1. Introduction
Patients with congenital univentricular heart undergoing the Fontan procedure (FP) are at high risk of liver disease. In these patients, chronically elevated right atrial pressure and tissue hypoxia may predispose to the development of hepatic masses, encompassed under the acronym FALD (Fontan-associated liver disease) and ranging from focal nodular hyperplasia (FNH)-like nodules and benign tumor as hepatic adenoma (HA) to malignant lesions including hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) [
1]. Cirrhosis, a potential prerequisite for HCC, may develop before the age of 25, approximately 10 to 15 years after Fontan operation. The incidence of cancer of 1.5 to 5% per year is estimated on the basis of previous studies [
2,
3,
4].
The etiology of neoplastic lesions, in particular of malignancies, is probably multifactorial: an underlying genetic predisposition, the presence of risk factors (aging, virus infections, alcohol assumption, steatosis), the abnormal physiology of chronically elevated central venous pressures. However, genetic and molecular studies of malignancy in the setting of FALD are lacking to date [
5].
The purpose of this study is to investigate the genetic and molecular profile of histologically proven hepatic lesions in a monocentric series of FALD in order to explore potential driver genes and the major dysregulated signaling pathways involved in the development of different types of lesions and identify possible biomarkers important for diagnostic and therapeutic purposes.
2. Methods
2.1. Case selection
Our institutional review board approved this single-institution retrospective review and waived the requirement for informed consent. Between January 1, 1990 and December, 2022, the Cardiac Surgery Department at Bambino Gesù Children's medical records were reviewed and 450 children, undergoing FP for palliative correction of congenital univentricular heart, were identified: 208 were over 18 years of age in 2022, with an average age of 28 years and a follow-up of 22 years after Fontan's operation. Most patients, 127/208 (61%), had no liver nodules during follow-up (performed with both imaging and alpha-fetoprotein serology - AFP-, equally negative); 50/208 (24%) patients had one nodule <1 cm in diameter which were followed up, but not biopsied, because AFP was low and their size did not change over time. However, an ultrasound examination of the liver was performed every 6 months. Finally, 31/208 (15%) patients had nodule/s >1 cm. Then, by reviewing the Pathology medical record, we identified all the patients with a history of FP who had undergone biopsy and/or resection of one or more focal liver lesions. Seven/208 (3%) with hepatic nodules were submitted to a histological examination. Other 5/208 (2%) patients received a liver biopsy to evaluate the fibrosis and architectural remodeling after FP. We have limited this report to lesions diagnosed on the basis of histopathologic sampling, as we made genetic/molecular analysis on hepatic tissue. Two patients were excluded from the study because the nodules were strongly suspicious for HCC on imaging, and the patients underwent chemoembolization without histological confirmation Interestingly, liver lesions in this population may have atypical imaging features that can lead to misdiagnosis [
6].
Medical records were reviewed for: sex (3/7 patients were female), age at FP, type of congenital heart disease (CHD), type of FP (1 patient had a lateral tunnel Fontan pathway and the remaining 6 had an extracardiac Fontan pathway), age at FALD (median age at the time of liver biopsy was 26 years, range: 20 years -34 years), interval from Fontan operation to biopsy (20 years, range: 10-28 years), lesion histopathologic diagnosis, AFP values closest prior to biopsy and follow-up (all data are summarized in
Table 1).
2.2. Histology
Liver biopsy or surgical sample were formalin-fixed and paraffin-embedded. Hematoxilin-Eosin (HE) and Masson’s trichrome stains to assess fibrosis were routinely made. Immunohistochemistry (IHC) with anti-β-catenin (Novocastra, NCL-L-B-CAT, 1:150 for 30 minutes), anti-glutamine syntetase (GS-6, Millipore, MAB302, 1:400 for 25 minutes), anti-glypican3 (Gly3, BioMosaics, B0025R, B0055R, 1:1000 for 30 minutes), anti-liver fatty acid bending protein (LFABP, Abcam, ab 7807, 1:100 for 30 minutes) and anti-SOX9 (Cell Signaling Technology, #82630, 1:100 for 30 minutes) were performed with Dako Omnis, Agilent Technology (Denmark).
