1. Introduction
Autophagy is a highly conserved intracellular self-degradation system in eukaryotic organisms, utilized to eliminate and recycle cellular components, thereby breaking down toxic substances or damaged organelles and reclaiming essential nutrients [
1,
2]. Currently, three types of autophagy have been mainly identified in plants: microautophagy, macroautophagy (referred to as "autophagy" hereafter), and mega-autophagy [
2]. Among them, autophagy is the most crucial and prevalent form in plants, involving the encapsulation of cellular materials within double-membrane structures known as autophagosomes. Subsequently, the outer membrane of the autophagosome fuses with a vacuolar membrane, leading to the transfer of cellular contents to the vacuole, where they are degraded by vacuolar proteases and hydrolases [
3].
The formation of autophagosomes is governed by a plethora of autophagy-related genes (ATGs). Initially discovered in yeast [
4], homologous ATG genes have been subsequently identified in animals and plants. Proteins encoded by ATG genes participate in various stages of the autophagy process [
5]. Among the numerous ATG proteins, the ubiquitin-like protein ATG8 plays a pivotal role in autophagy. It is involved in autophagosome formation, membrane elongation and fusion, degradation of cellular contents, and energy release [
6]. ATG8 protein binds to phosphatidylethanolamine in a ubiquitin-like conjugation reaction, serving as a marker for growing autophagosomes and complete autophagic bodies. On the one hand, loss of
ATG8 function prevents autophagosome formation in yeast and other fungi [
7]; however, this phenotype has not been observed in plants, which may be due to
ATG8 gene redundancy. On the other hand, overexpression of
ATG8 promotes autophagosome formation in Arabidopsis and rice [
8]. In addition to
ATG8 lipidation, ATG8 protein is deaffixed by
ATG4 and released from the autophagosome membrane, and the resulting released ATG8 protein is recycled to promote autophagosome formation in plants [
9]. Consequently, ATG8 protein is often utilized as a reliable marker to assess autophagy induction and progression [
10,
11].
Research has indicated the significant role of autophagy in fruit ripening [
9]. Transcriptome analysis of grape skin revealed elevated expression of
ATG18g,
ATG9,
ATG11, and
ATG2 during late stages of grape maturation [
13], implying the involvement of autophagy-related genes in the regulation of grape fruit aging. Subsequently, López-Vidal et al. investigated autophagy (ATG) components,
ATG4 activity, autophagy receptor
NBR1, protease
Lon1, and protease
Lon2 gene expression and protein content changes in two pepper varieties. Their findings confirmed the presence of autophagic structures in pepper fruit, highlighting their crucial role in fruit maturation and quality formation [
12]. Similarly, Sánchez-Sevilla et al. identified autophagy-related structures at the cellular level in different stages of strawberry fruit ripening and demonstrated that blocking autophagy significantly impairs strawberry fruit growth and maturation [
14]. These studies underscore the importance of autophagy in plant fruit development, yet the variations in autophagic flux during fruit ripening and its associated functional roles remain to be elucidated.
Tomato (
Solanum lycopersicum), one of the most esteemed vegetables globally, is renowned for its diverse flavors, forms, colors, and abundant nutritional value [
15]. During tomato fruit ripening, the color transitions from green to red, the flesh softens, and the content compounds increase [
16]. This process involves cell division, cell expansion, and a series of metabolic reactions [
17]. With continuous technological advancements, enhancing tomato fruit quality has become a significant focus. Considering the synthesis and cycling metabolism of nutrients during fruit development, acquiring more information about the role of autophagy in this process is imperative. In this study, through differential expression analysis of autophagy-related gene members of the
