1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic was followed by a global vaccination campaign on an unprecedented scale that turned the public attention towards the benefit-risk balance of the vaccines used for combating the disease.
Analysis of adverse events (AE), or in the case of vaccines, adverse events following immunization (AEFI), is an important component of the assessment of the benefit-risk balance. During the controlled conditions in the pre- and post-authorization clinical trials, there is enough information for each participant (doses taken, AE after each dose including no AE, as well as other variables like sex, age, severity, time after dose, etc.) to enable a proper comparison between control and treatment arms and unbiased conclusions for vaccines short term side-effects. Conversely, in spontaneous, continuous, post-marketing AE reporting much of the needed information is lacking and, moreover, additional hard-to-assess variables appear, like reporting rate and its variability over such factors as age, sex, education, location, etc. Although statistical approaches to the analysis of such data exist, with some methods more promising than others, these data are so limited that it is recommended they should be considered only for specific purposes like detecting very rare AE. Problems associated with the use of spontaneous reports in drug safety include (1) confounding by indication (i.e., patients taking a particular drug may have a disease that is itself associated with a higher incidence of the AE (e.g., anti-COVID-19 immunization after SARS-CoV-2 or another viral infection), (2) systematic underreporting, (3) questionable representativeness of patients, (4) effects of publicity in the media on numbers of reports, (5) extreme duplication of reports, (6) attribution of the event to a single drug when patients may be exposed to multiple drugs, and (7) extensive amounts of missing data. These limitations degrade the capacity for optimal data mining and analysis of AEs [
1,
2].
Time-independent risk prediction models for AEs belong mainly to two types: logistic regression and Poisson regression. Response variables for binary logistic regression are most often AE occurrence (no AE, AE) or severity (non-severe, severe). Severity might be divided to more than two levels and assessed with multinomial or ordinal logistic regression. These models require data from a clinical trial or at least from a clinical setting in which AE occurrence is followed up and severity of the adverse effect is assessed by one or more physicians as objectively as possible. Response variables for Poisson regression are usually the number of AE or number of individuals with AE. The data can also come from a population trial in which one has the information mentioned above about each individual.
Such models applied to data from spontaneous AE reporting would be misleading for the reasons outlined above. For example, severity of the AE is for the most part self-assessed by the reporting person and thus is subjective. The number of AE or number of reports depends on the total number of vaccine / drug doses taken, the reporting rate, its variation in different subgroups, media publicity, etc. On the other hand, spontaneous reporting has an important advantage over clinical trials, which is their greater coverage: whereas the typical sample size of experimental studies is <1000, spontaneous reports number several thousands to several tens (or even hundreds) of thousands depending on the drug(s) / vaccine(s) in consideration.
The pharmacovigilance database of the Bulgarian pharmaceutical regulator, the Bulgarian Drug Agency (BDA), records up to 20–25 AEs for each spontaneous report. The number of AEs in an individual report which we abbreviate as “adverse events count” (AEC) is here used as an indicator of drug risk that does not vary with the total number of reports or doses in different subgroups. It also obviates the need for a control group. In our opinion, this variable has the potential to alleviate many of the problems associated with spontaneously reported AE: (1) an eventual disease underlying the reported AEs may increase AEC but it can be discovered on follow-up and the report taken out from the analysis, (2) underreporting is not a problem since AEC does not depend on the number of reports, (3) unequal representation of some subgroups does not change the AEC reported from them, (4) AEC are not expected to be influenced much by media publicity at least not to such extent as number of reports, (5) report duplication would randomly increase the weights of some AEC values resulting in no systematic bias, (6) exposition to multiple drugs can increase AEC and, therefore, remains a problem; however, most AE reporting forms have an item for additional drugs and data can be culled for suspiciously high AEC with untypical AEs in presence of multiple drugs; another approach is to include the number of additional drugs as an independent variable in the model and assess its effect on AEC, (7) the amount of missing data depend on the number of independent variables included in the model; the bulk result from not filling an item in the AE reporting form. They are of the type missing-completely-at-random (MCAR) and do not affect the AEC distribution.
1.1. AER distributions
Looked at from a theoretical point of view, AEC provides a sufficient statistic as defined by Fisher [
3]. More specifically, the AEC random variable has a definite statistical distribution from which a likelihood function can be calculated. From the latter, when applied to the sample, maximal likelihood estimates for the parameters of the distribution are found and on these, a statistical model is built. The distribution parameters are a sufficient statistic because they contain all information extracted from the data and no other information is needed to build the model.
AEC are integer values resulting from counts. If they were distributed randomly and independently, they would follow the Poisson distribution:
where
y is the AEC count (
),
e is the Euler number (
) and ! is the factorial function. The positive real number
is the single distribution parameter that is equal to the mean and to the variance (
equidispersion).
It can be seen, however, that AEC violate both the conditions of randomness (they occur in clusters as several AE per report and not one at a time) and independence (the AE within the cluster (report) are correlated, i. e., they depend on each other). The result of such within-report correlation is that the variance of AEC would be larger than the mean, a phenomenon known as
overdispersion. The overdispersion may be modeled as follows [
4,
5]: first we assume that conditional on the
cluster (report) effect, denoted by
we have
Second, we assume that the random cluster effect has = 1, and
. Therefore
regardless of the distribution assigned to
. Here
is a random variable that summarizes the effect of our inability to model all pertinent independent variables and other sources of variation. In the above formula,
indicates the presence of overdispersion,
underdispersion, and
equidispersion. Under equidispersion, the Poisson model is the most commonly used model to analyze count data. In the other cases, it is assumed that the distribution of the response variable
Y has a discrete probability function
, conditional on a continuous random variable
u whose distribution is
. Then the marginal (or unconditional) probability function of
Y is given by
where
is the range of
u. The resulting distribution of
Y is called a continuous mixture of discrete distributions. For the clustered AEC data, the conditional distribution is the Poisson-like
where instead of
, there is
. Substituting (
5) in (
4), one obtains
To complete the model, one must specify
, the distribution of
u. The usual assumption is that
u follows a Gamma distribution
Substituting (
7) in (
6), solving the integral and rearranging, one obtains a negative binomial distribution as the final marginal distribution for AEC [
6]
where
and
are the parameters.
is the Gamma function, which is a special function replacing factorial expressions when the arguments are not integers. Equation (
8) is the negative binomial type I, NBI(
,
), probability distribution. Such a genesis of the negative binomial distribution as a mixture of Poisson and Gamma distributions is quite distinct from its classical development as the distribution of the number of failures until the
success in independent Bernoulli trials, and was originally developed by Greenwood and Yule [
7].
NBI(
,
) is quite flexible with many different parametrizations and has been used to model a variety of data with mild to moderate overdispersion from various fields of study: CD4 counts in HIV-infected women [
8], microbiome counts in mouse gut [
9], rainfall counts [
10], postfire conifer regeneration counts to examine seedling distributions [
11], data from single-cell RNA sequencing [
12] and dental caries count indices [
13], prediction of micronuclei frequency as a biomarker for genotoxic exposure and cancer risk [
14], a clinical trial to evaluate the effectiveness of a prehabilitation program in preventing functional disease among physically frail, community-living older persons [
15], investigating the social and demographic factors associated with public awareness of health warnings on the harmful effects of environmental tobacco smoke [
16], assessing highway crash frequency by injury severity [
17], improving planning and management of urban trail traffic [
18], modeling overdispersion in ecological count data, e. g., bird migration [
19], and other applications of NB in ecology and biodiversity reviewed in [
20], male satellites counts in the popular horseshoe crab data [
21], modeling the dependence of tropical storm counts in the North Atlantic basin on climate indices [
22], estimation of the reproduction number
for SARS 2003 coronavirus by the number of secondary cases [
23], predicting length of stay from an electronic patient record for patients with knee replacement [
24], or for elderly patients [
25], serial clustering of extratropical cyclones [
26], or of intense European storms [
27], digital gene expression counts [
28].
The variance of NBI(
,
) is
, hence, the overdispersion
is a quadratic in
as provisioned in formula (
3). Reparametrization of
to
in NBI(
,
) gives NBII(
,
) in which the variance is linear in
:
. For data that have a high initial peak, clumped heavily at 1 and 2, and that may be skewed to the far right as well as data that are highly Poisson overdispersed, another distribution for the random effect
u may provide a better fit. For such data, the Inverse Gaussian distribution is commonly used [
29]:
Substituting (
9) in (
6) and solving the integral for
u, one obtains the Poisson-inverse Gaussian distribution denoted by PIG(
,
) [
5]
for
, where
,
,
and
.
is the modified Bessel function of the second kind given by
The Bessel function
in general presents challenges in computation; however, when its order
is a half-integer, as it is in (
10), computations are simplified considerably as
and use is made of the recurrence relation
It can be shown with certain parametrizations of PIG that its overdispersion is cubic in
. Compared with NBI, with greater mean values of PIG come increasingly greater values of the variance. With a dispersion parameter >1, larger values of the mean of the response variable in a PIG regression provide for adjustment of larger amounts of overdispersion than does the NBI model. Simply put, PIG models can better deal with highly overdispersed data than can NBI regression [
29] (p. 163).