2.3. DNA mutational analysis
DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tumor tissue and non-lesional adjacent liver tissue using Maxwell CSC instrument (Promega, Madison, USA) with the Maxwell RSC DNA FFPE kit (Promega, Madison, USA) according to the manufacturer’s protocol; DNA concentrations were measured on a Qubit 2.0 Fluorometer (Thermofisher Scientific, Waltham, USA) using the Qubit dsDNA High Sensitivity.
For DNA library preparation and enrichment, the TruSight™ Oncology 500 Kit (Illumina) was used following the manufacturer’s instructions. Post-enriched libraries were quantified, pooled, and sequenced on a NextSeq 550 (Illumina Inc., San Diego, CA, USA). The quality of the NextSeq 550 (Illumina) sequencing runs was assessed with the Illumina Sequencing Analysis Viewer (Illumina). NGS data was analyzed with Illumina TruSight Oncology 500 Local App v2.1 and for interpretation and reporting, variant report files can be uploaded into the Pierian Clinical Genomics Workspace cloud (Pierian DX software CGW_V6.21.1).
2.4. RNA sequencing and analysis
RNA was extracted from FFPE tumor tissue using Maxwell CSC instrument (Promega, Madison, USA) with the Maxwell RSC RNA FFPE kit (Promega, Madison, USA) according to the manufacturer’s protocol; RNA concentrations were measured on a Qubit 2.0 Fluorometer (Thermofisher Scientific, Waltham, USA) using the Qubit RNA High and quality and quantity of RNA samples was ascertained with the use of Agilent 2200 Tapestation system (Agilent Technologies). Mean RNA integrity number (RIN) was 5.2 (range 3.0 to 6.9).
In order to identify genomic rearrangement, 300 nanograms of total RNA were used for NGS library preparation with SureSelect XT HS2 kit (Agilent Technologies) following the manufacturer's instructions. The sequencing run was performed in paired-end mode (2 X 151-bp reads) using the Illumina NextSeQ 550 platform and generating at least 30 million reads per sample. The resulting alignment files were then used by the STAR-Fusion and Arriba pipelines to identify any candidate fusion transcripts [
7,
8].
3. Results
