Submitted:
07 September 2023
Posted:
11 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
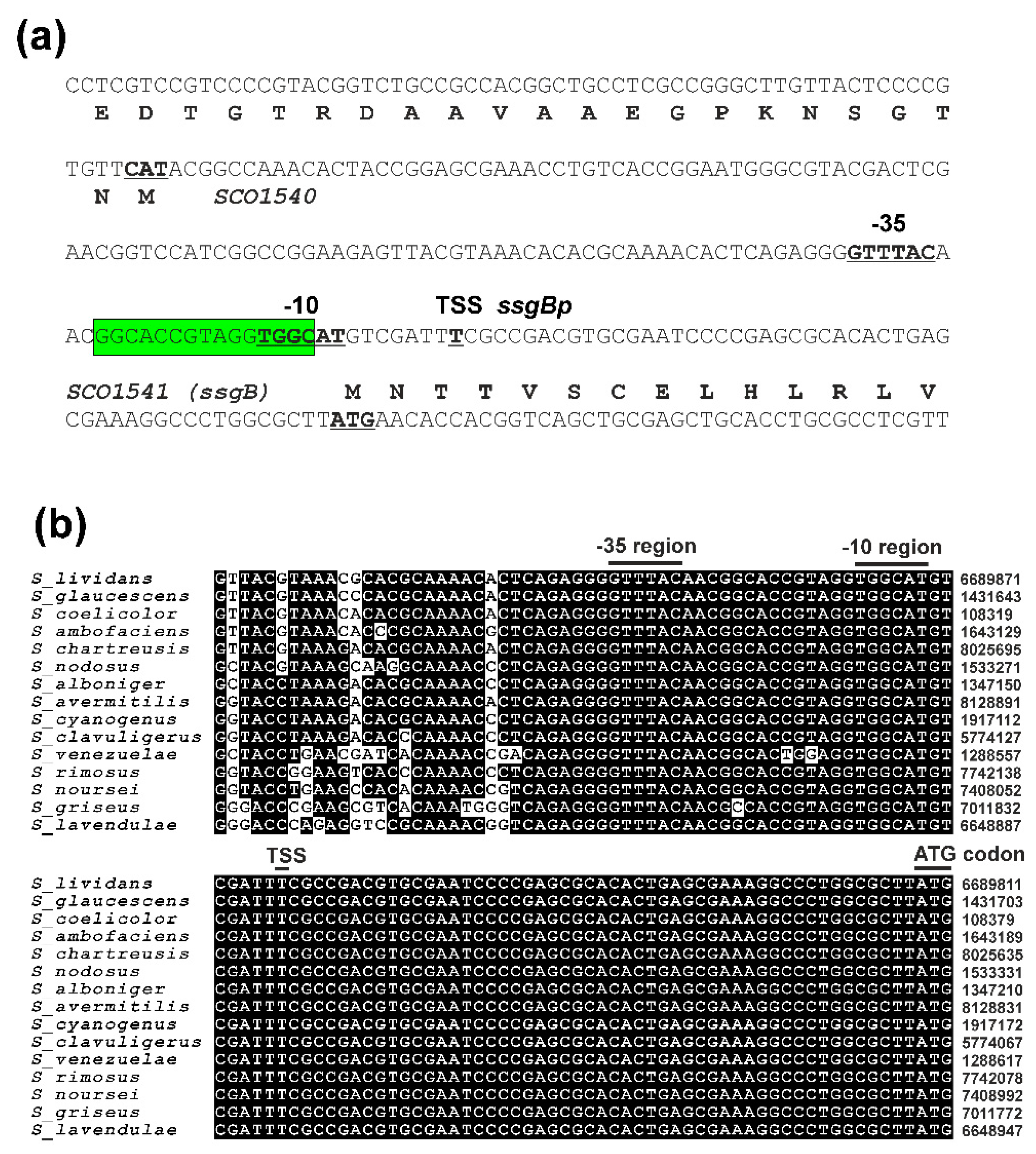
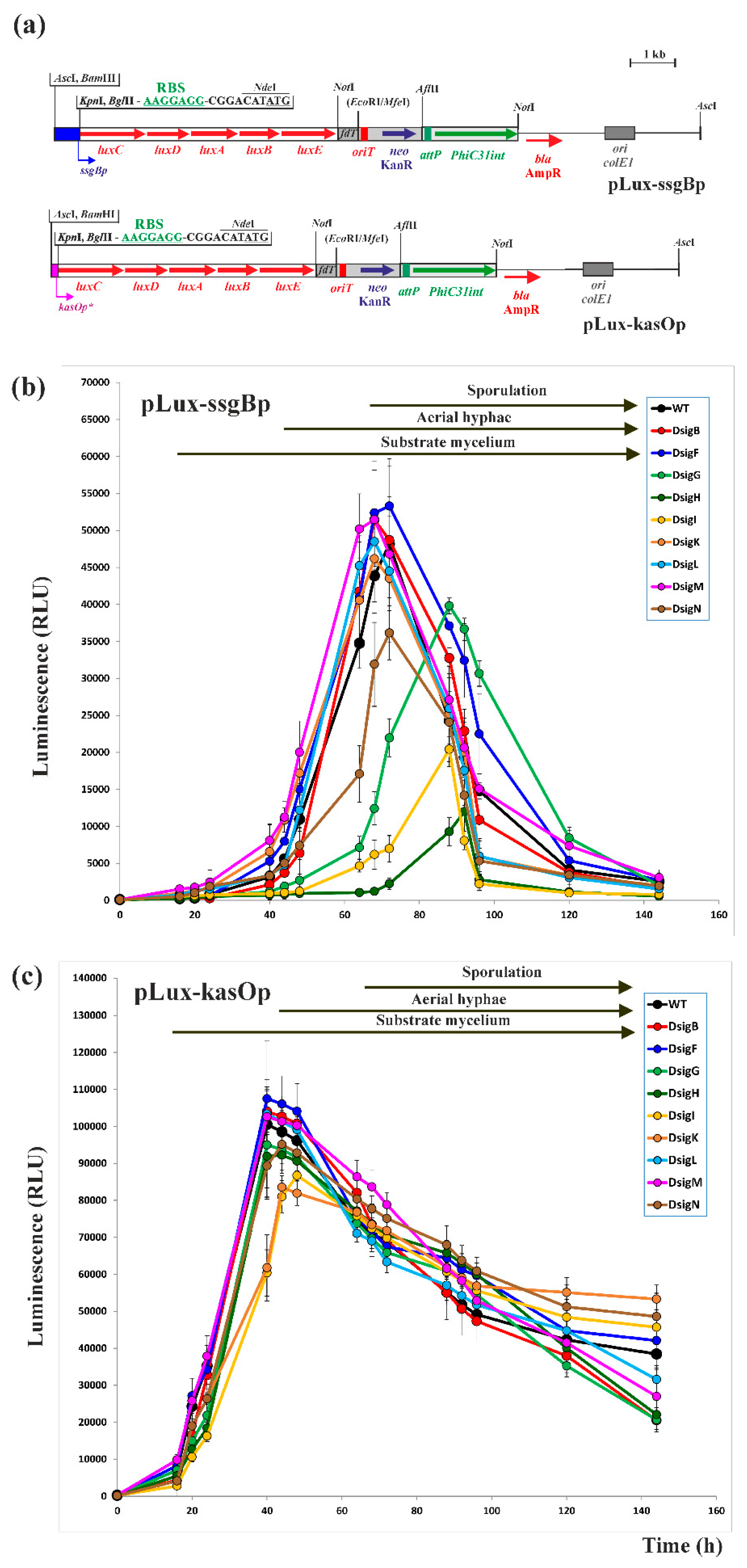
2.1. In vivo Cross-Recognition of the highly conserved ssgBp promoter by Nine SigB Homologues
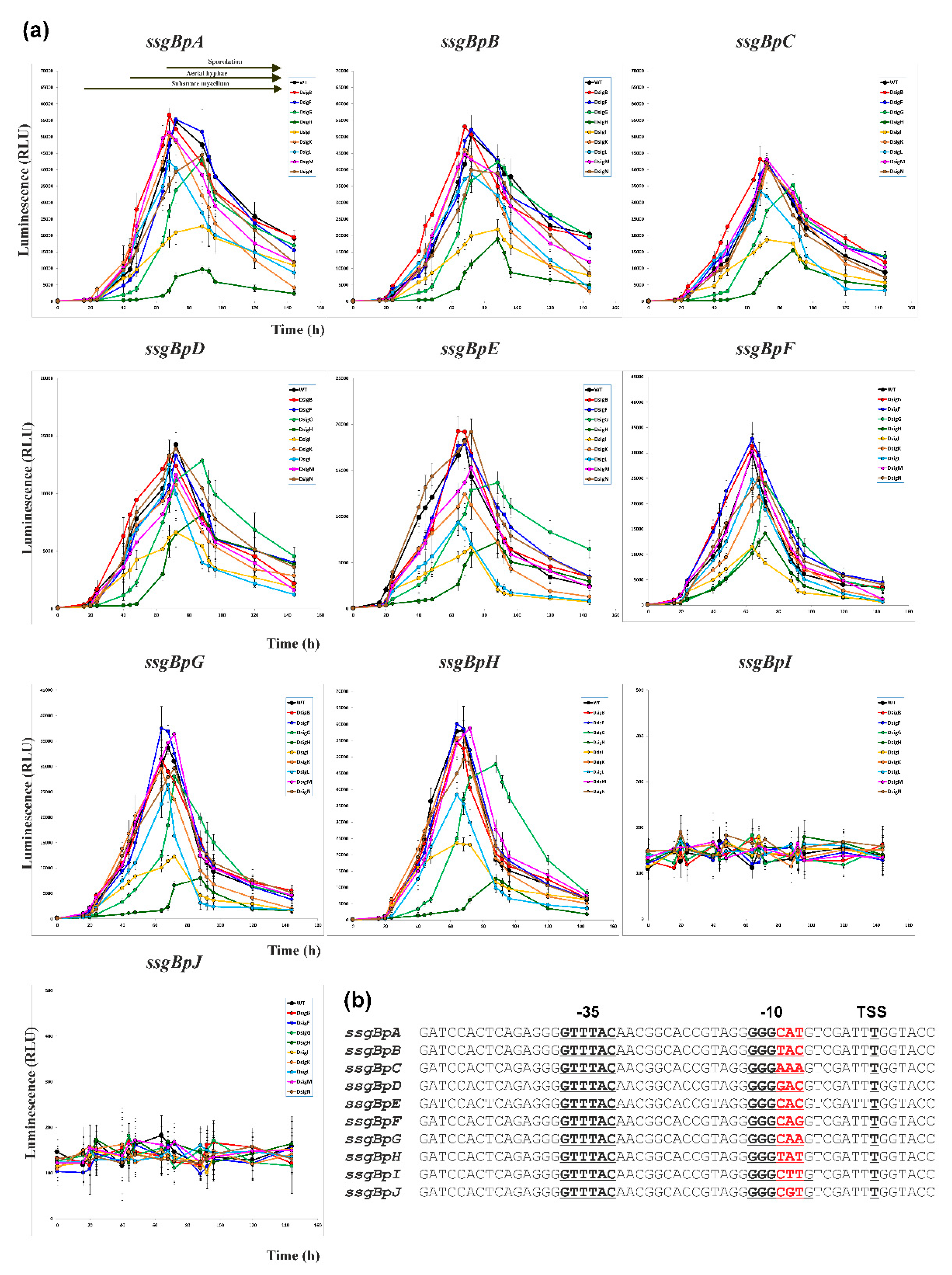
2.2. Mutagenesis of the ssgBp promoter to identify specific nucleotides for Nine SigB Homologues
2.3. In vivo Cross-Recognition of mutant ssgBp promoters by Nine SigB Homologues
3. Materials and Methods
3.1. Bacterial strains, culture conditions, and plasmids
3.2. Recombinant DNA techniques
3.3. Detection of E. coli clones containing ssgBp promoters and cross-recognition by nine SigB homologous sigma factors
3.4. Construction of deletion mutants in S. coelicolor A3(2)
3.5. Construction of a New luxCDABE-based luciferase reporter plasmid and Bioluminescence Measurement
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aa | amino acid(s) |
| Amp | ampicillin |
| Apr | apramycin |
| AprR | Apr resistance |
| B | Bacillus |
| CFU | colony forming units |
| Clm | chloramphenicol |
| E | Escherichia |
| h | hour(s) |
| IPTG | isopropyl-β-D-thiogalactopyranoside |
| Kan | kanamycin |
| KanR | kanamycin resistance |
| kb | kilobase |
| LB | Luria-Bertani (medium) |
| NA | nalidixic acid |
| nt | nucleotide(s) |
| PCR | polymerase chain reaction |
| RLU | relative luminescence unit(s) |
| S | Streptomyces |
| TSS | transcription start site |
| WT | wild type |
References
- Chater, K.F. Differentiation in Streptomyces: the properties and programming of diverse cell types. In Streptomyces: molecular biology and biotechnology; Dyson, P.J., Ed.; Caister Academic Press: Norfolk, UK, 2011; pp. 43–86. [Google Scholar]
- McCormick, J.R.; Flardh, K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 2012, 36, 206–231. [Google Scholar] [CrossRef] [PubMed]
- Schlimpert, S.; Wasserstrom, S.; Chandra, G.; Bibb, M.J.; Findlay, K.C.; Flärd, K.; Buttner, M.J. Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E6176–E6183. [Google Scholar] [CrossRef] [PubMed]