The PIG models are not as common as the NB models for several reasons: (1) in many cases, the NB models are adequate to handle the amount of overdispersion present in the data, (2) authors are not aware of the availability of PIG, and that a PIG model can fit their data better than a Poisson or NB model, (3) no commercial statistical software offers the PIG model as a component of its official offerings; the only software that supports PIG at present is the R package GAMLSS [
30].
Still, PIG models are not completely lacking and there is a variety of fields that these are used: PIG and NB distributions have been fitted to the popular horseshoe crab data and it was found that PIG provides a better fit [
21], identification of the relationship between farm management practices and risk of cow mastitis [
5], comparing PIG and NB for the analysis of automobile claim frequency data [
31], modeling overdispersed dengue data in the city of Campo Grande, MS, Brazil [
32], application of NBI, PIG, and Negative Binomial-Inverse Gaussian (NBIG) models to car insurance claim frequency [
33,
34], application of generalized PIG for analysis of bibliometric data sets [
35], modeling species abundance data [
36], application to the bonus-malus system in car insurance claims [
37], modeling infectious disease count data [
38], application to health services: number of hospital stays [
39], assessing microdata disclosure risk [
40], modeling the number of HIV and AIDS cases in two districts in Indonesia [
41], analysing long-term survival data in patients with skin cancer [
42], modeling the numbers of pauci-baciliary and multi-baciliary leprosy cases with geographical conditions factors in West Sumatera, Indonesia [
43], application to accident data for men in a soap factory and for women working with highly explosive shells [
44], number of falls in Parkinson’s disease patients over a 10-week period, analysing both the mean (
) and variability (
) parameters [
45], analysis of data from a Phase III cutaneous melanoma clinical trial (E1690 data) [
46], transportation origin–destination analysis by introducing predictory variables-based models which incorporate different transport modeling phases and also allow for direct probabilistic inference on link traffic based on Bayesian predictions [
47], analysis of motor vehicle crash data [
48].
AEC data structurally exclude zero counts as each report includes at least one AE. Conversely, Poisson, NB, and PIG distributions have a non-zero probability for a zero count and if one tries to fit them to the AEC data there would be some misfit just because these distributions do not accommodate the data property of zero exclusion. This situation is solved by “hurdle models”, so called because the deviating value (in this case, the zero count) is seen as a hurdle which is passed with some probability
, and the remaining probability
is reserved for the other values which are distributed according to the corresponding distribution. In the case of AEC,
, so that values higher than 0 occur almost surely. In this case, more appropriate are the so-called zero-altered or zero-adjusted (ZA) distributions, in which the 0 has a point mass distribution, known in physics as Dirac delta function. In other words, if the count is 0 then
otherwise
or, more precisely
The coefficient c is calculated from the parameters , , and is different for the different distributions but in any case, if then . Please note that the ZA distributions are already three-parametric with the additional parameter .
In this paper, we propose a model applied to AE data in the Bulgarian Drug Agency (BDA) pharmacovigilance database, whose dependent variable is the AEC. Two data sets: AEC due to vaccines against COVID-19 and AEC due to the administration of drugs other than COVID-19 vaccines were studied separately. To our knowledge, this is the first attempt to determine the distribution of AEC and use it in a model.
We also aim, using the increased sensitivity of the model to infer differences between the COVID-19 vaccines and the other drugs in terms of AEC also taking into account additional explanatory variables such as sex, age, vaccine, declared severity and sequence number of the report.
3. Discussion
To our knowledge, this is the first study attempting to determine the distribution of AEC and use it in a model. By systematically fitting different discrete distributions to AEC from COVID vaccine and other drugs data, it was found that they were best fitted by ZANBI and ZAPIG, respectively, as predicted on the theoretical basis presented in the Introduction. Furthermore, the AEC for the other drugs data, which have bigger overdispersion according to the Cameron-Trivedi test, is better fitted by ZAPIG while the AEC for COVID vaccines data are better fitted by ZANBI according to the theoretical conclusion that ZAPIG fits overdispersed data better than ZANBI. Nevertheless, in both data sets the difference in fit between the two distributions is small compared with the all other available in the GAMLSS package discrete distributions that were tested.
When fitted on the two data sets (COVID vaccines and other drugs), ZANBI and ZAPIG have only minor visible differences between each other. Judging by their parameters and moments, ZAPIG, indeed, handles better a larger overdispersion than ZANBI. For example, ZAPIG has smaller means but larger all other three moments (variance, skewness, and especially kurtosis). Fit deficiencies, in general, are most evident in kurtosis and skewness because they are the highest moments (powers of the distribution generating function), fourth and third, respectively, and any deviation would be magnified when multiplied several times.
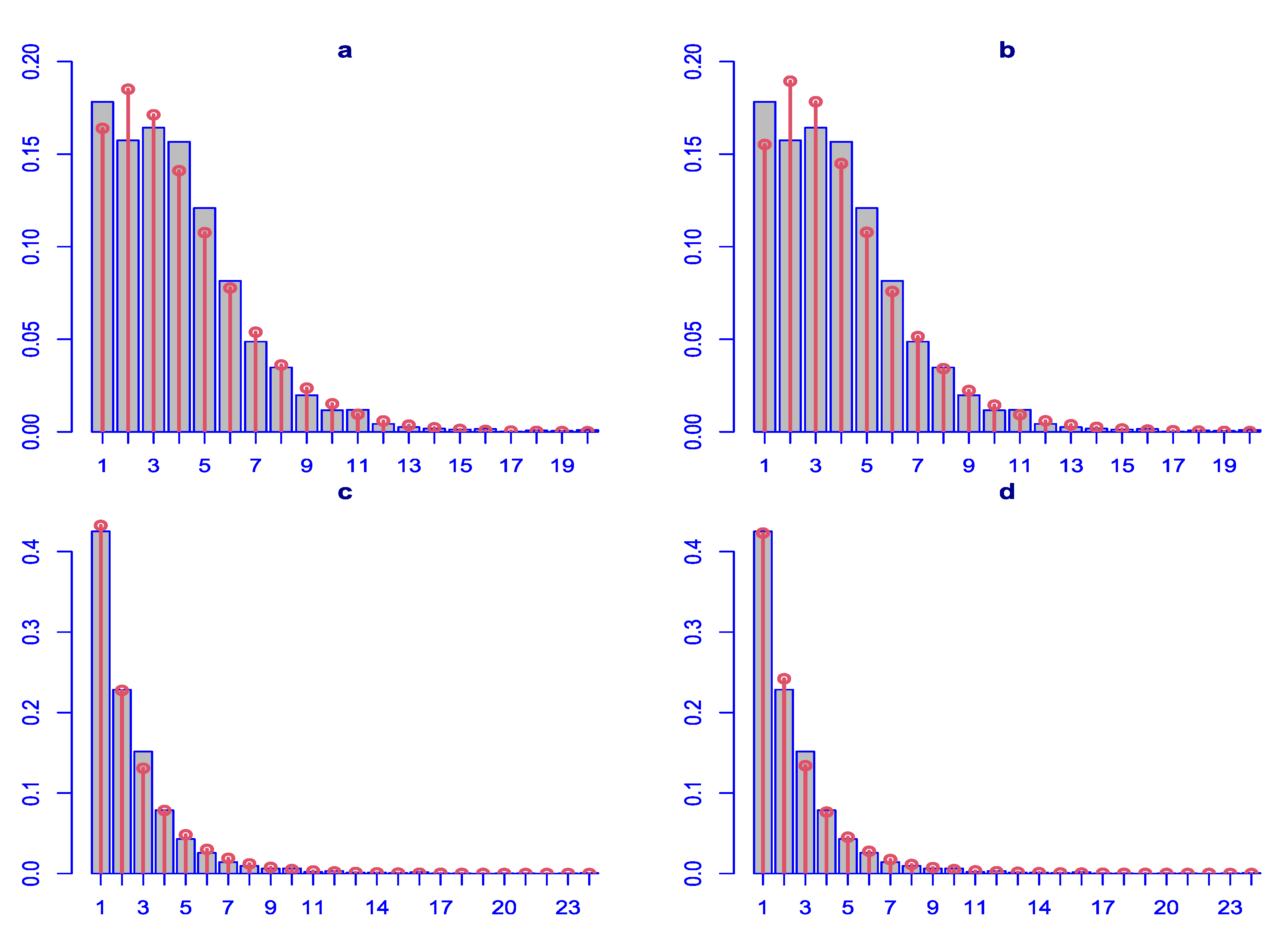
Differences between the two distributions are observed in the diagnostic residual plots with ZAPIG handling better data with higher overdispersion, as theoretically predicted. Thus, ZAPIG residuals have smaller excess kurtosis as evidenced by the smaller loops in the Q-Q plots, and, respectively, the lower right tails in the worm plots. The moments of the ZAPIG residual distributions are generally closer to the standard normal distribution than those of ZANBI.
Kurtosis and skewness show the biggest deviations from the standard normal in line with what was said above. Filliben coefficients, which reflect the degree of fit for the residuals, are almost the same for the ZANBI and ZAPIG distributions on the COVID vaccine data (difference in the sixth digit after the decimal point) and slightly different for the other drugs data (advantage for ZAPIG in the fourth digit after the decimal point). This confirms the results from the analysis of AIC and BIC criteria. As a whole, differences in diagnostic plots between the two distributions are minor compared to the differences between the two sets of data.