Seven patients underwent biopsy/surgical resection. One patient had multiple lesions targeted for biopsy with a total number of 9 nodules examined. Two patients with biopsied liver lesions subsequently underwent heart transplantation (HTx), 1 month and 2 years later, respectively. In addition, one of these patients also developed a subsequent lesion biopsied 5 years after HTx, probably as a result of earlier Fontan circulation even after HTx [
9,
10].
Non-lesional tissue was present in all but one cases and analyzed in pairs with neoplastic tissue.
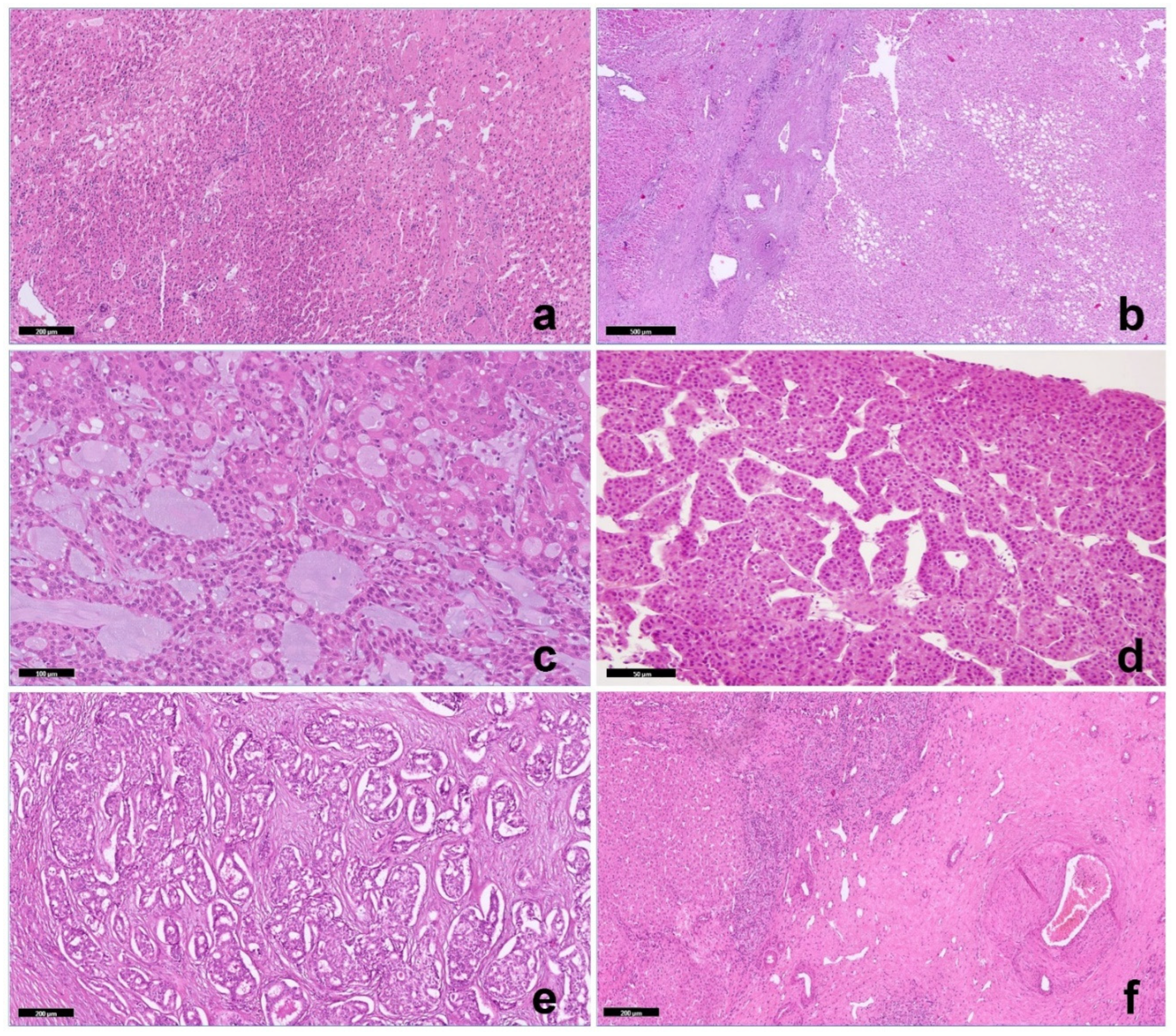
Biopsy of background (non-lesional) liver tissue showed changes consistent with Fontan pathophysiology in all cases, including sinusoidal dilatation with portal and pericentral fibrosis (
Figure 1a). One of the seven lesions was a nodular regenerative hyperplasia (NRH). Two lesions were hepatocellular adenomas (HA), one of the inflammatory subtype and one of the hepatocyte nuclear factor 1α [HNF1α] inactivated subtype (
Figure 1b). Four malignancies included a fibrolamellar carcinoma (FLC) (
Figure 1c) 2 trabecular HCC (
Figure 1d) and an intrahepatic cholangiocarcinoma (ICC,
Figure 1e) with an adjacent area of focal nodular hyperplasia, (
Figure 1f). in a patient with a previous RNH (7y before). FLC was composed of large polygonal cells with abundant eosinophilic cytoplasm, large vesicular nuclei, and large nucleoli in association with lamellar bands of fibrosis. In addition, an unusual appearance of nests of neoplastic hepatocytes or hepatocytes in a lace-like pattern were embedded in an abundant background of extracellular myxoid matrix. The 2 HCC were well to moderately differentiated carcinomas with a classic trabecular pattern.
Immunohistochemically, β-catenin was negative for nuclear accumulation in all cases. Glypican3 and GS were negative in HAs and in FLC, but diffusely positive in both trabecular HCC. SOX9 was multifocally positive only in trabecular HCC. The neoplastic cells in HNF1a-HA as in FLC were negative for LFABP, while its expression was preserved in inflammatory HA and trabecular HCCs.
