Submitted:
08 September 2023
Posted:
11 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell culture, treatments, and transfection
2.2. RT-PCR and quantitative PCR
2.3. Sequencing of the ANKRD1 open reading frame
2.4. Western blot
2.5. Immunoprecipitation
2.6. Immunocytochemistry and microscopy
2.7. Statistical analysis
3. Results
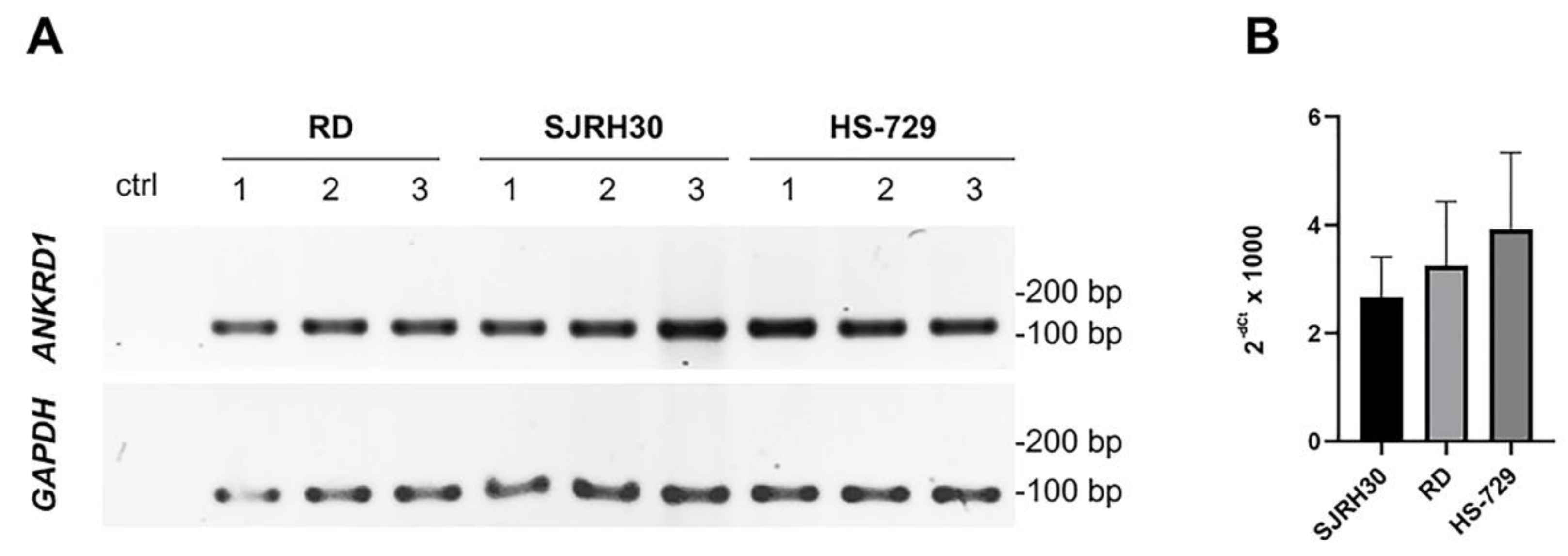
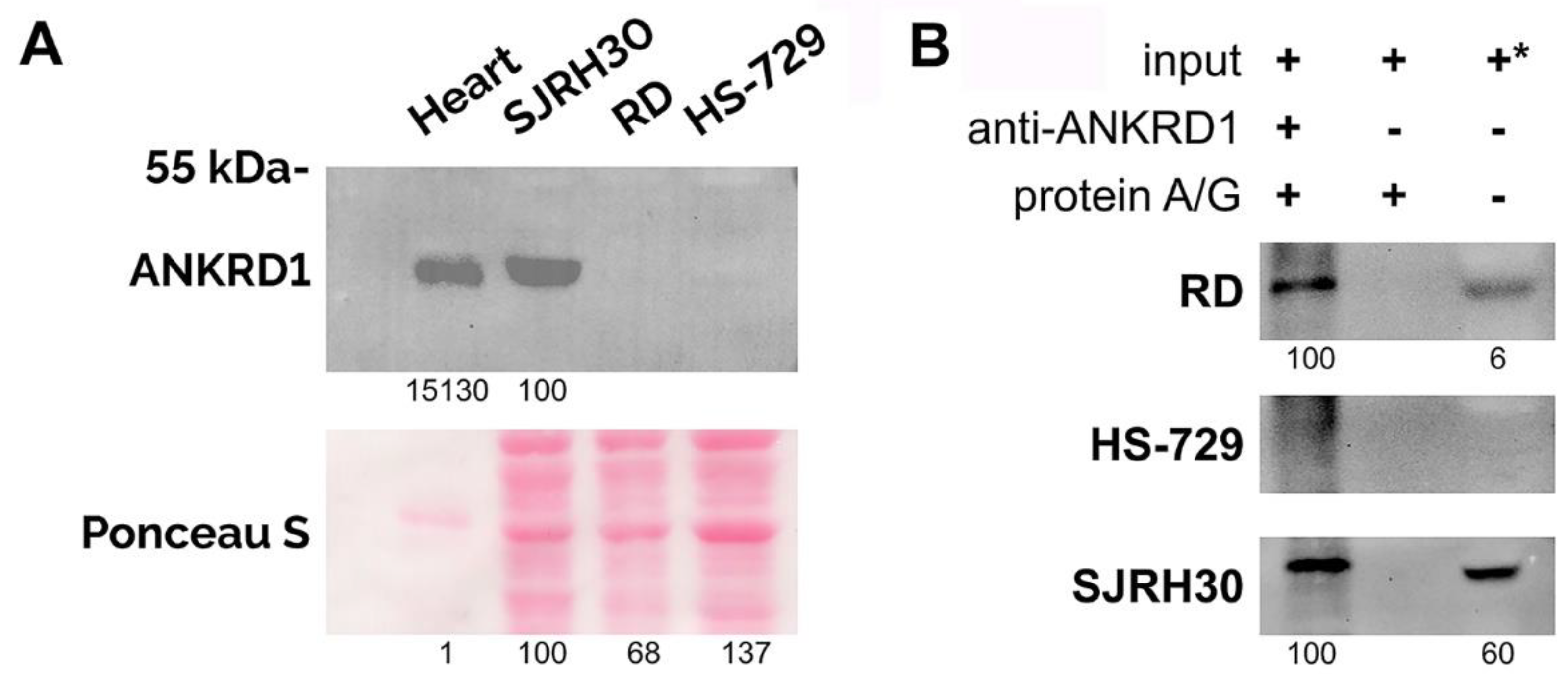
3.1. ANKRD1 transcript and protein are differently expressed in RMS cell lines
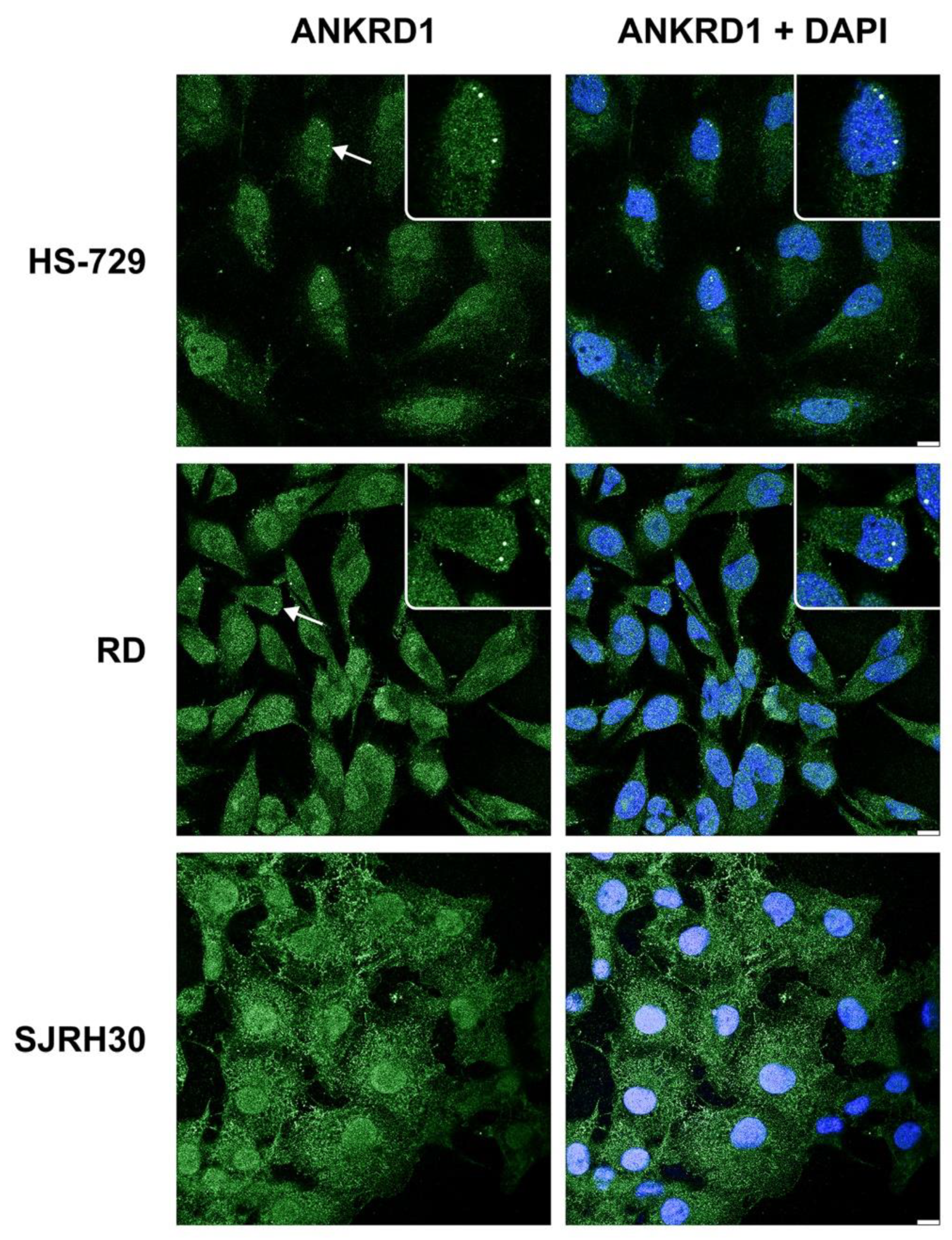
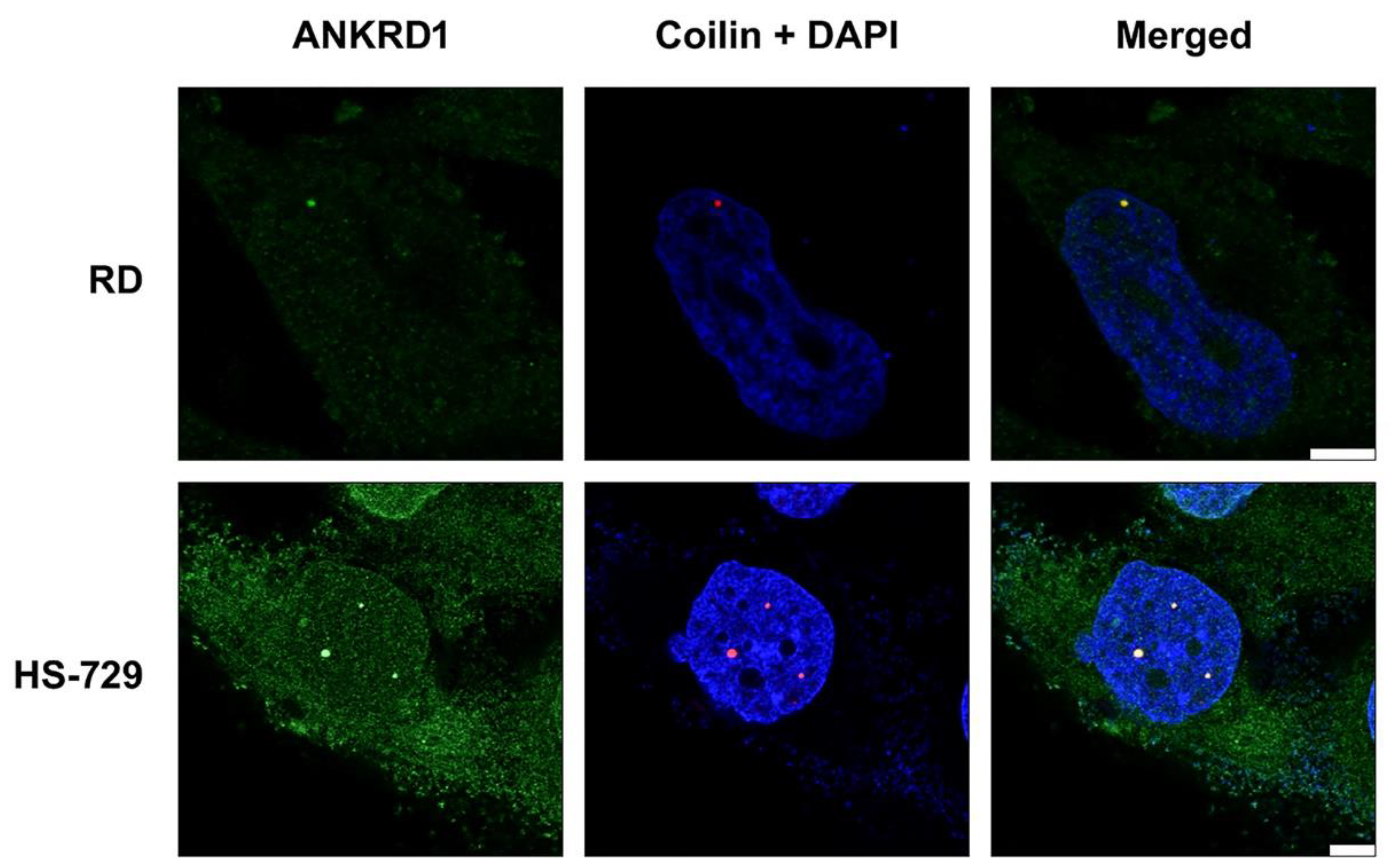
3.2. ANKRD1 is localized in the Cajal bodies of RMS cells’ nuclei
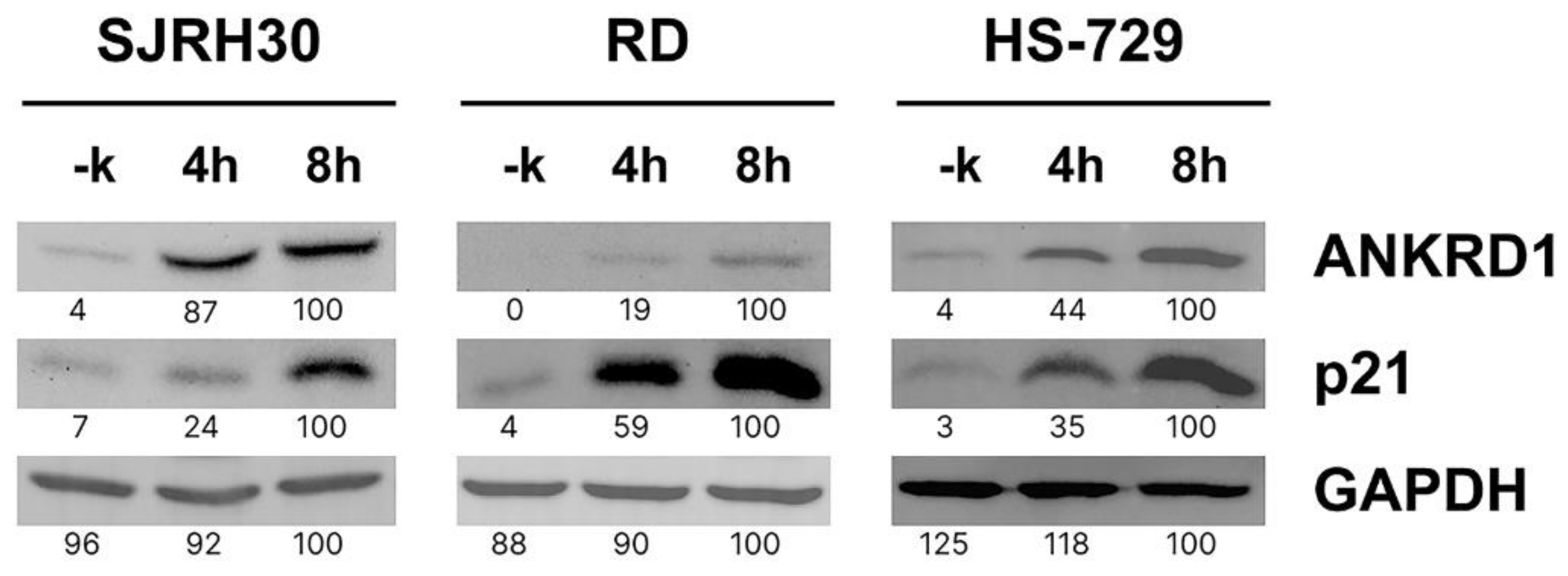
3.3. ANKRD1 is prone to proteasomal degradation in RMS cells
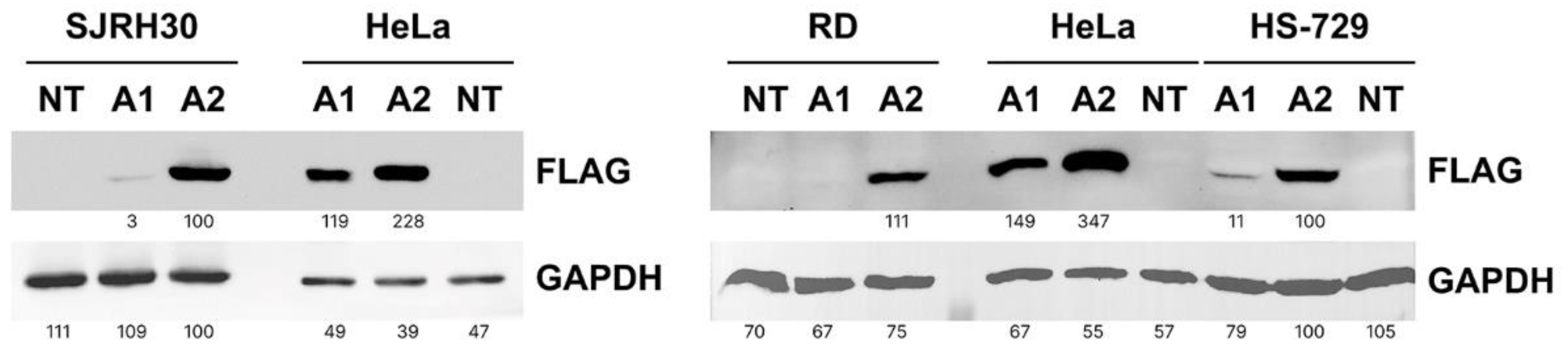
3.4. RMS cells do not tolerate overexpression of ANKRD1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Saab:, R.; Spunt, S.L.; Skapek, S.X. Myogenesis and rhabdomyosarcoma the Jekyll and Hyde of skeletal muscle. Curr Top Dev Biol 2011, 94, 197–234. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat Rev Dis Primers 2019, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.A.; Barr, F.G. Molecular diagnostics in the management of rhabdomyosarcoma. Expert Rev Mol Diagn 2017, 17, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Dziuba, I.; Kurzawa, P.; Dopierała, M.; Larque, A.B.; Januszkiewicz-Lewandowska, D. Rhabdomyosarcoma in children - current pathologic and molecular classification. Pol J Pathol 2018, 69, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, N.; Motoi, T.; Araki, N.; Ito, H.; Moriyama, M.; Yoshida, H. Expression of cardiac ankyrin repeat protein, CARP, in malignant tumors: diagnostic use of CARP protein immunostaining in rhabdomyosarcoma. Hum Pathol 2008, 39, 1673–1679. [Google Scholar] [CrossRef]