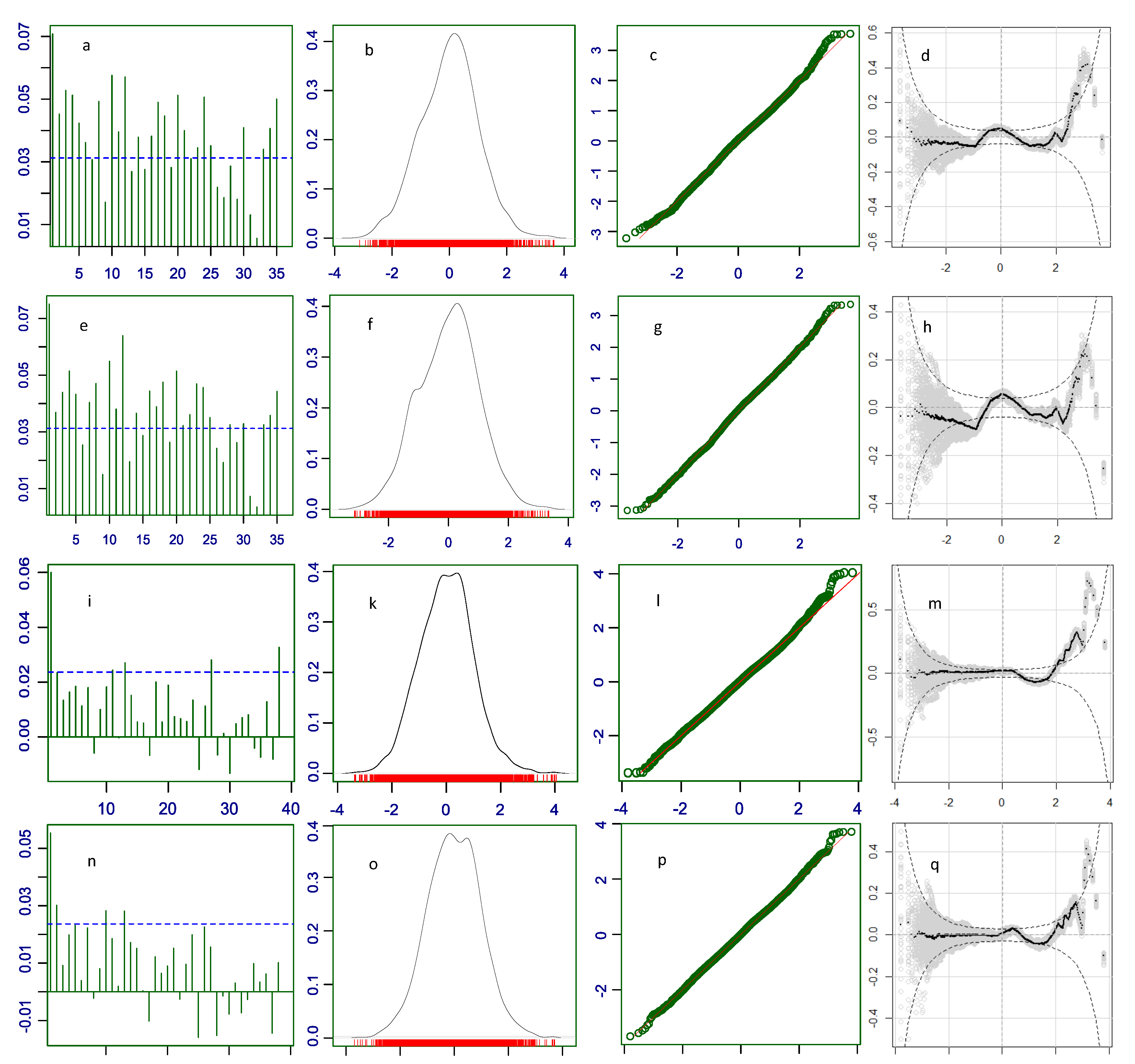
Part of the misfit of a distribution can be due to the effects of explanatory variables (fixed effects). Correlation within the same level of an explanatory variable is bigger than correlation among levels. For example, data obtained from subjects immunized with Comirnaty are stronger correlated than data from Comirnaty and those from other COVID-19 vaccines because of the differences of action and consequences of immunization with different vaccines. When explanatory variables are included into the model, their fixed effects are accounted for as separate coefficients, which reduces or eliminates the correlation within levels (autocorrelation). This is seen in our residual autocorrelation plots for the parametric models of the COVID vaccine data, which are highly autocorrelated. Low autocorrelation means more independence and randomness which are the implied conditions in the first equation in the systems of equations (
15) and (
19) in Materials and Methods. Because the distributions used to model the data, in our case ZANBI and ZAPIG, fit theoretically perfect random data, the more random are the real data, the better is the fit, that is, the higher is the likelihood that the distribution fits the data. Including the explanatory variables in the model makes data more random, simultaneously increasing likelihood and decreasing variance. Since the increased likelihood results in better fit, the full model has lower residual variance and AIC value than the parametric model.
These assertions are confirmed by our results. The high autocorrelation in the parametric model of the COVID vaccines is drastically reduced in the full model, which is supposedly due primarily to the inclusion of the vaccines fixed effects in the full model. Simultaneously, AIC and BIC decrease with inclusion of each new term; which is the primary criterion (AIC and BIC decrease) by which model selection is done. In principle, the AIC decrease and, correspondingly, likelihood increase, could continue for as many terms as we have with no guarantee that the more complex models are better than the simpler ones. BIC is preferred here since its second term is a penalty for model complexity that takes into account the balance between free and occupied degrees of freedom. The BIC drops from parametric to full model are considerable, 2754 and 5038 BIC units for the COVID vaccines and other drugs, respectively, which shows a high response to fixed effects and, therefore, a high sensitivity of the model.
During the model selection, ZANBI performed better than ZAPIG in that the former converged on more term combinations and had lower BIC even at the combinations where both distributions converged. Convergence depends on the model, distribution and its algorithmic implementation as well as the data. Increase in the sample size usually results in better convergence. It has been shown that dispersion is related to convergence though this relationship is still distribution-, model-, and data-dependent. For example, Fernandez and Vatcheva [
52] in a methodological paper examining data for hospital length of stay as well as simulated data with various dispersion measured through Pearson dispersion statistics, found that NB and zero-inflated NB (ZINB), but not Poisson and zero-inflated Poisson (ZIP), had less than 100% convergence rate occurring only in data with very low overdispersion. In our parametric models, all distributions had 100% convergence on both data sets. In the full models, NBI converged everywhere, and PIG did not converge only in the most complicated model – with all terms up to the 5-way interactions for both
and
parameters.
We found that both data sets have significant overdispersion according to Pearson dispersion statistic and Cameron-Trivedi tests, but they differ in the degree and direction of overdispersion. The Cameron-Trivedi test suggests that the other drugs data have slightly higher overdispersion than the COVID vaccines data, while the Pearson dispersion statistic suggests the opposite for most distributions. This discrepancy may be due to different assumptions and sensitivities of the two tests, or to model misspecification.
The results also show that different models and distributions have different effects on overdispersion. Zero-adjusted Poisson (ZAP) increases overdispersion, while pure negative binomial (NBI) and Poisson inverse Gaussian (PIG) reduce it too much, resulting in underdispersion. Zero-adjusted negative binomial (ZANBI) and zero-adjusted Poisson inverse Gaussian (ZAPIG) on the parametric models and ZANBI on the full models eliminate just enough of the overdispersion to make the models equidispersed. These results suggest that zero-inflation and excess dispersion should be accounted for separately and appropriately in count data models.
The results also indicate that the Pearson dispersion statistic is not a good indicator of model fit, as lower values do not necessarily imply better fit. The best models are those that have values close to one, indicating equidispersion. In this case, these are the parametric models of other drugs with ZAPIG and ZANBI and the full model of other drugs with ZANBI. The Pearson dispersion statistic also has a very short interval of non-significance around one, making it sensitive to small changes in values. The Cameron-Trivedi test may be more robust as it uses a wider interval based on the t-distribution.
Zero-adjustment was found to have a negative effect on convergence for both distributions though ZANBI converged on more full models than ZAPIG. This means that ZANBI can fit more complex models with multiple explanatory variables, while ZAPIG may struggle with complex models due to convergence issues. This can be explained with the fact that ZAPIG is more difficult to calculate and the necessity for the algorithms to use the recurrence relation (
13). This is supported by the observation that the ZAPIG algorithm is several times slower than the ZANBI one especially with the more complex models. Another reason for the worse performance of ZAPIG could be the implementation of the zero alteration to the algorithm. Using zero-truncated PIG (ZTPIG) rather than zero-altered PIG (ZAPIG) would likely perform better than ZAPIG or ZANBI in both parametric and full models even on highly overdispersed data and large samples.
Because of the ZAPIG non-convergence on full models, the best of them use ZANBI. Nevertheless, these models still have better fit than the respective parametric ZAPIG models as evident by their BIC criteria, diagnostic plots, and Filliben coefficients. Since one of the most important aims of modeling is to estimate the effects of the explaining variables, i.e., the fixed effects, it is interesting to explore the extent to which the replacement of ZAPIG by ZANBI would change the fixed effect estimates. We could not observe in practice the change of the fixed effects on replacement of distribution in the best models because ZAPIG did not converge with these term combinations. With our data, however, which have very close fits of ZAPIG and ZANBI and similar overdispersion, it is expected that change of distribution would minimally influence the fixed effects.