- Willemse, J.; Borst, J.W.; de Waal, E.; Bisseling, T.; van Wezel, G.P. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 2011, 25, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Bush, M.J.; Tschowri, N.; Schlimpert, S.; Flard, K.; Buttner, M.J. c-di-GMP signaling and the regulation of developmental transitions in streptomycetes. Nat. Rev. Microbiol. 2015, 13, 749–760. [Google Scholar] [CrossRef]
- Gallagher, K.A.; Schumacher, M.A.; Bush, M.J.; Bibb, M.J.; Chandra, G.; Holmes, N.A.; Zeng, W.; Henderson, M.; Zhang, H.; Findlay, K.C.; Brennan, R.G.; Buttner, M.J. c-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Mol Cell 2020, 77, 586–599. [Google Scholar] [CrossRef]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. ; Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Vohradsky, J.; Li, X.M.; Dale, G.; Folcher, M.; Nguyen, L. , Viollier, P.H.; Thompson, C.J. Developmental control of stress stimulons in Streptomyces coelicolor revealed by statistical analysis of global gene expression patters. J. Bacteriol. 2000, 182, 4979–4986. [Google Scholar] [CrossRef]
- Ayala, F.R.; Bartolini, M.; Grau, R. The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: an old friend with new functions. Front. Microbiol. 2020, 11, 1761. [Google Scholar]
- Kormanec, J.; Sevcikova, B.; Novakova, R.; Homerova, D.; Rezuchova, B.; Mingyar, E. The complex regulatory network in the regulation of stress-response sigma factors in Streptomyces coelicolor A3(2). In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; de Bruijn, F.J., Ed.; John Wiley & Sons, Inc: NJ, 2016; pp. 321–336. [Google Scholar]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; Bateman, A.; Brown, S.; Chandra, G.; Chen, C.; Collins, M.; Cronin, A.; Fraser, A.; Goble, A.; Hidalgo, J.; Hornsby, T.; Howarth, S.; Huang, C.H.; Kieser, T.; Larke, L.; Murphy, L.; Oliver, K.; O’Neil, S.; Rabbinowitsch, E.; Rajandream, M.A.; Rutherford, K.; Rutter, S.; Seeger, K.; Saunders, D.; Sharp, S.; Squares, R.; Squares, S.; Taylor, K.; Warren, T.; Wietzorrek, A.; Woodward, J.; Barrell, B.G.; Parkhill, J.; Hopwood, D.A. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Mittenhuber, G. A phylogenetic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. 2002, 4, 427–452. [Google Scholar]