ATG8 family during tomato fruit ripening, we demonstrate the existence of autophagic bodies within tomato fruit at the cellular level. Additionally, we observe changes in ATG8 protein content necessary for autophagosome formation. Overexpression of
SlATG8f leads to increased autophagic flux in tomato fruit, accelerating fruit ripening, upregulating ethylene-related genes, and enhancing fruit quality. Collectively, these findings suggest that autophagy is involved in regulating tomato fruit ripening and senescence, thereby enriching our understanding of the autophagic mechanisms in tomato fruit ripening and providing insights into key factors regulating fruit quality, ultimately contributing to the improvement of tomato varieties.
2. Results
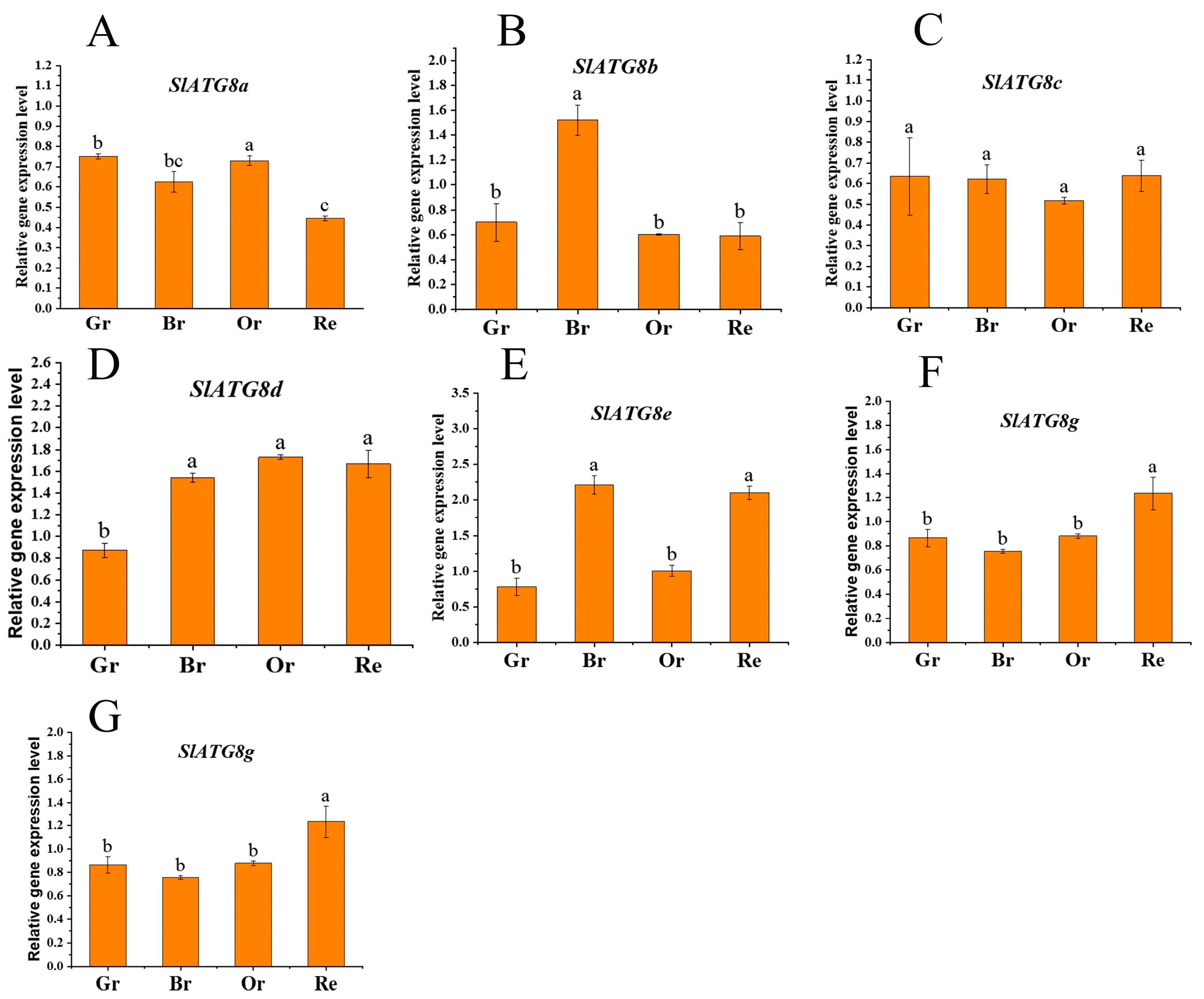
2.1. Expression Analysis of SlATG8 Family Members during Tomato Fruit Ripening
In order to ascertain the involvement of autophagy in tomato fruit ripening, this study conducted an analysis of the expression levels of the core autophagy gene family, the
ATG8 family members. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) was employed to quantify the transcript levels of
SlATG8 family members during different stages of tomato fruit maturation, including the green ripe, breaker, orange ripe, and red ripe stages (
Figure 1). The results revealed distinct expression patterns among the
SlATG8 family members.
SlATG8a and
SlATG8c exhibited relatively low expression levels during tomato fruit ripening.
SlATG8b showed elevated expression at the breaker stage, while its expression was low during the green ripe, orange ripe, and red ripe stages.
SlATG8d,
SlATG8e,
SlATG8f, and
SlATG8g displayed higher expression levels during the breaker, orange ripe, and red ripe stages compared to the green ripe stage. Taken together, the expression levels of
ATG8 family members during tomato fruit ripening were generally higher at the color transition stage or later ripening stages. Notably, the key gene of interest,
SlATG8f, exhibited an increasing expression trend from the green ripe stage to the breaker stage, followed by a gradual decrease from the breaker stage to the red ripe stage.
2.2. Overexpression of SlATG8f Promotes Pericarp Parenchyma Formation in Tomato Fruit
In this study, transgenic Micro-Tom tomato plants were generated with overexpression of the
SlATG8f coding sequence (CDS) under the control of the 35 S promoter. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) results demonstrated a substantial increase in
SlATG8f gene expression in the
SlATG8f-overexpressing (
OE) plants, reaching up to 16-fold higher levels compared to the wild-type (WT) plants (
Figure 2, A).
Phenotypic changes in fruit development were observed among different tomato plants during fruit ripening (
Figure 2, B). While there were no significant differences in fruit phenotypes between
SlATG8f OE and WT fruits, a notable dissimilarity was observed upon slicing the fruits. The pericarp parenchyma of
SlATG8f OE fruits exhibited accelerated development compared to WT fruits, with the pericarp parenchyma being more plump and filled in
SlATG8f OE fruits than in WT fruits. In the early stages of fruit development, pericarp parenchyma in WT fruits was relatively scant, whereas the pericarp tissue of
SlATG8f OE fruits was already completely filled. By the 35th day of fruit development, both
SlATG8f OE and WT fruits had entered the liquefaction phase. However,
SlATG8f OE fruits displayed a higher degree of liquefaction in their pericarp parenchyma. In the late stages of fruit development, both
SlATG8f OE and WT fruits were fully matured, and their pericarp parenchyma had undergone significant liquefaction, with no conspicuous differences between them. Nonetheless, the pericarp parenchyma tissue of
SlATG8f OE fruits remained fuller compared to WT fruits.
