with Nitazoxanide. PLoS Negl Trop Dis 2013, 7, e2553. [CrossRef]
- Pedroza-Gómez, Y.J.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Scarcella, S.; Reynaud, E.; Sanchez-Carbente, M. del R.; Narváez-Padilla, V.; Miranda-Miranda, E. Transcriptome-Based Identification of a Functional Fasciola Hepatica Carboxylesterase B. Pathogens 2021, 10, 1454. [CrossRef]
- Matoušková, P.; Vokřál, I.; Lamka, J.; Skálová, L. The Role of Xenobiotic-Metabolizing Enzymes in Anthelmintic Deactivation and Resistance in Helminths. Trends in Parasitology 2016, 32, 481–491. [CrossRef]
- Scarcella, S.; Miranda-Miranda, E.; Cossío-Bayúgar, R.; Ceballos, L.; Fernandez, V.; Solana, H. Increase of Carboxylesterase Activity in Fasciola hepatica Recovered from Triclabendazole Treated Sheep. Molecular and Biochemical Parasitology 2012, 185, 151–153. [CrossRef]
-
Casarett & Doull’s Toxicology: The Basic Science of Poisons; Klaassen, C.D., Ed.; 9th ed.; McGraw Hill Medical: New York, NY., 2019; ISBN 978-1-259-86374-5.
- Fissiha, W.; Kinde, M.Z. Anthelmintic Resistance and Its Mechanism: A Review. IDR 2021, Volume 14, 5403–5410. [CrossRef]
- Mordvinov, V.; Pakharukova, M. Xenobiotic-Metabolizing Enzymes in Trematodes. Biomedicines 2022, 10, 3039. [CrossRef]
- Wang, L.-L.; Huang, Y.; Lu, X.-P.; Jiang, X.-Z.; Smagghe, G.; Feng, Z.-J.; Yuan, G.-R.; Wei, D.; Wang, J.-J. Overexpression of Two α-Esterase Genes Mediates Metabolic Resistance to Malathion in the Oriental Fruit Fly, Bactrocera Dorsalis (Hendel). Insect Mol Biol 2015, 24, 467–479. [CrossRef]
- Hosokawa, M. Structure and Catalytic Properties of Carboxylesterase Isozymes Involved in Metabolic Activation of Prodrugs. Molecules 2008, 13, 412–431. [CrossRef]
- Miranda-Miranda, E.; Scarcella, S.; Reynaud, E.; Narváez-Padilla, V.; Neira, G.; Mera-y-Sierra, R.; Aguilar-Díaz, H.; Cossio-Bayugar, R. A Single Nucleotide Polymorphism Translates into a Radical Amino Acid Substitution at the Ligand-Binding Site in Fasciola hepatica Carboxylesterase B. Genes 2022, 13, 1899. [CrossRef]
- Field, L.M.; Blackman, R.L.; Tyler-Smith, C.; Devonshire, A.L. Relationship between Amount of Esterase and Gene Copy Number in Insecticide-Resistant Myzus Persicae (Sulzer). Biochem J 1999, 339 ( Pt 3), 737–742.
- Hemingway; Karunaratne Mosquito Carboxylesterases: A Review of the Molecular Biology and Biochemistry of a Major Insecticide Resistance Mechanism. Med Vet Entomol 1998, 12, 1–12. [CrossRef]
- Newcomb, R.D.; Campbell, P.M.; Ollis, D.L.; Cheah, E.; Russell, R.J.; Oakeshott, J.G. A Single Amino Acid Substitution Converts a Carboxylesterase to an Organophosphorus Hydrolase and Confers Insecticide Resistance on a Blowfly. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 7464–7468. [CrossRef]
- Wang, K.; Huang, Y.; Li, X.; Chen, M. Functional Analysis of a Carboxylesterase Gene Associated With Isoprocarb and Cyhalothrin Resistance in Rhopalosiphum Padi (L.). Front. Physiol. 2018, 9, 992. [CrossRef]
- Scarcella, S.; Solana, M.V.; Fernandez, V.; Lamenza, P.; Ceballos, L.; Solana, H. Increase of Gluthatione S-Transferase, Carboxyl Esterase and Carbonyl Reductase in Fasciola hepatica Recovered from Triclabendazole Treated Sheep. Molecular and Biochemical Parasitology 2013, 191, 63–65. [CrossRef]
- Davis, C.N.; Winters, A.; Milic, I.; Devitt, A.; Cookson, A.; Brophy, P.M.; Morphew, R.M. Evidence of Sequestration of Triclabendazole and Associated Metabolites by Extracellular Vesicles of Fasciola hepatica. Sci Rep 2020, 10, 13445. [CrossRef]
- Boyko, K.M.; Kryukova, M.V.; Petrovskaya, L.E.; Nikolaeva, A.Y.; Korzhenevsky, D.A.; Novototskaya-Vlasova, K.A.; Rivkina, E.M.; Dolgikh, D.A.; Kirpichnikov, M.P.; Popov, V.O. Crystal Structure of PMGL2 Esterase from the Hormone-Sensitive Lipase Family with GCSAG Motif around the Catalytic Serine. PLoS ONE 2020, 15, e0226838. [CrossRef]
- Miranda-Miranda, E.; Cossio-Bayugar, R.; Aguilar-Díaz, H.; Narváez-Padilla, V.; Sachman-Ruíz, B.; Reynaud, E. Transcriptome Assembly Dataset of Anthelmintic Response in Fasciola hepatica. Data in Brief 2021, 35, 106808. [CrossRef]
- Peterson, G.L. A Simplification of the Protein Assay Method of Lowry et al. Which Is More Generally Applicable. Analytical Biochemistry 1977, 83, 346–356. [CrossRef]
- Bardi, L.; Dell’oro, V.; Delfini, C. Rapid Spectrophotometric Method to Determine Esterase Activity of Yeast Cells in an Aqueous Medium. Journal of the Institute of Brewing 1993, 99, 385–388. [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Research 2021, 49, D480–D489. [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Research 2022, 50, D439–D444. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Research 2022, 50, W159–W164. [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein–Ligand Binding Site Recognition Using Complementary Binding-Specific Substructure Comparison and Sequence Profile Alignment. Bioinformatics 2013, 29, 2588–2595. [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Research 2021, 49, W431–W437. [CrossRef]
- Partridge, F.A.; Forman, R.; Bataille, C.J.R.; Wynne, G.M.; Nick, M.; Russell, A.J.; Else, K.J.; Sattelle, D.B. Anthelmintic Drug Discovery: Target Identification, Screening Methods and the Role of Open Science. Beilstein J. Org. Chem. 2020, 16, 1203–1224. [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).