- Miller, M.K.; Bang, M.L.; Witt, C.C.; Labeit, D.; Trombitas, C.; Watanabe, K.; Granzier, H.; McElhinny, A.S.; Gregorio, C.C.; Labeit, S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J Mol Biol 2003, 333, 951–964. [Google Scholar] [CrossRef]
- Ling, S.S.M.; Chen, Y.T.; Wang, J.; Richards, A.M.; Liew, O.W. Ankyrin Repeat Domain 1 Protein: A Functionally Pleiotropic Protein with Cardiac Biomarker Potential. Int J Mol Sci 2017, 18, 1362. [Google Scholar] [CrossRef]
- Jeyaseelan, R.; Poizat, C.; Baker, R.K.; Abdishoo, S.; Isterabadi, L.B.; Lyons, G.E.; Kedes, L. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J Biol Chem 1997, 272, 22800–22808. [Google Scholar] [CrossRef]
- Kuo, H.; Chen, J.; Ruiz-Lozano, P.; Zou, Y.; Nemer, M.; Chien, K.R. Control of segmental expression of the cardiac-restricted ankyrin repeat protein gene by distinct regulatory pathways in murine cardiogenesis. Development 1999, 126, 4223–4234. [Google Scholar] [CrossRef]
- Ishiguro, N.; Baba, T.; Ishida, T.; Takeuchi, K.; Osaki, M.; Araki, N.; Okada, E.; Takahashi, S.; Saito, M.; Watanabe, M.; et al. Carp, a cardiac ankyrin-repeated protein, and its new homologue, Arpp, are differentially expressed in heart, skeletal muscle, and rhabdomyosarcomas. Am J Pathol 2002, 160, 1767–1778. [Google Scholar] [CrossRef]