- Potuckova, L.; Kelemen, G.H.; Findlay, K.C.; Lonetto, M.A.; Buttner, M.J.; Kormanec, J. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Mol. Microbiol. 1995, 17, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.A.; Thibessard, A.; Hunter, J.I.; Kelemen, G.H. A novel compartment, the ‘subapical stem’of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 2007, 64, 719–737. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.-M.; Zhou, Z.; Hou, X.-P.; Guan, W.-J.; Li, Y.-Q. Reciprocal regulation between sigK and differentiation programs in Streptomyces coelicolor. J. Bacteriol. 2009, 91, 6473–6481. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Karoonuthaisiri, N.; Kim, H.S.; Park, J.H.; Cha, C.J.; Kao, C.M.; Roe, J.H. A master regulator σB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 2005, 57, 1252–1264. [Google Scholar] [CrossRef]
- Homerova, D.; Sevcikova, B.; Rezuchova, B.; Kormanec, J. Regulation of an alternative sigma factor σI by a partner switching mechanism with an anti-sigma factor PrsI and an anti-anti-sigma factor ArsI in Streptomyces coelicolor A3(2). Gene 2012, 492, 71–80. [Google Scholar] [CrossRef]
- Kormanec, J.; Homerova, D.; Barak, I.; Sevcikova, B. A new gene, sigG, encoding a putative alternative sigma factor of Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 1999, 172, 153–158. [Google Scholar] [CrossRef]
- Strakova, E.; Zikova, A.; Vohradsky, J. Interference of sigma factor controlled networks by using numerical modeling applied to microarray time series data of the germinating prokaryote. Nucleic Acids Res. 2013, 42, 748–763. [Google Scholar] [CrossRef]
- Kelemen, G.H.; Viollier, P.; Tenor, J.L.; Marri, L.; Buttner, M.J.; Thompson, C.J. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2). Mol. Microbiol. 2001, 40, 804–814. [Google Scholar] [CrossRef]
- Kormanec, J.; Sevcikova, B.; Halgasova, N.; Knirschova, R.; Rezuchova, B. Identification and transcriptional characterization of the gene encoding the stress- response sigma factor σH in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2000, 189, 31–38. [Google Scholar]
- Sevcikova, B.; Benada, O.; Kofronova, O.; Kormanec, J. Stress-response sigma factor σH is essential for morphological differentiation of Streptomyces coelicolor A3(2). Arch. Microbiol. 2001, 177, 98–106. [Google Scholar] [CrossRef]
- Viollier, P.H.; Kelemen, G.H.; Dale, G.E.; Nguyen, K.T.; Buttner, M.J.; Thompson, C.J. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Mol. Microbiol. 2003. 47, 699–714. [CrossRef]