A total of 19 single nucleotide variants (SNV) and 3 copy number variations (CNV) from 500 genes were identified in 5 samples. The most commonly altered genes in our cohort were FGFRs: 1/1 ICC (P3) and 2/3 HCCs (P1 and 2) showed loss of one copy of the FGFR3 gene, one of this HCC (P2) showed in addition a FGFR2 variant. Furthermore, the same patients (P1 to 3) showed variant on GNAS. 1 HCC (P4) showed a hotspot mutation on CTNNB1 and NRAS; the only ICC (P3) displayed a variant of unknown significance (VUS) on CTNNB1.
TMB was usually low (TMB < 5) and only one case showed an intermediate TMB (TMB ranging from 5 to 10).
The non-lesional liver displayed similar genomic profile of tumor tissue in two cases (P2 and 3); only one case (P1) had a non-lesional tissue profile without CNVs or variants. DNA from non-lesional tissue in P4 and P5 were not available.
RNA sequencing, performed in 4 cases, P1 to 3 and P5 (1 FLC, 1 HCC, 1 ICC, 1 HA), identified
DNAJB1-PRKACA in-frame fusion in FLC. RNA was not available in the P4 case. One NRH and the inflammatory HA were not included in the molecular study.
Table 2 summarizes the molecular results.
4. Discussion
Routine screening for liver disease in Fontan patients remains a challenge. Noninvasive measurements of the hepatic function do not show abnormalities until advanced stages of hepatic fibrosis. Determining which lesions are potentially benign, reactive or malignant is one of the difficulties in FALD evaluation. Liver tumors develop approximately 10 to 20 years after FP, frequently in a cirrhotic background and with elevated AFP.
In the present cohort of 208 univentricular heart submitted to FP, 6 neoplastic nodules were identified, with a biopsy performed only in 5. The histologic diagnosis of HA (1), FLC (1), HCC (2) and ICC (1) confirmed the heterogeneity of tumors arising in FALD, although all occurred in the context of a cirrhotic background. In all HCC and ICC immunostaining for β-catenin was negative (absence of nuclear accumulation). GS, Glypican3 and SOX9 were negative in FLC and focal or diffusely positive in trabecular HCC.
CTNNB1 point mutation in exon 3 was present in one trabecular HCC, in line with the reported mutation rate of 35% in HCC outside the context of FALD [
11].
FGFR3 allele loss was detected in 2 HCCs as well as in 1 ICC, although this loss did not lead to lack of FGFR3 expression. The only identification in
FGFR3 copy number alteration does not exclude the possibility of a gross loss of a chromosomal region containing important genes. No variants in either
TERT promoter or
TP53 were identified in the HCCs as well as no
TP53, CDKN2A/B, KRAS, ARID1A and
IDH1 variations or
FGFR2 fusion were present in the ICC.
Fibroblast growth factors (FGFs) in the liver have been shown to promote regeneration. Fibroblast growth factors bind to distinct receptors mediating different effects. Overall, overexpression of FGF receptors (FGFRs) seems to drive HCC development and progression [
12]. While
FGFR3 amplification has been reported in HCC and plays an important role in HCC development and progression and FGFR3 expression is elevated in human cirrhotic livers, the allele loss in 2 HCC and 1 CC in FALD is intriguing and deserves further investigation. Although in one case RNAseq demonstrated reduction of
FGFR3 expression, it is difficult to know if loss of one allele plays a role in the pathogenesis. However, the recurrence of the alteration in 3 cases may be significant in terms of molecular alterations potentially predisposing to tumor development, especially because it was also identified in non-tumor tissue.