- Bang, M.L.; Mudry, R.E.; McElhinny, A.S.; Trombitás, K.; Geach, A.J.; Yamasaki, R.; Sorimachi, H.; Granzier, H.; Gregorio, C.C.; Labeit, S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J Cell Biol 2001, 153, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Kojic, S.; Radojkovic, D.; Faulkner, G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Crit Rev Clin Lab Sci 2011, 48, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Henderson, B.R.; Emmanuel, C.; Harnett, P.R.; deFazio, A. Inhibition of ANKRD1 sensitizes human ovarian cancer cells to endoplasmic reticulum stress-induced apoptosis. Oncogene 2015, 34, 485–495. [Google Scholar] [CrossRef]
- Scurr, L.L.; Guminski, A.D.; Chiew, Y.E.; Balleine, R.L.; Sharma, R.; Lei, Y.; Pryor, K.; Wain, G.V.; Brand, A.; Byth, K.; et al. Ankyrin repeat domain 1, ANKRD1, a novel determinant of cisplatin sensitivity expressed in ovarian cancer. Clin Cancer Res 2008, 14, 6924–6932. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Seike, M.; Chiba, M.; Takahashi, S.; Nakamichi, S.; Matsumoto, M.; Takeuchi, S.; Minegishi, Y.; Noro, R.; Kunugi, S.; et al. Ankyrin Repeat Domain 1 Overexpression is Associated with Common Resistance to Afatinib and Osimertinib in EGFR-mutant Lung Cancer. Sci Rep 2018, 8, 14896. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, Q.; Tang, Y.; Liu, Z.; Sun, G.; Lu, Z.; Chen, Y. Identification and validation of a cigarette smoke-related five-gene signature as a prognostic biomarker in kidney renal clear cell carcinoma. Sci Rep 2022, 12, 2189. [Google Scholar] [CrossRef]
- Yin, P.; Tong, C. LncRNA RGMB-AS1 up-regulates ANKRD1 Through Competitively Sponging miR-3614-5p to Promote OSA Cell Proliferation and Invasion. Arch Med Res 2022, 53, 131–137. [Google Scholar] [CrossRef]