- Lee, E.J.; Cho, Y.H.; Kim, H.S.; Ahn, B.E.; Roe, J.H. Regulation of σB by an anti- and anti-anti-sigma factor in Streptomyces coelicolor in response to osmotic stress. J. Bacteriol. J. Bacteriol. 2004, 186, 8490–8498. [Google Scholar] [CrossRef] [PubMed]
- Gaskell, A.A.; Crack, J.C.; Kelemen, G.H.; Hutchings, M.I.; Le Brun, N.E. RmsA is an anti-sigma factor that modulates its activity through [2Fe-2S] cluster cofactor. J. Biol. Chem. 2007, 282, 31812–31820. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, B.; Kormanec, J. Activity of the Streptomyces coelicolor stress-response sigma factor σH is regulated an anti-sigma factor. FEMS Microbiol. Lett. 2002, 209, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Viollier, P.H.; Weihofen, A.; Folcher, M.; Thompson, C.J. Post-translational regulation of the Streptomyces coelicolor stress responsive sigma factor, SigH, involves translational control, proteolytic processing, and an anti-sigma factor homolog. J. Mol. Biol. 2003, 325, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, B.; Rezuchova, B.; Homerova, D.; Kormanec, J. The anti-anti sigma factor BldG is involved in activation of stress-response sigma factor σH in Streptomyces coelicolor A3(2). J. Bacteriol. 2010, 192, 5674–5681. [Google Scholar] [CrossRef]
- Sevcikova, B.; Rezuchova, B.; Mingyar, E.; Homerova, D.; Novakova, R.; Feckova, L.; Kormanec, J. Pleiotropic anti-anti-sigma factor BldG is phosphorylated by several anti-sigma factor kinases in the process of activating multiple sigma factors in Streptomyces coelicolor A3(2). Gene 2020, 755, 144883. [Google Scholar] [CrossRef]
- Sevcikova, B.; Rezuchova, D.; Mazurakova, V.; Homerova, D.; Novakova, R.; Feckova, L.; Kormanec, J. Cross-recognition of promoters by the nine SigB homologues present in Streptomyces coelicolor A3(2). Int. J. Mol. Sci. 2021, 22, 7849. [Google Scholar] [CrossRef]
- Kormanec, J.; Sevcikova, B. The stress-response sigma factor σH controls the expression of ssgB, a homologue of the sporulation-specific cell division gene ssgA in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 2002, 267, 536–543. [Google Scholar] [CrossRef]