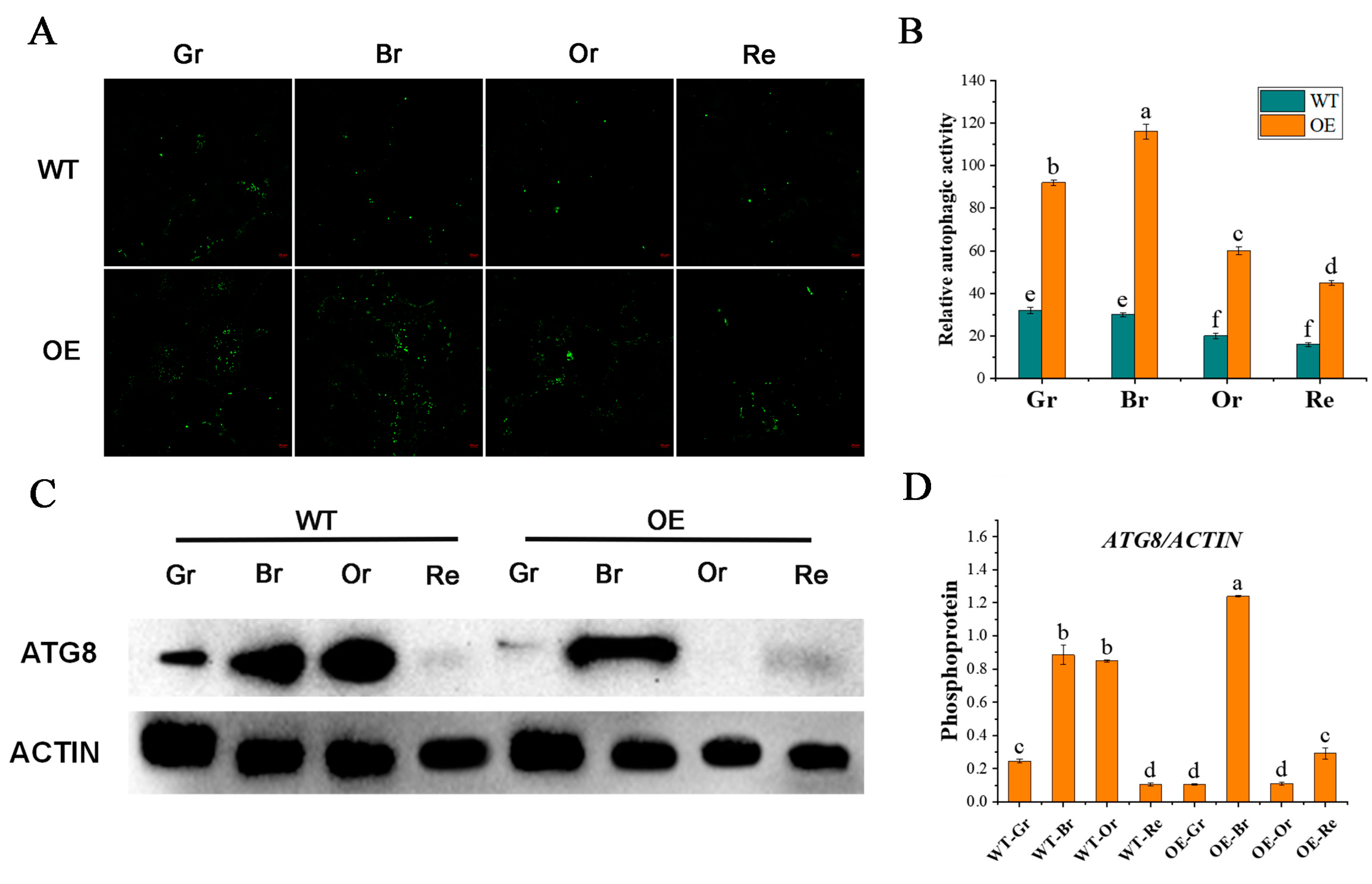
2.3. Overexpression of SlATG8f Enhances Autophagic Activity in Tomato Fruit
To investigate whether
SlATG8f enhances autophagic activity in tomato fruit, the study employed MDC staining and protein immunoblotting (Western blotting) to assess changes in autophagic activity between
SlATG8f-overexpressing (
OE) and wild-type (WT) fruits at different stages of ripening (
Figure 3). MDC staining results revealed (
Figure 3, A, B) that compared to WT fruits,
SlATG8f OE fruits exhibited a higher intensity of green fluorescence corresponding to MDC-stained structures, indicative of autophagosomes. While the number of autophagosomes decreased from the green ripe stage to the red ripe stage in WT fruits, the number of autophagosomes gradually increased from the green ripe stage to the breaker stage in
SlATG8f OE fruits, peaking at the breaker stage before declining.
Protein immunoblotting was employed to analyze ATG8 protein levels during tomato fruit ripening, and the quantification of the corresponding bands was performed (
Figure 3, C, D). The results demonstrated that the ATG8 protein band intensity was strongest in the breaker stage of
SlATG8f OE fruits, whereas it was weakest in the green ripe, orange ripe, and red ripe stages, almost undetectable. In contrast, the ATG8 protein band signal was strong in the breaker and orange ripe stages of WT fruits. The grayscale values of the extracted ATG8 protein bands indicated that in WT fruits, ATG8 protein expression gradually increased from the green ripe stage to the breaker stage, followed by a decrease from the breaker stage to the orange ripe stage, with the lowest expression in the red ripe stage. In
SlATG8f OE plants, ATG8 protein expression was significantly enhanced at the breaker stage, while it remained relatively low during the green ripe, orange ripe, and red ripe stages. These findings suggest that
SlATG8f might be involved in the formation of autophagosomes during the breaker stage of tomato fruit.