It may be speculated that hypoxic damage, related to the type of surgery, may cause
FGFR3 variants, or also the fusion, being these areas of DNA fragility in hepatocytic precursors. This is also supported by experimental models in mouse. The functional role of FGFR3 and its isoforms has not yet been investigated in the context of hepatic fibrosis. To elucidate the significance of
FGFR3 allele loss in 2 HCC and 1 ICC, it might be important to consider the cytoprotective function of FGFR3 in hepatocytes. Experimentally, mice lacking FGFR3 showed increased liver necrosis, particularly in response to CCl4 treatment, and enhanced fibrosis. Furthermore, FGFR3 in hepatocytes can directly limit fibrosis by suppressing the expression of pro-fibrotic molecules, such as Loxl4 and Tff3, which were expressed at higher levels in the hepatocytes of Alb-R3 mice [
13]. Thus, we could hypothesize that the loss of
FGFR3 may be related to the cirrhotic background rather than to the neoplastic transformation in our cases.
Another common alteration in our series was
GNAS variants. The
GNAS gene encodes the alpha-subunit of the stimulatory G protein, which regulates neurotransmitters and many hormones through generating cAMP. Even if
GNAS mutations are highly associated with McCune-Albright syndrome,
GNAS-activating mutations have been reported also in a subgroup of inflammatory liver adenoma and seldom in HCC with signal transducer and activator of transcription 3 (STAT3) activation [
14]. Besides,
GNAS-activating mutations seem to be associated with a fibrotic pattern of tumors [
15]. A congestive and fibrotic remodeling was present in all cases but only three patients showed variant of uncertain significance (VUS) on
GNAS. The role of
GNAS variants in the fibrogenesis of these lesions needs to be further investigated.
HCCs have generally low (<5muts/Mb) TMB and very rarely are hypermutated [
16]. Tumor mutation burden (TMB) is an emerging biomarker that is also predictive of response to immune checkpoint inhibitors and patient with higher TMB were more likely to get benefits from immunotherapy. TMB (total number of somatic nonsynonymous mutations present in the coding region expressed as mutations per megabase in a tumor – muts/Mb) [
17] is currently graduated as low, intermediate, and high. Many genomic alterations have been found associated to different TMB, though there is no single oncogenic driver mutation in the majority of HCCs. In our cohort 1 trabecular HCC presented intermediate (7.8/Mb) TMB and the patient presented a rapid progression of the tumor and died 1 year after the diagnosis.
Interestingly, a FLC with the pathognomonic recurrent fusion transcript DNAJB1-PRKACA, absent in adjacent non-neoplastic liver tissue, was identified in the current series. As far as we know, this is the first FLC reported in FALD. Whether this is a mere coincidence or whether the fusion could be related to the potential role of hypoxic factors and other injuries is debatable.
5. Conclusion
The Fontan procedure promotes a congestive condition in the liver, therefore HCC/ICC in this context may have peculiar histological, immunohistochemical and molecular characteristics and cannot be treated using the same protocols as post-viral infection, alcohol abuse or of NASH-related HCC [
18]. Despite the limited number of cases, our series supports the possibility that these tumors show peculiar and unique characteristics, especially in malignancies and highlight the need for a liver biopsy in case of suspicious imaging.
More in-depth investigations into the pathophysiology of FALD are desirable to define a new targeted therapeutic approach in the context of prospective multicenter studies.