Such an expectation is additionally supported by theory. From formula (6) which represents the two distributions before mixing, it is seen that only the first, Poisson-like, distribution of formula (
5) contains the
parameter. The second distribution,
, which could be Gamma (for NB) or inverse Gaussian (for PIG) is a distribution for
u, which is more connected to
than to
. True, the distinction between the two initial distributions is to a large extent obliterated after mixing but nevertheless the possibility to have alternative distributions for
suggests that the choice of
is not critical for the fixed effects.
This assumption is rigorously proven by the theoretical study of Weems and Smith [
53] assessing the robustness of the fixed-effect estimators when fitting Poisson mixed models. The nominal distribution
F (e.g., PIG) when contaminated with a proportion
of the contaminating distribution
G (e.g., NB) becomes a contaminated distribution
. Then the Gâteaux derivative in the direction of
G of the functional form
connecting the explanatory and response variables of the
model, becomes
. The Gâteaux derivative can be viewed as a measure of the sensitivity of
T to small perturbations of
F and by definition it follows that
T is robust against distribution misspecification if
is bounded for all
G. For PIG, this definition can be restated by replacing
with the integral of the conditional expected score function
, where
and
. Weems and Smith [
53] proved that the integral of the conditional expected score matrix is uniformly bounded and found the bounds by using properties of Bessel functions and by relating the Bessel functions to the PIG probabilities. Numerical integration in R showed that the upper and lower bounds for
and
are similar and the distance between the upper and lower bounds for
increases at a faster rate than the distance between the bounds corresponding to the regression parameters which suggests that
may be more sensitive to PIG misspecification than
. The authors investigated further with simulation study done with the gamlss package the performance of maximum likelihood estimates (MLEs) when the assumed PIG distribution is misspecified in different ways (
and
). The simulations generated 1000 iid covariates, random effects, and sample responses. Monte Carlo estimates were obtained for
, and
, and their statistical behavior is examined. The bias of
and
is very small, regardless of the true value of
, both when there is a small amount of contamination and when there is complete PIG misspecification. Their standard errors also increase gradually as
increases, but their behavior is not affected by the PIG misspecification. Conversely, the bias of
increases rapidly as
increases, and there is a considerable bias of
for
under complete PIG misspecification. The Q-Q plots suggest normality of the estimates, but there is some deviation from a straight line in the tails of the distribution for
. Overall, Weems and Smith [
53] conclude that
is not affected in both parametric (
) and full (
) models, while both bias and standard error of
are significantly affected upon misspecification of the mixing distribution which in the present case involves the replacement of PIG by NBI in the full models.
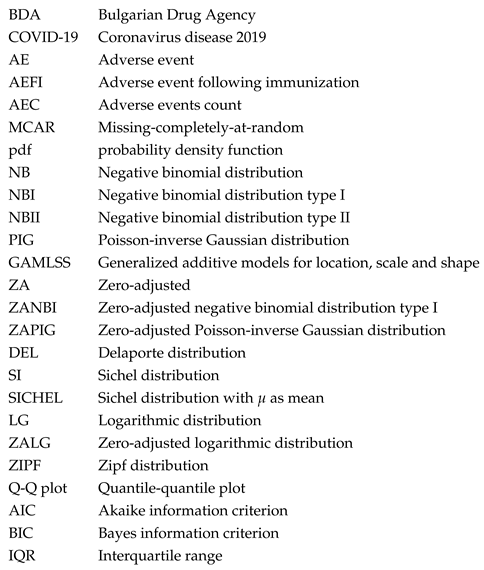
While the differences between ZANBI and ZAPIG on the same data set are minimal, the differences between the two data sets are significant and easily observable in the histograms of the experimental data (
Figure 1). In particular, the peak in the theoretical distributions of COVID vaccines is shifted to the right with a clearly delineated left shoulder while in the other drugs data, the peak is at the very beginning and there is no left shoulder. This is reflected in differences between the parameters and moments of the distributions between the two data sets. The most important and of greatest practical implication is the difference between locations (means) of the two data sets: 1.60 and 2.19 times larger mean for COVID vaccines compared to other drugs for ZANBI and ZAPIG, respectively. Such large difference in means, taken directly, shows that COVID vaccines, on average, cause much more AECs than the other drugs combined. This can be explained by the reactogenicity of vaccines, which is connected to a large-scale systemic immunological response. The vaccines against diseases other that COVID in the other drugs data probably have AECs comparable to those in COVID vaccines and this could be a subject for a further study.
Variance is larger and skewness is smaller in COVID vaccines approximately in the same ratio interval as the mean while excess kurtosis is 3.63 and 4.06 bigger in the other drugs for ZANBI and ZAPIG, respectively. These differences are carried over in the diagnostic residuals plots, especially in the autocorrelation (discussed above), density, and worm plots.
Fixed effects (i.e., the effects of explanatory variables on the response variable AEC that are established in the full models) are practically important and the theoretical conclusion outlined above that they are robust in relation to contaminating distributions makes their estimates stable and accurate. We chose to estimate the effects of five explanatory variables: three factors (Severity, Vaccine, Sex) and two covariates (Age, Sequence_number) because these are more complete and easily processed from the data. Age and Sex are the most frequently used demographic characteristics; the Vaccine factor with levels Comirnaty, Spikevax, Vaxzevria and Jcovden is estimated only in COVID vaccine data; and the Sequence_number used in combination with one or more factors gives a time-dependent multiple regression.
The factor Severity with two levels (Severe, Non-Severe) in the data refers for the whole record and not for each AE contained in the record. Since patients submit about 90% of the reports, this explanatory variable suffers from high subjectivity. Despite this limitation, the presence of the Severity estimates for each report makes it possible to conclude about the correlation between event severity and AEC number. The following results in our study point unambiguously to a positive correlation between AEC and severity:
- (1)
In the total other drug reports, 45.28% were reported as severe, but this percentage rose to 75.08% in the AEC outliers (7–9 AEC) and 84.68% in the extreme points ( AEC).
- (2)
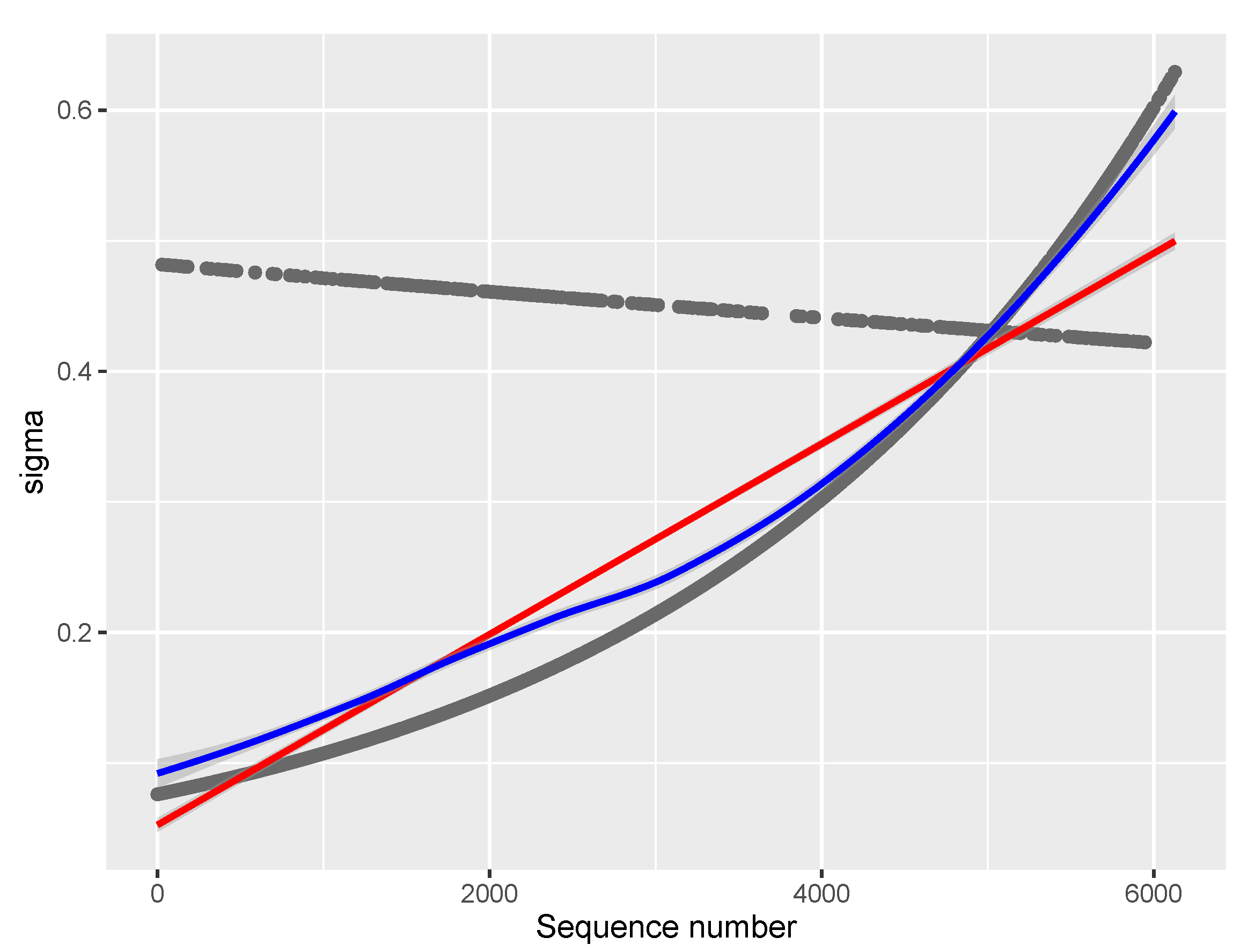
Severity has a significant positive effect on and for COVID vaccine data, as well as on in other drugs data. The latter effect interacts with report Sequence: severe AEs contribute most to in the beginning with a small linear decline while non-severe increase to become dominant factor for toward the end of the period.