- Park, J.H.; Liu, L.; Kim, I.H.; Kim, J.H.; You, K.R.; Kim, D.G. Identification of the genes involved in enhanced fenretinide-induced apoptosis by parthenolide in human hepatoma cells. Cancer Res 2005, 65, 2804–2814. [Google Scholar] [CrossRef]
- Jiménez, A.P.; Traum, A.; Boettger, T.; Hackstein, H.; Richter, A.M.; Dammann, R.H. The tumor suppressor RASSF1A induces the YAP1 target gene. Oncotarget 2017, 8, 88437–88452. [Google Scholar] [CrossRef]
- Kojic, S.; Nestorovic, A.; Rakicevic, L.; Belgrano, A.; Stankovic, M.; Divac, A.; Faulkner, G. A novel role for cardiac ankyrin repeat protein Ankrd1/CARP as a co-activator of the p53 tumor suppressor protein. Arch Biochem Biophys 2010, 502, 60–67. [Google Scholar] [CrossRef]
- Tang, J.; Tian, Z.; Liao, X.; Wu, G. SOX13/TRIM11/YAP axis promotes the proliferation, migration and chemoresistance of anaplastic thyroid cancer. Int J Biol Sci 2021, 17, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhou, J.; Tang, J.; Zhou, F.; He, Z.; Liu, T. MINDY1 promotes bladder cancer progression by stabilizing YAP. Cancer Cell Int 2021, 21, 395. [Google Scholar] [CrossRef] [PubMed]
- Jasnic-Savovic, J.; Nestorovic, A.; Savic, S.; Karasek, S.; Vitulo, N.; Valle, G.; Faulkner, G.; Radojkovic, D.; Kojic, S. Profiling of skeletal muscle Ankrd2 protein in human cardiac tissue and neonatal rat cardiomyocytes. Histochem Cell Biol 2015, 143, 583–597. [Google Scholar] [CrossRef]
- Kojic, S.; Medeot, E.; Faulkner, G. Characterization of Antibodies Directed Against the Ankrd2 Human Muscle Protein. Archive of Biological Science 2009, 61, 9. [Google Scholar] [CrossRef]
- Phillips, D.C.; Hunt, J.T.; Moneypenny, C.G.; Maclean, K.H.; McKenzie, P.P.; Harris, L.C.; Houghton, J.A. Ceramide-induced G2 arrest in rhabdomyosarcoma (RMS) cells requires p21Cip1/Waf1 induction and is prevented by MDM2 overexpression. Cell Death Differ 2007, 14, 1780–1791. [Google Scholar] [CrossRef]