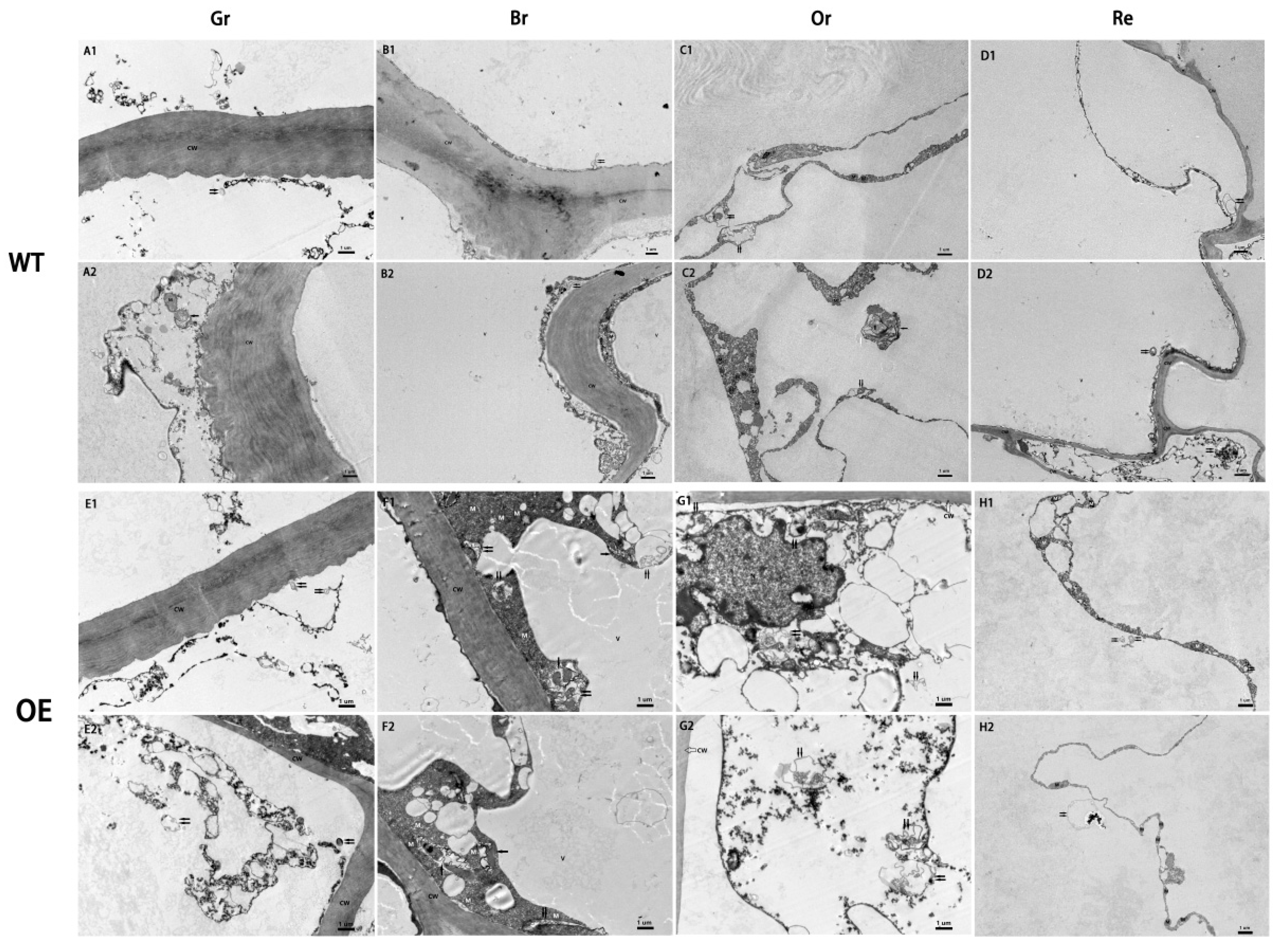
To gain deeper insights into whether overexpression of
ATG8f promotes an increase in autophagic flux during tomato fruit ripening, the study conducted an analysis of autophagic body structural changes using electron microscopy across four stages of fruit maturation (
Figure 4). The results revealed distinct patterns of autophagic activity among the different fruit ripening stages. In wild-type (WT) fruits, there was a relatively high number of autophagic bodies during the orange ripe stage, while autophagic activity was low in the other stages (
Figure 4, A, B, C, D).
SlATG8f OE fruits exhibited the highest autophagic activity during the breaker stage, displaying the highest number of visible autophagic bodies (
Figure 4, F). The following stage with notable autophagic activity was the orange ripe stage (
Figure 4, G), while autophagic activity was lower in the green ripe and red ripe stages (
Figure 4, E, H). Compared to WT fruits,
SlATG8f OE fruits contained a greater abundance of autophagic structures within the cells, particularly evident during the breaker and orange ripe stages of
SlATG8f OE fruits. Notably, not only were numerous autolysosomal structures observed, but also early autophagosome structures were evident (
Figure 4, F, G; indicated by double arrows and single arrows, respectively). These findings underscore that overexpression of
SlATG8f significantly enhances autophagic flux during tomato fruit ripening.
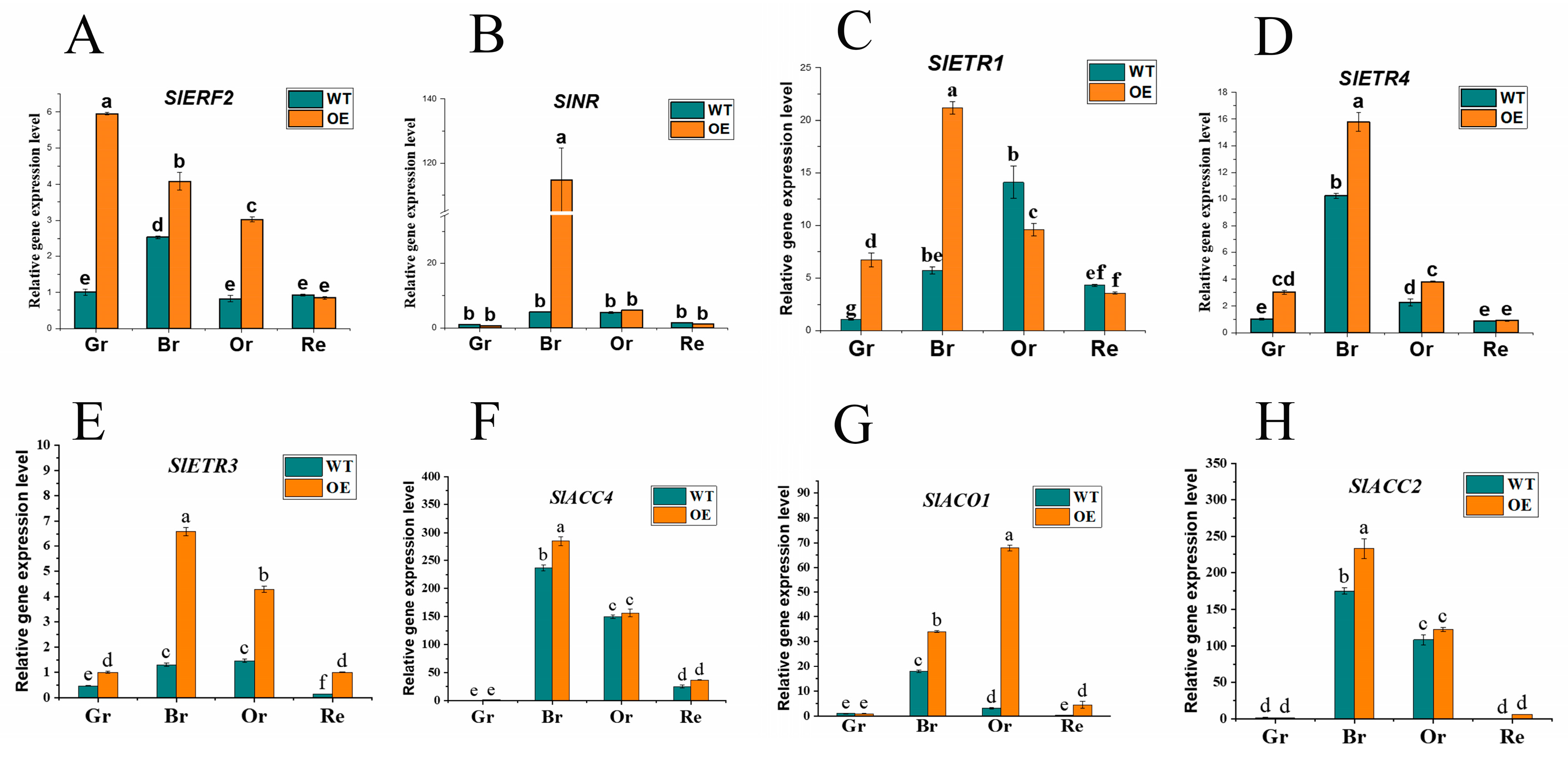
2.4. Overexpression of SlATG8f Facilitates Tomato Fruit Ripening
In earlier investigations, it was observed that the degree of liquefaction of pericarp parenchyma in
SlATG8f OE plants was higher than in WT plants at the 35th day of fruit development. Based on this finding, the study hypothesized that
SlATG8f might promote tomato fruit ripening. To validate this hypothesis, a selection of genes associated with ethylene signaling was screened, including ethylene signal transduction gene
ERF2; ethylene receptor genes
NR,
ETR1,
ETR3,
ETR4; and ethylene synthesis genes
ACC2,
ACC4,
ACO1. Their expression level changes during fruit ripening were analyzed (
Figure 5).