- (3)
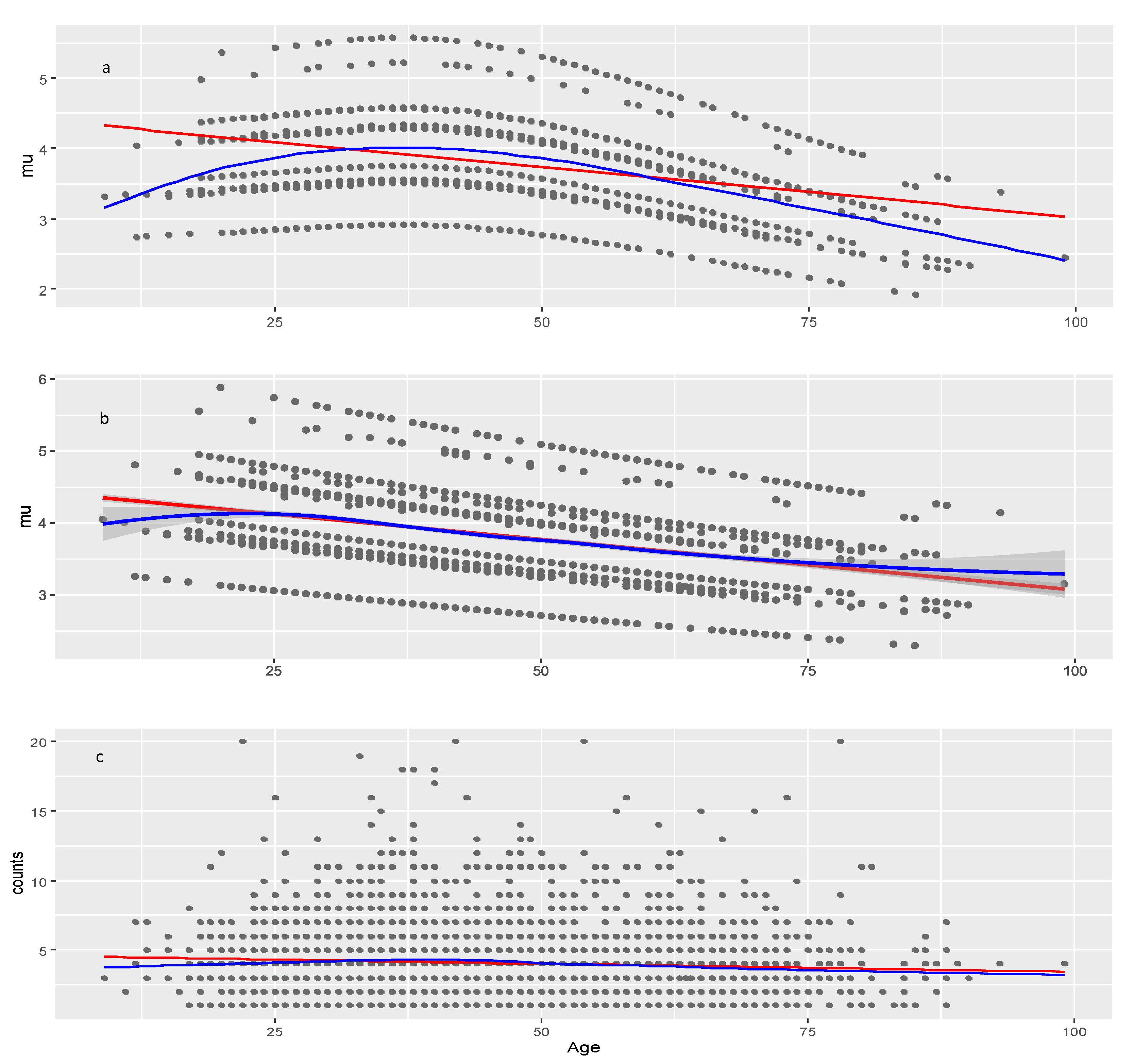
In the age-dependent multiple regression plot for in COVID vaccines, severity was found to be the most important factor determining the high AEC number.
While further research is needed to fully understand the relationship between AEC and AE severity, these findings have important implications for pharmacovigilance. They give another meaning for AEC, which can be looked at as a proxy for AE severity in addition to its primary meaning as a measure for the multiplicity of symptoms.
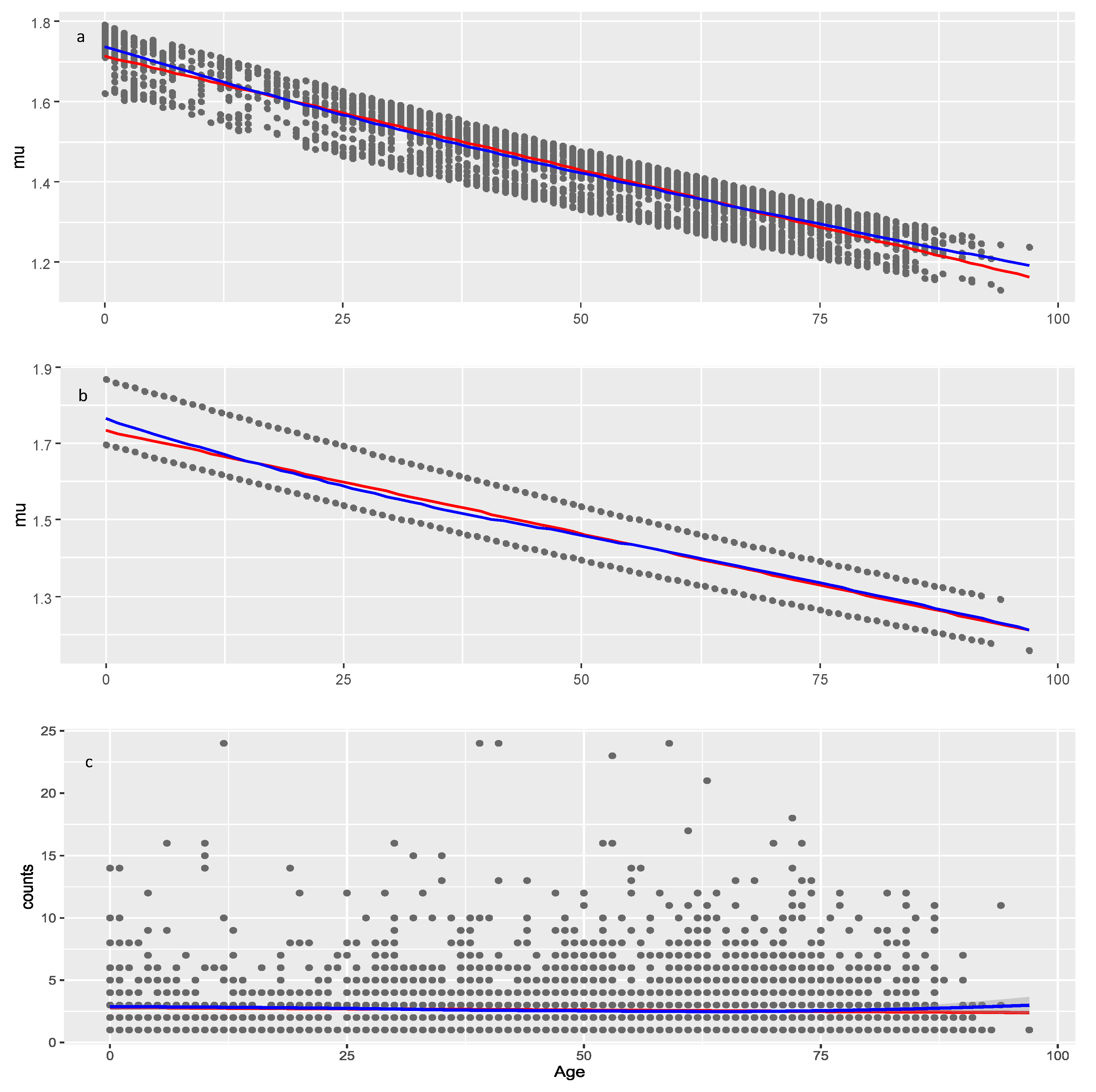
The effect of Age on
is negative and significant in both data sets suggesting that younger individuals may be more likely to experience numerous adverse events compared to older individuals, which pertains not only to COVID vaccines but to all medicinal products. Lack of model improvement with smoothers on Age shows that decrease of AEC with age is approximately linear. The significance of Age is seen from the result that on average it decreases AEC with about 1.3 (4.4–3.1) and 0.51 (1.72–1.21) in COVID vaccines and other drugs, respectively (red line on
Figure 5b and
Figure 7b). The multiple regressions on these figures show also that the Age effect is strongly modified by the different levels of the factors Severity, Sex, and Type of Vaccine in COVID vaccines and less affected by Severity in other drugs. The slopes of all lines in these multiple regressions are approximately equal so they do not intersect which means that there are no interactions among factors and between any of the factors and the Age covariate. This is confirmed in the full model table (
Table 6), in which there are no significant interactions for the
parameter.