- Kikuchi, M.; Yamashita, K.; Waraya, M.; Minatani, N.; Ushiku, H.; Kojo, K.; Ema, A.; Kosaka, Y.; Katoh, H.; Sengoku, N.; et al. Epigenetic regulation of ZEB1-RAB25/ESRP1 axis plays a critical role in phenylbutyrate treatment-resistant breast cancer. Oncotarget 2016, 7, 1741–1753. [Google Scholar] [CrossRef]
- Hui, B.; Ji, H.; Xu, Y.; Wang, J.; Ma, Z.; Zhang, C.; Wang, K.; Zhou, Y. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis 2019, 10, 207. [Google Scholar] [CrossRef]
- Badi, I.; Cinquetti, R.; Frascoli, M.; Parolini, C.; Chiesa, G.; Taramelli, R.; Acquati, F. Intracellular ANKRD1 protein levels are regulated by 26S proteasome-mediated degradation. FEBS Lett 2009, 583, 2486–2492. [Google Scholar] [CrossRef]
- Samaras, S.E.; Chen, B.; Koch, S.R.; Sawyer, D.B.; Lim, C.C.; Davidson, J.M. 26S proteasome regulation of Ankrd1/CARP in adult rat ventricular myocytes and human microvascular endothelial cells. Biochem Biophys Res Commun 2012, 425, 830–835. [Google Scholar] [CrossRef]
- Piazzi, M.; Kojic, S.; Capanni, C.; Stamenkovic, N.; Bavelloni, A.; Marin, O.; Lattanzi, G.; Blalock, W.; Cenni, V. Ectopic Expression of Ankrd2 Affects Proliferation, Motility and Clonogenic Potential of Human Osteosarcoma Cells. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Bersani, F.; Taulli, R.; Accornero, P.; Morotti, A.; Miretti, S.; Crepaldi, T.; Ponzetto, C. Bortezomib-mediated proteasome inhibition as a potential strategy for the treatment of rhabdomyosarcoma. Eur J Cancer 2008, 44, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Péron, J.; Polivka, V.; Chabaud, S.; Poupart, M.; Ceruse, P.; Ramade, A.; Girodet, D.; Zrounba, P.; Fayette, J. An effective and well-tolerated strategy in recurrent and/or metastatic head and neck cancer: successive lines of active chemotherapeutic agents. BMC Cancer 2014, 14, 504. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Zhu, Y.; Sabo, Y.; Goff, S.P. Embryonic Cells Redistribute SUMO1 upon Forced SUMO1 Overexpression. mBio 2019, 10. [Google Scholar] [CrossRef]
- Sommer, P.; Le Rouzic, P.; Gillingham, H.; Berry, A.; Kayahara, M.; Huynh, T.; White, A.; Ray, D.W. Glucocorticoid receptor overexpression exerts an antisurvival effect on human small cell lung cancer cells. Oncogene 2007, 26, 7111–7121. [Google Scholar] [CrossRef]
- Van Meter, M.; Mao, Z.; Gorbunova, V.; Seluanov, A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011, 10, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, J.G.; Chen, S.T.; Tafani, M.; Snyder, J.W.; Farber, J.L. The overexpression of Bax produces cell death upon induction of the mitochondrial permeability transition. J Biol Chem 1998, 273, 7770–7775. [Google Scholar] [CrossRef]
- Druskovic, M.; Suput, D.; Milisav, I. Overexpression of caspase-9 triggers its activation and apoptosis in vitro. Croat Med J 2006, 47, 832–840. [Google Scholar]
- Zhu, J.; Cai, Y.; Xu, K.; Ren, X.; Sun, J.; Lu, S.; Chen, J.; Xu, P. Beclin1 overexpression suppresses tumor cell proliferation and survival via an autophagy-dependent pathway in human synovial sarcoma cells. Oncol Rep 2018, 40, 1927–1936. [Google Scholar] [CrossRef]