- Keijser, B.J.F.; Noens, E.E.E.; Kraal, B.; Koerten, H.K.; van Wezel, G.P. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 2003, 225, 59–67. [Google Scholar] [CrossRef]
- Sevcikova, B.; Kormanec, J. The ssgB gene, encoding a member of the regulon of stress-response sigma factor σH, is essential for aerial mycelium formation in Streptomyces coelicolor A3(2). Arch. Microbiol. 2003, 180, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Willemse, J.; Voskamp, P.; Li, X.; Prota, A.E.; Lamers, M.; Pannu, N.; Abrahams, J.P.; van Wezel, G.P. Ectopic positioning of the cell division plane is associated with single amino acid substitutions in the FtsZ-recruiting SsgB in Streptomyces. Open Biol. 2021, 11, 200409. [Google Scholar] [CrossRef] [PubMed]
- Buttner, M.J.; Chater, K.F.; Bibb, M.J. C Cloning, Disruption, and Transcriptional Analysis of Three RNA Polymerase Sigma Factor Genes of Streptomyces coelicolor A3(2). J. Bacteriol. 1990, 172, 3367–3378. [Google Scholar] [CrossRef] [PubMed]
- den Hengst, C.D.; Tran, N.T.; Bibb, M.J.; Chandra, G.; Leskiw, B.K.; Buttner, M.J. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 2010, 78, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Craney, A.; Hohenauer, T.; Xu, Y.; Navani, N.K.; Li, Y.; Nodwell, J. A synthetic luxCDABE gene cluster optimized for expression in high-GC bacteria. Nucleic Acids Res. 2007, 35, e46. [Google Scholar] [CrossRef]
- Kelemen, G.H.; Brown, G.L.; Kormanec, J.; Potuckova, L.; Chater, K.F.; Buttner, M.J. The position of the sigma-factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2). Mol. Microbiol. 1996, 21, 593–603. [Google Scholar] [CrossRef]
- Sun, J.; Kelemen, G.H.; Fernandez-Abalos, J.M.; Bibb, M.J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology-SGM 1999, 145, 2221–2227. [Google Scholar] [CrossRef]
- Bush, M.J.; Chandra, G.; Al-Bassam, M.M.; Findlay, K.C.; Buttner, M.J. BldC delays entry into development to produce a sustained period of vegetative growth in Streptomyces venezuelae. mBio 2019, 10, e02812–18. [Google Scholar] [CrossRef]
- Kim, S.H.; Traag, B.A.; Hasan, A.H.; McDowall, K.J.; Kim, B-G. ; van Wezel, G.P. Transcriptional analysis of the cell division-related ssg genes in Streptomyces coelicolor reveals direct control of ssgR by AtrA. Antonie van Leeuwenhoek 2015, 108, 201–213. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Wang, J.; Xiang, S.; Feng, X.; Yang, K. An engineered strong promoter for streptomycetes. Appl. Environ. Microbiol. 2013, 79, 4484–4492. [Google Scholar] [CrossRef]