The results indicated that in
SlATG8f OE fruits, the expression of
SlERF2 was upregulated during the green ripe, breaker, and orange ripe stages (
Figure 5, A).
SlNR was significantly upregulated during the breaker stage of
SlATG8f OE fruits (
Figure 5, B). The expression levels of
SlETR1 and
SlETR4 were elevated during the green ripe and breaker stages of
SlATG8f OE fruits, but their expression levels were not notably high during the orange ripe and red ripe stages (
Figure 5, C, D).
SlAETR3 exhibited increased expression during the breaker, orange ripe, and red ripe stages of
SlATG8f OE fruits (
Figure 5, E).
SlACO1 showed significant upregulation during the breaker and orange ripe stages of
SlATG8f OE fruits (
Figure 5, G). However, the expression levels of
SlACC4 and
SlACC2 during the breaker and orange ripe stages of
SlATG8f OE fruits did not differ significantly from those of the wild type (
Figure 5, F, H). These findings suggest that
SlATG8f might influence ethylene production by regulating the expression of
SlACO1, thereby promoting fruit ripening.
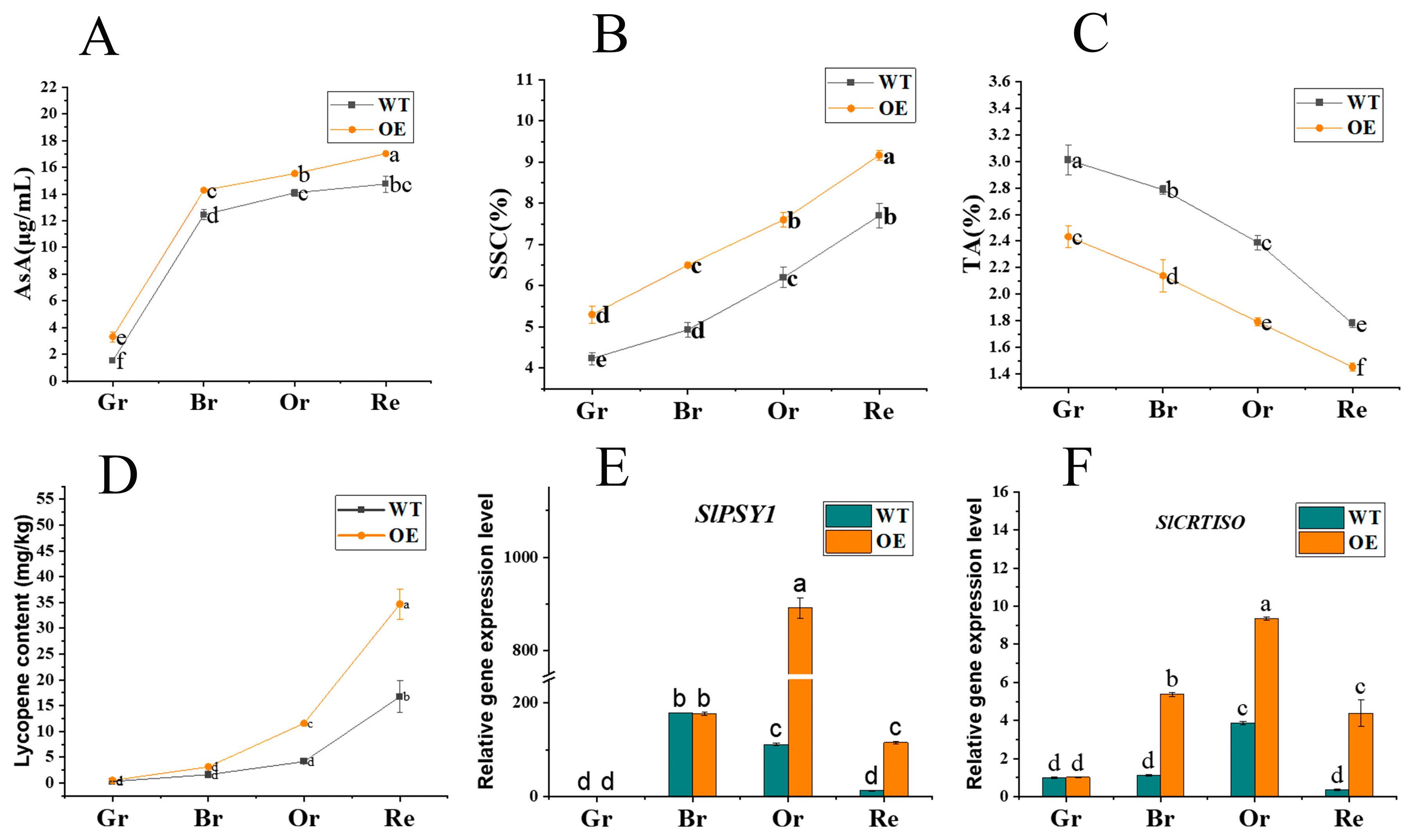
2.5. Overexpression of SlATG8f Improvements of tomato fruit quality
To further comprehend whether the promotion of fruit ripening by
SlATG8f overexpression influences fruit quality and flavor, the study evaluated the content of ascorbic acid (AsA), soluble solids content (SSC), titratable acidity (TA), and tomato lycopene levels. The results revealed notable variations in these components. At all ripening stages of
SlATG8f OE fruits, the content of both ascorbic acid and soluble solids was higher compared to WT fruits (
Figure 6, A, B). Conversely, the titratable acidity content was consistently lower in
SlATG8f OE fruits compared to WT (
Figure 6, C). During the orange ripe and red ripe stages of fruit development,
SlATG8f OE fruits exhibited significantly higher levels of tomato lycopene content compared to WT fruits (
Figure 6, D). The key gene involved in tomato lycopene synthesis,
SlPSY1, was notably upregulated during the orange ripe stage of
SlATG8f OE fruits (
Figure 6, E). Additionally, the expression of the tomato carotenoid isomerase gene,
SlCRTISO, was elevated during the breaker, orange ripe, and red ripe stages of
SlATG8f OE fruits compared to WT fruits (
Figure 6, F). These findings led to the hypothesis that overexpression of
SlATG8f could enhance tomato fruit quality, potentially mediating the biosynthesis pathway of tomato lycopene and thereby influencing the color change of tomato fruits.
3. Discussion
Autophagy is a crucial process in eukaryotic cells for the degradation and selective elimination of cytoplasmic components, playing a significant role in bulk and selective degradation of targeted substances [
18]. In plant cells, autophagy involves the transfer of damaged or redundant macromolecules and organelles to vacuoles for degradation, which holds key roles in maintaining cellular homeostasis, growth and development, as well as responding to environmental stress [
19]. However, the role of autophagy in fruit development and ripening remains largely unexplored, and the analysis of ATG genes or ATG proteins in this context is seldom reported. Bernard et al. suggested that the transcriptional upregulation of ATG genes enhances autophagy [
20]. In our study, the expression of tomato
SlATG8s was detected during fruit ripening, indicating the presence of autophagy activity in these fruits.
SlATG8b,
SlATG8d,
SlATG8e, and
SlATG8f genes exhibited higher expression levels during the breaker stage,
SlATG8d gene showed higher expression during the orange ripe stage, and
SlATG8d,
SlATG8e, and
SlATG8g genes showed higher expression during the red ripe stage. This suggests that these genes might activate distinct regulatory mechanisms at different stages of tomato fruit ripening. In Arabidopsis, nine