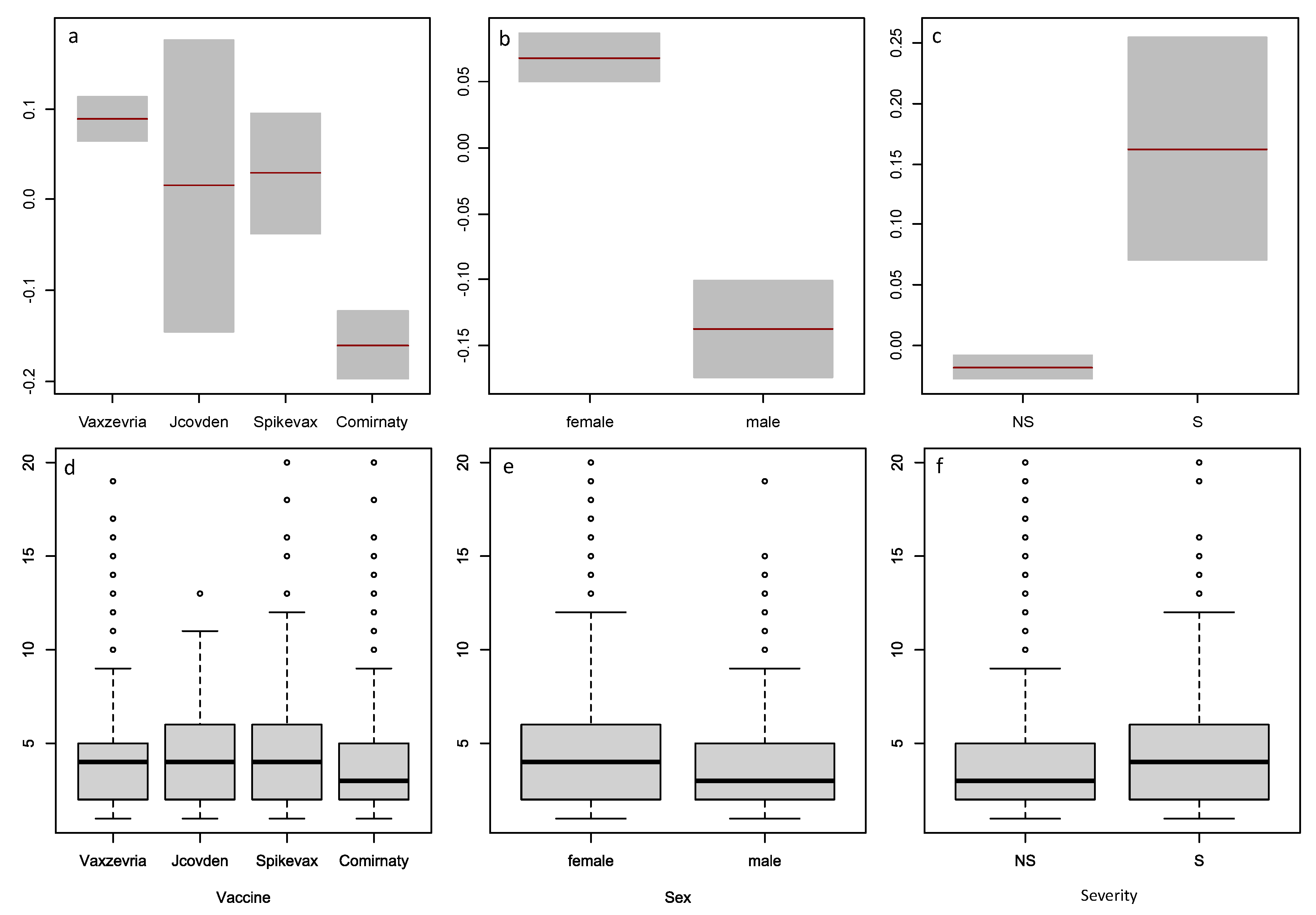
Regarding the effect of Sex, males have significantly less AEC than females with COVID vaccines while this effect is not significant with the other drugs data. The higher number of reports from women supports the finding that females have more AEC (last column of
Table 7).
The type of COVID vaccine also has an effect on
for COVID vaccine data. Taking Vaxzevria as a reference level, only Comirnaty causes significantly lower number of AEC. The other two vaccines, Spikevax and Jcovden, are not significantly different from Vaxzevria. No significant interactions of Vaccines’ type with other factors were observed which can be seen from the non-intersecting lines in
Figure 5.
When using parametric statistics assuming normality, the raw AEC data and the modeled values of the parameter have different means and standard deviations across the subgroups of vaccine type, sex, and AE severity. The raw data have higher means and larger variability than the modeled values, which may reflect the overdispersion and skewness of the data. The modeled values are based on the ZANBI distribution, which accounts for zero-inflation and excess dispersion in count data. The results also indicate that using parametric statistics to compare the raw data and the modeled values may not be appropriate, as the last assume a normal or symmetric distribution, which is not the case for the raw data.
With our data, both modeling and non-parametric methods lead to similar conclusions about the effects of vaccine type, sex, and severity on AEC. Vaxzevria, Jcovden, and Spikevax have higher AEC than Comirnaty; women and severe case reports have higher AEC than men and non-severe cases. However, these effects are more significant in the model than in the box plots, as shown by the confidence intervals and p-values. This suggests that the model is more sensitive and precise than the non-parametric statistics in detecting differences between subgroups.
We have modeled
in addition to
in the NB- and PIG-based models. This approach provides a more flexible model than one assuming a constant dispersion parameter
. It allows for the possibility that the dispersion of the dependent variable may vary according to the values of one or more covariates. For example, it may be that the variance of the dependent variable increases or decreases as the value of an explanatory variable increases. Modeling
can also improve the fit and efficiency of the model, as well as provide more accurate standard errors and confidence intervals for the coefficients. Heller et al. [
45] in a simulation study confirmed that estimates in the
model could be severely biased if the
model is misspecified. However, using the (
) parametrization,
model estimates are robust to misspecification of the
model. This potentially has implications not only for PIG regression, but also for regression models for any response distribution, in which the scale parameter is not orthogonal to the mean.
3.1. Limitations
We prefer referring to the independent variables as “explanatory variables” rather than “predictory variables” because our modeled data have a limited predictive power. On one hand, the sample size is insufficient to establish permanent trends and differences in levels of explanatory variables, and, on the other, data are local pertaining to only one country (Bulgaria) with its specifics in health system, hospital care, vaccination coverage and profile of vaccinated population, ethnic and sex specifics, etc.
Some groups are too small to infer conclusions and their modeled values are imbalanced such as persons immunized with Jcovden and Spikevax and their subgroups. Furthermore, even trends obtained on groups with moderate size, such as the gradual decrease of AEC with age for all groups, may not occur in other countries or in summary AE databases, such as EudraVigilance or VAERS because of the above mentioned specifics of our modeled data.
The choice of groups (explanatory variables) was limited by their availability in the data. The set of explanatory variables used in the present study are clearly insufficient to encompass all the dependencies and interactions existing in the data. These may possibly be explained or modified by including additional explanatory variables such as dose sequence (first vs. second vs. booster doses), reporter (patient, medical personnel), medical office visits, hospitalized (yes, no), time after immunization, immunization venue, etc.
The causal link between AEC and AE severity remains unclear due to the lack of research on the topic. The present paper found a tentative positive correlation between self-reported AE severity and AEC number. However, due to the above data limitations and the uncertainty in the severity designation, these results are to be considered as preliminary. When speaking about this correlation, we are fully aware that it holds in statistical sense, i.e., only if averaged on a large number of reports. This certainly does not exclude cases of a single AE that is more severe than multiple AEs in different organs and systems. Although severity in the other drugs data correlated with higher AEC, this result was not significant presumably because of the high variability generated by the great variety of drugs used. Mixed-effects models in which the drugs are modeled as a random effect can be used for a further study. Better conclusions could be made from clinical studies where the severity of each AE can be assessed objectively by one or more skilled physicians.
These different limitations and the circumstance that this is the first model using AEC number as a dependent variable makes this study more a proof-of-concept than a systematic investigation of all factors and covariates affecting the AEC number.
Figure 1.
Observed and theoretical distributions from the parametric models. The panels are: (a) ZANBI on COVID vaccines data; (b) ZAPIG on COVID vaccines data; (c) ZANBI on other drugs data; (d) ZAPIG on other drugs data.
Figure 1.
Observed and theoretical distributions from the parametric models. The panels are: (a) ZANBI on COVID vaccines data; (b) ZAPIG on COVID vaccines data; (c) ZANBI on other drugs data; (d) ZAPIG on other drugs data.
Figure 2.