- Kormanec, J.; Sevcikova, B. Identification and transcriptional analysis of a cold shock inducible gene, cspA, in Streptomyces coelicolor A3(2). Mol. Gen. Genet. 2000, 264, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingstone, R.E.; Moore, D.O.; Seidman, J.S.; Smith, J.A.; Struh, K. Current Protocols in Molecular Biology; Wiley: New York, 1995. [Google Scholar]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces genetics; The John Innes Foundation: Norwich UK,, 2000. [Google Scholar]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Redenbach, M.; Kieser, H.M.; Denapaite, D.; Eichner, A.; Cullum, J.; Kinashi, H.; Hopwood, D.A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3 (2) chromosome. Mol. Microbiol. 1996, 21, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, G.H.; Brian, P.; Flard, K.; Chamberlin, L.; Chater, K.F.; Buttner, M.J. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 1998, 180, 2515–2521. [Google Scholar] [CrossRef]
- Novakova, R.; Homerova, D.; Csolleiova, D.; Rezuchova, D.; Sevcikova, B.; Javorova, R.; Feckova, L.; Kormanec, J. A stable vector for efficient production of heterologous proteins and secondary metabolites in streptomycetes. Appl. Microbiol. Biotechnol. 2022, 106, 7285–7299. [Google Scholar] [CrossRef]
- Novakova, R.; Sevcikova, B.; Kormanec, J. A method for the identification of promoters recognized by RNA polymerase containing a particular sigma factor: Cloning of a developmentally regulated promoter and corresponding gene directed by the Streptomyces aureofaciens sigma factor RpoZ. Gene 1998, 208, 43–50. [Google Scholar] [CrossRef]
- Sevcikova, B.; Mazurakova, V.; Kormanec, J. Characterization of the alternative sigma factor σG in Streptomyces coelicolor A3(2). Folia Microbiol. 2005, 50, 47–58. [Google Scholar] [CrossRef]
- Kormanec, J.; Sevcikova, B. Stress-response sigma factor σH directs expression of the gltB gene encoding glutamate synthase in Streptomyces coelicolor A3(2). Biochim. Biophys. Acta 2002, 1577, 149–154. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