- Mansour, M.A.; Rahman, M.; Ayad, A.A.; Warrington, A.E.; Burns, T.C. P21 Overexpression Promotes Cell Death and Induces Senescence in Human Glioblastoma. Cancers (Basel) 2023, 15. [Google Scholar] [CrossRef]
- Shen, L.; Chen, C.; Wei, X.; Li, X.; Luo, G.; Zhang, J.; Bin, J.; Huang, X.; Cao, S.; Li, G.; et al. Overexpression of ankyrin repeat domain 1 enhances cardiomyocyte apoptosis by promoting p53 activation and mitochondrial dysfunction in rodents. Clin Sci (Lond) 2015, 128, 665–678. [Google Scholar] [CrossRef]
- Kojic, S.; Medeot, E.; Guccione, E.; Krmac, H.; Zara, I.; Martinelli, V.; Valle, G.; Faulkner, G. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J Mol Biol 2004, 339, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Staněk, D. Cajal bodies and snRNPs - friends with benefits. RNA Biol 2017, 14, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 2013, 4, 17–34. [Google Scholar] [CrossRef]
- Zou, Y.; Evans, S.; Chen, J.; Kuo, H.C.; Harvey, R.P.; Chien, K.R. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development 1997, 124, 793–804. [Google Scholar] [CrossRef]
- Oda, Y.; Kohashi, K.; Yamamoto, H.; Tamiya, S.; Kohno, K.; Kuwano, M.; Iwamoto, Y.; Tajiri, T.; Taguchi, T.; Tsuneyoshi, M. Different expression profiles of Y-box-binding protein-1 and multidrug resistance-associated proteins between alveolar and embryonal rhabdomyosarcoma. Cancer Sci 2008, 99, 726–732. [Google Scholar] [CrossRef]
- Bogolyubova, I.O.; Lyabin, D.N.; Bogolyubov, D.S.; Ovchinnikov, L.P. Immunocytochemical study of YB-1 nuclear distribution in different cell types. Tissue Cell 2014, 46, 457–461. [Google Scholar] [CrossRef]
- Chansky, H.A.; Hu, M.; Hickstein, D.D.; Yang, L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res 2001, 61, 3586–3590. [Google Scholar]