Diagnostic plots for the residuals of the parametric distributions. First row (a, b, c, d): ZANBI on COVID vaccines data; second row (e, f, g, h): ZAPIG on COVID vaccines data; third row (i, k, l, m): ZANBI on other drugs data; fourth row (n, o, p, q): ZAPIG on other drugs data. Plots are the following types: first column (a, e, i, n): autocorrelation function (ACF); second column (b, f, k, o): probability density function (pdf); third column (c, g, l, p): Q-Q plots; fourth column (d, h, m, q): worm plots (black points) with bootstrap (gray points).
Figure 2.
Diagnostic plots for the residuals of the parametric distributions. First row (a, b, c, d): ZANBI on COVID vaccines data; second row (e, f, g, h): ZAPIG on COVID vaccines data; third row (i, k, l, m): ZANBI on other drugs data; fourth row (n, o, p, q): ZAPIG on other drugs data. Plots are the following types: first column (a, e, i, n): autocorrelation function (ACF); second column (b, f, k, o): probability density function (pdf); third column (c, g, l, p): Q-Q plots; fourth column (d, h, m, q): worm plots (black points) with bootstrap (gray points).
Figure 3.
Diagnostic plots for the residuals of the GAMLSS models with explanatory variables. First row (
a,
b,
c,
d): ZANBI on COVID vaccines data; second row (
i,
k,
l,
m): ZANBI on other drugs data. Plots are the following types: first column (
a,
i): autocorrelation function (ACF); second column (
b,
k): probability density function (pdf); third column (
c,
l): Q-Q plots; fourth column (
d,
m): worm plots (black points) with bootstrap (gray points). Letters for plots in this figure correspond to those in
Figure 2.
Figure 3.
Diagnostic plots for the residuals of the GAMLSS models with explanatory variables. First row (
a,
b,
c,
d): ZANBI on COVID vaccines data; second row (
i,
k,
l,
m): ZANBI on other drugs data. Plots are the following types: first column (
a,
i): autocorrelation function (ACF); second column (
b,
k): probability density function (pdf); third column (
c,
l): Q-Q plots; fourth column (
d,
m): worm plots (black points) with bootstrap (gray points). Letters for plots in this figure correspond to those in
Figure 2.
Figure 4.
Comparison of the factors Vaccine, Sex, and Severity by non-parametric statistics and from estimates for in Model 15.6. Variables are: Vaccine (a and d), Sex (b and e), Severity (c and f); term plots are top row (a, b, c), box plots are bottom row (d, e, f).
Figure 4.
Comparison of the factors Vaccine, Sex, and Severity by non-parametric statistics and from estimates for in Model 15.6. Variables are: Vaccine (a and d), Sex (b and e), Severity (c and f); term plots are top row (a, b, c), box plots are bottom row (d, e, f).
Figure 5.
Models of the parameter for the COVID vaccine data. a, model with penalized B-splines on age (pb(Age)); b, model linear in age (Model 15.6); c, raw data (observed AER counts). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Figure 5.
Models of the parameter for the COVID vaccine data. a, model with penalized B-splines on age (pb(Age)); b, model linear in age (Model 15.6); c, raw data (observed AER counts). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Figure 6.
Model of the parameter for the COVID vaccine data (Model 15.6). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Figure 6.
Model of the parameter for the COVID vaccine data (Model 15.6). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Figure 7.
Models of the parameter for the other drugs data. a, model with two covariates: Age and Sequence_number (29.4.1); b, multiple regression model (29.4) with covariate Age and factor Seriousness; c, raw data (observed AER counts). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Figure 7.
Models of the parameter for the other drugs data. a, model with two covariates: Age and Sequence_number (29.4.1); b, multiple regression model (29.4) with covariate Age and factor Seriousness; c, raw data (observed AER counts). Red line: linear least squares regression; blue line: loess (a non-linear regression smoother) applied on raw or modeled data at the moment of plotting.
Table 1.
Distributions with the smallest information criteria
Table 1.
Distributions with the smallest information criteria
| COVID vaccines |
|---|
| Information criteria |
ZANBI *
|
ZAPIG |
DEL |
SI |
SICHEL |
| Akaike ( AIC) |
17235.9 |
17246.74 |
17635.49 |
17658.71 |
17658.71 |
| Bayes ( BIC) |
17254.8 |
17265.58 |
17657.04 |
17677.55 |
17685.83 |
| other drugs |
| Information criteria |
ZAPIG |
ZANBI |
LG |
ZALG |
ZIPF |
| Akaike ( AIC) |
22877.62 |
22909.63 |
23127.03 |
23129.03 |
24771.36 |
| Bayes ( BIC) |
22898.13 |
22930.14 |
23133.86 |
23142.70 |
24778.19 |
Table 2.
Parameters and moments from the parametric models
Table 2.
Parameters and moments from the parametric models
| Parameter/moment |
COVID vaccines |
Other drugs |
| |
ZANBI |
ZAPIG |
ZANBI |
ZAPIG |
|
3.61265 |
3.68168 |
1.15929 |
1.58990 |
|
0.29823 |
0.27991 |
1.76746 |
1.07132 |
|
1.57e-10 |
3.87e-09 |
1.60e-10 |
1.60e-10 |
|
c coefficient |
1.09429 |
1.00000 |
2.13765 |
1.05786 |
| mean |
3.95328 |
3.68168 |
2.47816 |
1.68190 |
| variance |
6.86600 |
7.47576 |
4.28757 |
4.39195 |
| skewness |
1.29504 |
1.31084 |
2.36928 |
2.60040 |
| excess kurtosis |
2.32031 |
2.88184 |
8.42981 |
11.69541 |
Table 3.
Moments and goodness of fit for the diagnostic residuals distributions
Table 3.
Moments and goodness of fit for the diagnostic residuals distributions
| Moment |
COVID vaccines |
Other drugs |
| |
ZANBI |
ZAPIG |
ZANBI |
ZAPIG |
| mean |
– 0.00313 |
0.00145 |
0.00214 |
0.00489 |
| variance |
1.01015 |
1.01015 |
0.98431 |
0.99056 |
| skewness |
– 0.02186 |
– 0.03680 |
0.07862 |
0.02204 |
| kurtosis |
3.10751 |
2.92447 |
3.28073 |
3.13219 |
| Filliben coefficient |
0.9989902 |
0.9989915 |
0.9990338 |
0.9996147 |
Table 4.
Steps in the selection process of GAMLSS models with explanatory variables
Table 4.
Steps in the selection process of GAMLSS models with explanatory variables
| Model *
|
Terms **
|
BIC |
Distribution |
Parameter |
Converge |
| COVID vaccines |
| Model 1 |
1 |
14651.28 |
ZANBI |
|
Yes |
| Model 2 |
1 |
14659.57 |
ZAPIG |
|
Yes |
| Model 3 |
2 |
14642.24 |
ZANBI |
|
Yes |
| Model 4 |
2 |
14642.67 |
ZAPIG |
|
Yes |
| Model 5 |
1, 2 |
14644.42 |
ZANBI |
|
Yes |
| Model 6 |
1, 2 |
14644.59 |
ZAPIG |
|
Yes |
| Model 7 |
1, 2, 1×2 |
14652.17 |
ZANBI |
|
Yes |
| Model 8 |
1, 2, 1×2 |
14650.66 |
ZAPIG |
|
Yes |
| Model 9 |
3 |
14611.18 |
ZANBI |
|
Yes |
| Model 10 |
3 |
14647.72 |
ZAPIG |
|
Yes |
| Model 11 |
2, 3 |
14602.40 |
ZANBI |
|
Yes |
| Model 12 |
2, 3 |
14682.75 |
ZAPIG |
|
No |
| Model 13 |
2, 3, 2×3 |
14609.92 |
ZANBI |
|
Yes |
| Model 14 |
2, 3, 4 |
14583.51 |
ZANBI |
|
Yes |
| Model 15 |
2, 3, 4, 5 |
14553.20 |
ZANBI |
|
Yes |
| Model 16 |
2, 3, 4, 5, 2×4 |
14560.68 |
ZANBI |
|
Yes |
| Model 17 |
2, 3, 4, 5, 4×5 |
14576.83 |
ZANBI |
|
Yes |
| Model 18 |
pb(2), 3, 4, 5 |
14556.41 |
ZANBI |
|
Yes |
| Model 19 |
cs(2,df=2), 3, 4, 5 |
14557.85 |
ZANBI |
|
Yes |
| Model 15.1 |
1 |
14509.69 |
ZANBI |
|
Yes |
| Model 15.2 |
1, 2 |
14510.32 |
ZANBI |
|
Yes |
| Model 15.3 |
1, 3 |
14516.84 |
ZANBI |
|
Yes |
| Model 15.4 |
1, 4 |
14504.85 |
ZANBI |
|
Yes |
| Model 15.5 |
1, 4, 5 |
14508.38 |
ZANBI |
|
Yes |
| Model 15.6 |
1+4+1×4 |
14500.97 |
ZANBI |
|
Yes |
| Other drugs |
| Model 20 |
1 |
18036.65 |
ZANBI |
|
No |
| Model 21 |
1 |
19610.37 |
ZAPIG |
|
No |
| Model 22 |
2 |
18032.81 |
ZANBI |
|
No |
| Model 23 |
2 |
18114.44 |
ZAPIG |
|
Yes |
| Model 24 |
3 |
18039.84 |
ZANBI |
|
No |
| Model 25 |
3 |
18042.80 |
ZAPIG |
|
No |
| Model 26 |
4 |
17899.01 |
ZANBI |
|
No |
| Model 27 |
4 |
20016.47 |
ZAPIG |
|
No |
| Model 28 |
1, 2 |
18036.91 |
ZANBI |
|
No |
| Model 29 |
2, 4 |
17885.05 |
ZANBI |
|
Yes |
| Model 30 |
2, 3, 4 |
17893.51 |
ZANBI |
|
Yes |
| Model 31 |
1, 2, 4 |
17890.33 |
ZANBI |
|
Yes |
| Model 32 |
pb(2), 4 |
17894.02 |
ZANBI |
|
Yes |
| Model 33 |
cs(2,df=2), 4 |
17893.08 |
ZANBI |
|
No |
| Model 29.1 |
1 |
17888.98 |
ZANBI |
|
Yes |
| Model 29.2 |
2 |
17882.25 |
ZANBI |
|
No |
| Model 29.3 |
3 |
17891.85 |
ZANBI |
|
Yes |
| Model 29.4 |
4 |
17861.46 |
ZANBI |
|
Yes |
| Model 29.5 |
2, 4 |
17865.46 |
ZANBI |
|
No |
| Model 29.4.1 |
1, 2 |
17859.86 |
ZANBI |
|
Yes |
Table 5.