Author Contributions
“Conceptualization, P.F. and R.A; methodology, I.G., C.T., MG.G., R.P., G.B., M.S., G.M., A.P., L.M., A.C.,MC.G; validation and formal analysis I.G and C.T., investigation P.F., MG.G. and R.P., resources R.A. and AOM; writing—original draft preparation, P.F. and I.G.; writing—review and editing, R.A.; supervision AOM. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported also by the Italian Ministry of Health with "Current Research funds”
Acknowledgments
The Authors thank Maria Chiara Benedetti for technical assistance with histologic and immunohistochemical staining and Chiara Puggioni for technical assistance in the acid nucleic extractions and NGS library preparations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Engelhardt EM, Trout AT, Sheridan RM, Veldtman GR, Dillman JR. Focal liver lesions following Fontan palliation of single ventricle physiology: a radiology-pathology case series. Congenit Heart Dis. 2019;14(3):380–88. [CrossRef]
- Asrani SK, Asrani NS, Freese DK, et al. Congenital heart disease and the liver. Hepatology 2012; 56: 1160-9. [CrossRef]
- Kiesewetter CH, Sheron N, Vettukattil JJ, et al. Hepatic changes in the failing Fontan circulation. Heart 2007; 93: 579-84. [CrossRef]
- Asrani SK, Warnes CA, Karmath PS. Hepatocellular Carcinoma after Fontan procedure. New England Journal of Medicine 2013; 368(18): 1756-7. [CrossRef]
- Hilscher MB, Wells ML, Venkatesh SK, Cetta F, Kamath PS. Fontan-associated liver disease. Hepatology. 2022 May;75(5):1300-1321. [CrossRef]
- Haeffele C, Aggarwal A, Lutchman G, Veldtman GR, Wu FM, Lui GK. Fontan Liver Lesions: Not Always HCC. JACC Case Rep. 2019 Aug 21;1(2):175-178. [CrossRef]
- Sahraeian SME, Mohiyuddin M, Sebra R, Tilgner H, Afshar PT, Au KF, Bani Asadi N, Gerstein MB, Wong WH, Snyder MP, Schadt E, Lam HYK. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017 Jul 5;8(1):59. [CrossRef]
- Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kilkku O. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data, bioRxiv, Nov. 2014. [CrossRef]
- Gordon-Walker TT, Bove K, Veldtman G Fontan-associated liver disease: A review. J Cardiol. 2019 Sep;74(3):223-232. [CrossRef]
- Emamaullee J, Zaidi AN, Schiano T, Kahn J, Valentino PL, Hofer RE, Taner T, Wald JW, Olthoff KM, Bucuvalas J, Fischer R. Fontan-Associated Liver Disease: Screening, Management, and Transplant Considerations. Circulation. 2020 Aug 11;142(6):591-604. [CrossRef]
- Xu C, Xu Z, Zhang Y, Evert M, Calvisi DF, Chen X. β-Catenin signaling in hepatocellular carcinoma. J Clin Invest. 2022 Feb 15;132(4):e154515. [CrossRef]
- Seitz T, Hellerbrand C. Role of fibroblast growth factor signalling in hepatic fibrosis. Liver Int. 2021 Jun;41(6):1201-1215. [CrossRef]
- Fearon AE, Slabber CF, Kuklin A, Bachofner M, Tortola L, Pohlmeier L, Pantasis S, Hornemann T, Chen L, Kopf M, Werner S Fibroblast growth factor receptor 3 in hepatocytes protects from toxin-induced liver injury and fibrosis. iScience. 2021 Sep 16;24(10):103143. [CrossRef]
- Ding H, Zhang X, Su Y, Jia C, Dai C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell Mol Biol Lett. 2020 Feb 24;25:8. [CrossRef]
- Nault JC, Fabre M, Couchy G, Pilati C, Jeannot E, Tran Van Nhieu J, Saint-Paul MC, De Muret A, Redon MJ, Buffet C, Salenave S, Balabaud C, Prevot S, Labrune P, Bioulac-Sage P, Scoazec JY, Chanson P, Zucman-Rossi J. GNAS-activating mutations define a rare subgroup of inflammatory liver tumors characterized by STAT3 activation. J Hepatol. 2012 Jan;56(1):184-91. [CrossRef]
- Wong M, Kim JT, Cox B, Larson BK, Kim S, Waters KM, Vail E, Guindi M. Evaluation of tumor mutational burden in small early hepatocellular carcinoma and progressed hepatocellular carcinoma. Hepat Oncol. 2021 Aug 3;8(4): HEP39. [CrossRef]
- Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017 Apr 19;9(1):34. [CrossRef]
- Daniels CJ, Bradley EA, Landzberg MJ, Aboulhosn J, Beekman RH 3rd, Book W, Gurvitz M, John A, John B, Marelli A, Marino BS, Minich LL, Poterucha JJ, Rand EB, Veldtman GR. Fontan-Associated Liver Disease: Proceedings from the American College of Cardiology Stakeholders Meeting, October 1 to 2, 2015, Washington DC. J Am Coll Cardiol. 2017 Dec 26;70(25):3173-3194. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).