ATG8 homologous genes have been identified, encoding proteins whose C-terminal cleavage is
ATG4-dependent, followed by the formation of
ATG8-PE complexes [
21].
ATG8-PE complexes promote the initiation of autophagosomes [
22]. However, in our study, changes in the expression of ATG8 protein levels were detected using Western blotting, while the expression of
ATG8-PE complexes was not assessed, suggesting the need for further investigation.
The presence of multiple autophagosome vesicles in tomato fruit vacuoles indicates the pivotal role of autophagy in fruit ripening. Through transmission electron microscopy, an increased number of vesicles was observed in
SlATG8f OE fruits compared to WT, with the peak vesicle count occurring during the turning stage, a result corroborated by MDC staining. As fruit undergoes various metabolic shifts during the transition from green to red, including the transformation of chloroplasts to chromoplasts [
23], it suggests that different autophagy mechanisms might take place to sustain energy for fruit maturation. Prior research on peppers and strawberries indicated higher autophagy activity in yellow peppers compared to red peppers, as well as two strong autophagy fluxes during the white and ripe stages of strawberry maturation [
12,
14]. Our findings also suggest that the peak autophagy activity during the breaker and orange ripe stages of tomato fruit ripening, enhanced autophagy activity during the breaker stage in the
SlATG8f OE genotype, is advantageous for energy balance during fruit maturation.
Autophagy primarily participates in plant aging and senescence [
24]. The process of fruit ripening is also an aging process, dependent on cellular vitality and gene regulation [
25]. Among these, ethylene-related genes play pivotal roles in tomato fruit ripening [
26]. Our research revealed increased liquefaction of the locule in
SlATG8f OE fruits during the breaker stage, accompanied by upregulated expression of ethylene-related genes. This, in conjunction with the high expression of ATG8 protein during the breaker stage of
SlATG8f OE fruits, further confirms the promoting role of autophagy in tomato fruit ripening. Simultaneously, autophagy induced during fruit ripening enhances fruit coloration, increases levels of ascorbic acid and soluble solids, reduces acidity, and improves fruit texture and quality – possibly arising from the dual effect of autophagy. Previous studies reported that autophagy promotes strawberry fruit ripening while delaying senescence [
27]. Our results suggest the presence of autophagy during tomato fruit ripening, with the peak autophagy activity during the breaker and orange ripe stages;
SlATG8f overexpression effectively enhances autophagy activity during the breaker stage, promoting fruit maturation and enhancing fruit quality. These findings have significant implications for understanding the role of autophagy in tomato fruit growth and development, exploring the regulatory mechanisms of autophagy in fruit, and advancing tomato breeding strategies.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Micro-Tom tomato seeds were disinfected with 0.3 % sodium hypochlorite for 10 minutes, followed by three rinses with sterile water. After soaking in water at 55 °C for 15 minutes and then in 28 °C water for 6 h, the seeds were germinated in darkness at 28 °C. Once seeds were visibly germinated, they were sown in trays containing soilless growth medium. The growth chamber conditions were set at 18 h light/6 h dark with a constant temperature of 25 °C. Fruit samples were collected at different ripening stages: green ripe (day 30), breaker (day 35), orange ripe (day 40), and red ripe (day 45). For each stage, 4-5 fruits were sampled, and the middle portion of the peel and flesh was harvested, frozen in liquid nitrogen, and stored at -80 °C. Gr: green ripe; Br: breaker; Or: orange ripe; Re: red ripe.