Moments and goodness of fit for the diagnostics of the best full models
Table 5.
Moments and goodness of fit for the diagnostics of the best full models
| Moment |
COVID vaccines |
Other drugs |
| |
(Model 15.4) |
(Model 29.4.1) |
| mean |
– 0.00574 |
0.00187 |
| variance |
1.01269 |
1.01149 |
| skewness |
– 0.00122 |
– 0.01752 |
| kurtosis |
3.16622 |
3.15641 |
| Filliben coefficient |
0.9990017 |
0.9995885 |
Table 6.
Statistics of the best full models for COVID vaccines and other drugs
Table 6.
Statistics of the best full models for COVID vaccines and other drugs
| |
Estimate |
Std. Error |
t value |
p value |
Significance |
| |
Model 15.6 (COVID vaccines) |
|
parameter
|
|
|
|
|
|
| Intercept |
1.68278 |
0.04475 |
37.608 |
<2e–16 |
*** |
| Age |
– 0.00487 |
0.00093 |
– 5.243 |
1.68e–07 |
*** |
| Sex: male |
– 0.20626 |
0.02737 |
– 7.535 |
6.28e–14 |
*** |
| Severity: Severe |
0.18047 |
0.05422 |
3.329 |
0.000883 |
*** |
| Vaccine: Jcovden |
– 0.07376 |
0.07277 |
– 1.014 |
0.310837 |
|
| Vaccine: Spikevax |
– 0.06029 |
0.04124 |
– 1.462 |
0.143916 |
|
| Vaccine: Comirnaty |
– 0.24956 |
0.02962 |
– 8.426 |
<2e–16 |
*** |
|
parameter
|
|
|
|
|
|
| Intercept |
– 2.575e+00 |
1.021e–01 |
– 25.214 |
<2e–16 |
*** |
| Sequence_number |
3.447e–04 |
5.753e–05 |
5.992 |
2.29e–09 |
*** |
| Seriousness: Serious |
1.846e–00 |
2.276e–01 |
8.109 |
7.11e–16 |
*** |
| Sequence_number×Serious |
– 3.670e–04 |
1.487e–04 |
– 2.468 |
0.0136 |
* |
| |
Model 29.4.1 (other drugs) |
|
parameter
|
|
|
|
|
|
| Intercept |
6.936e–01 |
1.046e–01 |
6.630 |
3.71e–11 |
*** |
| Age |
– 3.806e–03 |
8.279e–04 |
– 4.598 |
4.38e–06 |
*** |
| Sequence_number |
– 9.303e–06 |
5.834e–06 |
– 1.595 |
0.111 |
|
|
parameter
|
|
|
|
|
|
| Intercept |
– 0.39184 |
0.06498 |
– 6.03 |
1.75e–09 |
*** |
| Severity: Severe |
1.15604 |
0.08258 |
14.00 |
<2e–16 |
*** |
Table 7.
Parametric statistics of the factors that significantly affect
Table 7.
Parametric statistics of the factors that significantly affect
| Levels |
Raw data |
Modeled values |
N |
| |
Mean |
Std. deviation |
Mean |
Std. deviation |
|
| Vaxzevria |
4.25460 |
2.66433 |
4.08621 |
0.58845 |
1740 |
| Jcovden |
4.37323 |
2.76218 |
4.01120 |
0.57521 |
142 |
| Spikevax |
4.31649 |
2.64401 |
3.98038 |
0.59216 |
376 |
| Comirnaty |
3.65524 |
2.65239 |
3.29001 |
0.59439 |
1050 |
| Female |
4.29031 |
2.65346 |
4.03942 |
0.59382 |
2208 |
| Male |
3.64727 |
2.65225 |
3.37426 |
0.59445 |
1100 |
| Non-severe |
4.00973 |
2.65287 |
3.76593 |
0.59432 |
2979 |
| Severe |
4.68085 |
2.64759 |
4.29183 |
0.59326 |
1540 |
| Total |
4.07648 |
2.65287 |
3.81823 |
0.59432 |
3308 |
Table 8.
Factor level combinations for the lines in the
parameter in
Figure 5 for the non-linear model of COVID vaccine data
Table 8.
Factor level combinations for the lines in the
parameter in
Figure 5 for the non-linear model of COVID vaccine data
| Line No |
Age |
Vaccine |
Sex |
Seriousness |
|
Repeats |
| 1 |
50 |
Vaxzevria |
female |
S |
5.37038 |
2 |
| 2 |
49 |
Jcovden |
female |
S |
5.08064 |
1 |
| 3 |
62 |
Spikevax |
female |
S |
4.55633 |
1 |
| 4 |
48 |
Vaxzevria |
male |
S |
4.46271 |
3 |
| 5 |
50 |
Vaxzevria |
female |
NS |
4.34896 |
19 |
| 6 |
32 |
Jcovden |
male |
S |
4.32715 |
1 |
| 7 |
47 |
Spikevax |
male |
S |
4.21830 |
2 |
| 8 |
50 |
Comirnaty |
female |
S |
4.18823 |
1 |
| 9 |
50 |
Jcovden |
female |
NS |
4.09347 |
1 |
| 10 |
50 |
Spikevax |
female |
NS |
4.08612 |
3 |
| 11 |
50 |
Vaxzevria |
male |
NS |
3.56603 |
10 |
| 12 |
51 |
Comirnaty |
male |
S |
3.40940 |
1 |
| 13 |
50 |
Comirnaty |
female |
NS |
3.39165 |
13 |
| 14 |
50 |
Jcovden |
male |
NS |
3.35653 |
1 |
| 15 |
50 |
Spikevax |
male |
NS |
3.35050 |
1 |
| 16 |
50 |
Comirnaty |
male |
NS |
2.78106 |
4 |
Table 9.
Overdispersion in the parametric and full models with different distributions
Table 9.
Overdispersion in the parametric and full models with different distributions
| Model |
Data |
Distribution |
Pearson disp. statistic |
p-value |
| Parametric |
COVID vaccines |
Poisson |
1.512 |
0.0000 |
| Parametric |
other drugs |
Poisson |
1.368 |
0.0000 |
| Parametric |
COVID vaccines |
ZAP |
1.704 |
0.0000 |
| Parametric |
other drugs |
ZAP |
1.852 |
0.0000 |
| Parametric |
COVID vaccines |
NBI |
0.937 |
0.9955 |
| Parametric |
other drugs |
NBI |
0.842 |
1.0000 |
| Parametric |
COVID vaccines |
PIG |
0.920 |
0.9996 |
| Parametric |
other drugs |
PIG |
0.766 |
1.0000 |
| Parametric |
COVID vaccines |
ZANBI |
1.022 |
0.1803 |
| Parametric |
other drugs |
ZANBI |
0.997 |
0.5606 |
| Parametric |
COVID vaccines |
ZAPIG |
1.063 |
0.0062 |
| Parametric |
other drugs |
ZAPIG |
0.997 |
0.5529 |
| Model 15.6 |
COVID vaccines |
Poisson |
1.437 |
0.0000 |
| Model 29.4.1 |
other drugs |
Poisson |
1.360 |
0.0000 |
| Model 15.6 |
COVID vaccines |
ZAP |
1.641 |
0.0000 |
| Model 29.4.1 |
other drugs |
ZAP |
1.843 |
0.0000 |
| Model 15.6 |
COVID vaccines |
NBI |
0.937 |
0.9954 |
| Model 29.4.1 |
other drugs |
NBI |
0.862 |
1.0000 |
| Model 15.6 |
COVID vaccines |
PIG |
0.914 |
0.9998 |
| Model 29.4.1 |
other drugs |
PIG |
0.818 |
1.0000 |
| Model 15.6 |
COVID vaccines |
ZANBI |
1.019 |
0.2243 |
| Model 29.4.1 |
other drugs |
ZANBI |
1.001 |
0.4706 |
| Model 15.6 |
COVID vaccines |
ZAPIG *
|
1.961 |
0.0000 |
| Model 29.4.1 |
other drugs |
ZAPIG *
|
4.154 |
0.0000 |