- Rapp, T.B.; Yang, L.; Conrad, E.U.; Mandahl, N.; Chansky, H.A. RNA splicing mediated by YB-1 is inhibited by TLS/CHOP in human myxoid liposarcoma cells. J Orthop Res 2002, 20, 723–729. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, L.; Li, X.; Xu, W.; Yang, S.; Song, J.; Zhang, W.; Zhan, J.; Luo, J.; Zhang, H. Oxidative stress-CBP axis modulates MOB1 acetylation and activates the Hippo signaling pathway. Nucleic Acids Res 2022, 50, 3817–3834. [Google Scholar] [CrossRef]
- Beneventi, G.; Munita, R.; Cao Thi Ngoc, P.; Madej, M.; Cieśla, M.; Muthukumar, S.; Krogh, N.; Nielsen, H.; Swaminathan, V.; Bellodi, C. The small Cajal body-specific RNA 15 (SCARNA15) directs p53 and redox homeostasis via selective splicing in cancer cells. NAR Cancer 2021, 3, zcab026. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Sleeman, J.E. The Cajal body and the nucleolus: "In a relationship" or "It's complicated"? RNA Biol 2017, 14, 739–751. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer sequence (5’-3’) | Direction | Application | Position within ORF (bp) |
|---|---|---|---|---|
| GAPDH | GTGAAGGTCGGAGTCAACG | Forward | RT-PCR | 10-28 |
| GAPDH | TGAGGTCAATGAAGGGGTC | Reverse | RT-PCR | 103-121 |
| ANKRD1 | AGTAGAGGAACTGGTCACTGG | Forward | RT-PCR, Sequencing | 15-35 |
| ANKRD1 | TGGGCTAGAAGTGTCTTCAGAT | Reverse | RT-PCR, Sequencing | 131-152 |
| ANKRD1 | ACGCCAAAGACAGAGAAGGA | Forward | Sequencing | 737-756 |
| ANKRD1 | GCTGCAGCGATGATGGTACTGAAAGTA | Forward | Sequencing, ORF amplification | 1-18 |
| ANKRD1 | TAGGATCCTCAGAATGTAGCTATGCG | Reverse | Sequencing, ORF amplification | 943-960 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