4.2. Quantitative Real-Time PCR (qRT-PCR)
Quantitative real-time PCR was employed to study the expression of
SlATG8s and ethylene-related genes in tomato fruits. Frozen tomato fruit samples were used for RNA extraction with a TIANGEN kit (DP432, TIANGEN), and cDNA synthesis was performed using a reverse transcription kit (KR118, TIANGEN). Specific primers were designed using Primer 5 software, with the Actin gene selected as the internal reference (
Supplementary Table S1). QRT-PCR was conducted using a 7500 Real-Time PCR System (Applied Biosystems, USA) and SYBR Green Supermix (RK02001, Bio Marker). The experiments were carried out with three biological replicates and three technical replicates.
4.3. Transmission Electron Microscopy Observation
Tomato fruits were soaked in a 100 nmol/L rapamycin solution for 12 h to induce autophagy. Fruit tissues were rapidly cut into 1-2 mm3 pieces, fixed in electron microscopy fixative solution (G1102, Servicebio) at 4 °C for 2-4 h, and then dehydrated using a series of acetone gradients. Dehydrated tissues were infiltrated and embedded in mixtures of dehydrating agents and epoxy resin in ratios of 3:1, 1:1, and 1:3. Ultrathin sections (50 nm) were obtained using an ultrathin slicer, and these sections were placed on copper grids. The sections were stained with uranyl acetate for 10-15 minutes and then with lead citrate for 1-2 minutes at room temperature. Transmission electron microscopy was performed using a JEM-1400PLUS instrument.
4.4. Dansylcadaverine (MDC) Staining
Tomato fruits were soaked in a 100 nmol/L rapamycin solution for 12 h to induce autophagy. Tomato fruit sections were prepared according to a previous protocol [
28]. The sections were immediately stained with 100 μM MDC (30432, Sigma-Aldrich) under vacuum and dark conditions for 30 minutes and then washed twice with phosphate-buffered saline (PBS, P1020, Solarbio). Autophagosome structures were observed using an LSM-780 confocal microscope (Carl Zeiss) with excitation at 405 nm and emission ranging from 400 to 580 nm.
4.5. Protein Blot (Western Blotting)
Tomato fruits were soaked in a 100 nmol/L rapamycin solution for 12 h to induce autophagy. Fruit tissues were frozen in liquid nitrogen and used for total protein extraction using a kit (PTE011, Coolaber). SDS-PAGE gel electrophoresis was performed (P1200, Solarbio) to separate the proteins. The membrane was blocked with 5 % skim milk for 2 h and then washed with TBST. The PVDF membrane was cut into strips and incubated with primary antibodies (1:1000) overnight at 4 °C. After washing with TBST three times, the membrane was incubated with secondary antibodies (1:1000) against rabbit ATG8a polyclonal antibody (Abcam, ab77003) for 1 hour at room temperature. Chemiluminescence was detected using a chemiluminescence detection kit (Thermo Fisher Scientific, 34080).
4.6. Detection of Lycopene Content, Ascorbic Acid, Soluble Solids (SSC), and Titratable Acidity (TA)
The lycopene content of tomato fruits was determined according to Sun et al.'s method. Five grams of tomato flesh and peel tissue were frozen and ground into powder. The powder was mixed with 50 mL of hexane-acetone-ethanol (2:1:1, v/v) and shaken. After adding 15 mL of water and allowing for phase separation, the organic phase (hexane) absorbance was measured at 503 nm to calculate lycopene concentration. The molar extinction coefficient was 17.2 L mol
-1·m
-1, and the units were expressed as mg·kg
−1 based on fresh weight [
29]. Ascorbic acid was measured using a kit (A009-1-1, Nanjing Jiancheng Bioengineering Institute). SSC and TA content measurements were performed using a PAL-BX/ACD1 sugar acidity meter (ATAGO, Tokyo, Japan) for SSC (%) and TA (%) in fruit. The experiments included three biological replicates and three technical replicates.
5. Conclusions
In this study, quantitative analysis of the expression of the ATG8 gene family members during tomato fruit ripening revealed that the expression levels of most ATG8 family members were higher during the fruit color transition period or late stages of fruit ripening. Among them, the expression of SlATG8f gradually increased from the green ripe stage to the breaker stage and then gradually decreased from the breaker stage to the red ripe stage. In the SlATG8f overexpression line, the locular gel tissue of tomato fruits appeared more plump compared to the wild type (WT), and the liquefaction rate of the locular gel tissue was faster, indicating that the SlATG8f gene positively regulates tomato fruit development. MDC staining and electron microscopy observations revealed that the number of autophagosomes in SlATG8f OE fruits was significantly higher than in WT, especially during the breaker stage, with a substantial increase in the number of autophagosomes and autolysosomes in the fruit flesh cells, indicating enhanced autophagic activity. Western blot results demonstrated a significant increase in ATG8 protein expression during the breaker stage in SlATG8f OE fruits, and the expression level of SlATG8f correlated positively with autophagic activity. Overexpression of SlATG8f induced the formation of autophagosomes in tomato fruit cells, enhancing autophagic activity. The increased autophagic flux due to SlATG8f overexpression accelerated fruit ripening and upregulated the expression of ethylene-related genes regulating fruit ripening. Taken together, our findings suggest that autophagy is involved in regulating tomato fruit ripening and senescence. These discoveries contribute to a deeper understanding of the role of autophagy in the process of tomato fruit ripening, elucidate key factors regulating fruit quality, and provide a theoretical basis for improving tomato varieties.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
C.W., T.M.-L., writing—original draft, formal analysis, data curation, software, methodology, and validation. Z.H., software and validation. W.X., writing—review and editing, funding acquisition, project administration, resources, and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31760594 and 32260754), the Special Project for Cultivating Innovative Talents of Thousand Levels in Guizhou Province ((2018)02), and the Guizhou University of Science and Technology Research Start-up Fund for New Faculty ((2016)48).
Data Availability Statement
The authors will supply the relevant data in response to reasonable requests.
Acknowledgments
The authors are grateful to the National and Local Vegetable Engineering Center (Guizhou) and the Laboratory of the Department of Horticulture (Agricultural College of Guizhou University) for their support in this project.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Michaeli, S.; Galili, G.; Genschik, P., Fernie, A.R., Avin-Wittenberg, T. Autophagy in Plants--What's New on the Menu? Trends in plant science. 2016, 21(2), 134–144.
- Marshall, R.S.; Vierstra, R.D. Autophagy: The Master of Bulk and Selective Recycling. Annual review of plant biology. 2018, 69, 173–208. [CrossRef]
- Wen, X.; Klionsky, D.J. An overview of macroautophagy in yeast. Journal of molecular biology. 2016, 428, 1681–1699. [CrossRef]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS letters. 1993, 333(1-2), 169–174. [CrossRef]
- Meijer, W.H.; van der Klei, I.J.; Veenhuis, M.; Kiel, J. A. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007, 3(2), 106–116. [CrossRef]
- Hafrén, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proceedings of the National Academy of Sciences of the United States of America. 2017, 114(10), E2026–E2035. [CrossRef]
- Ren, W.; Liu, N.; Sang, C.; Shi, D.; Zhou, M.; Chen, C.; Qin, Q.; Chen, W. The Autophagy Gene BcATG8 Regulates the Vegetative Differentiation and Pathogenicity of Botrytis cinerea. Appl Environ Microbiol. 2018, 84(11), e02455-17. [CrossRef]
- Zhen, X.; Xu, F.; Zhang, W.; Li, N.; Li, X. Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS One. 2019, 14(9), e0223011. [CrossRef]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004, 16(11), 2967-83. [CrossRef]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: lessons from yeast help define the process in plants. Current opinion in plant biology. 2005, 8(2), 165–173. [CrossRef]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a Conserved Mechanism for Protein Degradation, Responds to Heat, and Other Abiotic Stresses in Capsicum annuum L. Frontiers in plant science. 2016, 7, 131. [CrossRef]
- López-Vidal, O.; Olmedilla, A.; Sandalio, L.M.; Sevilla, F.; Jiménez, A. Is Autophagy Involved in Pepper Fruit Ripening? Cells. 2020, 9(1), 106.
- Ghan, R.; Petereit, J.; Tillett, R.L.; Schlauch, K.A.; Toubiana, D.; Fait, A.; Cramer, G.R. The common transcriptional subnetworks of the grape berry skin in the late stages of ripening. BMC plant biology. 2017, 17(1), 94. [CrossRef]
- Sánchez-Sevilla, J.F.; Botella, M.A.; Valpuesta, V.; Sanchez-Vera, V. Autophagy Is Required for Strawberry Fruit Ripening. Frontiers in plant science. 2021, 12, 688481. [CrossRef]
- Szabo, K.; Cătoi A.F.; Vodnar D.C. Bioactive Compounds Extracted from Tomato Processing by-Products as a Source of Valuable Nutrients. Plant Foods Hum Nutr. 2018, 73: 268–277. [CrossRef]
- Ronen, G.; Cohen, M.; Zamir, D.; Hirschberg, J. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. The Plant journal : for cell and molecular biology. 1999, 17(4), 341–351. [CrossRef]
- Cheniclet, C.; Rong, W.Y.; Causse, M.; Frangne, N.; Bolling, L.; Carde, J.P.; Renaudin, J.P. Cell expansion and endoreduplication show a large genetic variability in pericarp and contribute strongly to tomato fruit growth. Plant physiology. 2005, 139(4), 1984–1994. [CrossRef]
- Signorelli, S.; Tarkowski, Ł.P.; Van den Ende, W.; Bassham, D. C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends in plant science. 2019, 24(5), 413–430. [CrossRef]
- Qi, H.; Xia, F.N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. Journal of integrative plant biology. 2021, 63(1), 161–179. [CrossRef]
- Bernard, A.; Jin, M.; González-Rodríguez, P.; Füllgrabe, J.; Delorme-Axford, E.; Backues, S.K.; Joseph, B.; Klionsky, D.J. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Current biology. 2015, 25(5), 546–555.
- Woo, J.; Park, E.; Dinesh-Kumar, S.P. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proceedings of the National Academy of Sciences of the United States of America. 2014, 111(2), 863–868. [CrossRef]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. The Plant cell. 2004, 16(11), 2967–2983. [CrossRef]
- McAtee, P.; Karim, S.; Schaffer, R.; David, K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Frontiers in plant science. 2013, 4, 79. [CrossRef]
- López-Vidal, O.; Olmedilla, A.; Sandalio, L.M.; Sevilla, F.; Jiménez, A. Is Autophagy Involved in Pepper Fruit Ripening? Cells. 2020, 9(1), 106.
- Sánchez-Sevilla, J.F.; Botella, M.A.; Valpuesta, V.; Sanchez-Vera, V. Autophagy Is Required for Strawberry Fruit Ripening. Frontiers in plant science. 2021, 12, 688481. [CrossRef]
- Wang, H.; Schippers, J.H.M. The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants. Genes. 2019, 10(4), 267. [CrossRef]
- Thomas H. Senescence, ageing and death of the whole plant. The New phytologist. 2013, 197(3), 696–711. [CrossRef]
- Shinozaki, Y.; Hao, S.; Kojima, M.; Sakakibara, H.; Ozeki-Iida, Y.; Zheng, Y.; Fei, Z.; Zhong, S.; Giovannoni, J.J.; Rose, J.K.; Okabe, Y.; Heta, Y.; Ezura, H.; Ariizumi, T. Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. The Plant journal : for cell and molecular biology. 2015, 83(2), 237–251. [CrossRef]
- Sun, Q.; Zhang, N.; Wang, J.; Zhang, H.; Li, D.; Shi, J.; Li, R.; Weeda, S.; Zhao, B.; Ren, S.; Guo, Y. D. Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. Journal of experimental botany. 2015, 66(3), 657–668. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).