1. Introduction
Interest and curiosity for the predictability of human lifespan date back to ancient times. The ability of rare individuals to reach advanced age, and even to live 100 years or more, is already attested in the writings of the Roman era, Middle Ages and Renaissance as well as nowadays [
1,
2]. However, the interest is the systematic study of centenarians, regarded as a model of successful aging, emerged and grew just a few decades ago, fueling the hope of discovering major biological factors affecting the human life span. In laboratory animals, due to their relatively shorter lifespan compared to humans, it is easier to study causal factors of longevity under controlled conditions. The role of genes especially attracted great interest, and the combined efforts of numerous international research teams have ascertained that the heritable component of lifespan is nearly 70% in the yeasts [
3], 25-50% in nematodes, flies, and mice [
4]. There are no
a priori reasons why this should not apply to humans whose genome harbors many genes orthologous to those identified in animals (for a review see [
5]). During the 1990s the heritability of human longevity was estimated through twins and families studies to be nearly 30% [
6,
7,
8], hence most of the lifespan variance appears clearly non-heritable. Nonetheless, a large body of research in ethnically different populations sought to identify “longevity genes” with the hope of manipulating them through pharmacological interventions in order to ensure a longer lifespan. In the first decade of this century, the diffusion of genome-wide association studies (GWASs), driven by the identification of millions of single nucleotide polymorphisms (SNPs) across the human genome, dramatically changed the scenario [
9]. The basic assumption is that a relatively small number of these SNPs can capture common genetic variation in long-lived and short-lived individuals via linkage disequilibrium. In this type of studies, major drawbacks were the small sample size of centenarian cohorts usually available, especially men, and the wide genetic variability of most populations where centenarians come from, which may generate a considerable genetic background noise. The first problem has been partially solved by creating international consortia recruiting a large number of long-lived individuals [
10,
11], and more recently, by studying centenarians' offspring [
12,
13], relatively more numerous and easier to recruit, who show a higher probability to become long-lived themselves compared to the general population. On the other hand, the problem of background genetic variability is more challenging and may more easily be tackled with the study of genetically homogeneous populations. This is the main reason for the present investigation.
Over the past twenty years, various genetic studies have been carried out on oldest Sardinians in relation to longevity: initially, case-control studies compared centenarians from all Sardinian villages with sexagenarians recruited in surrounding areas in relation to polymorphisms of p53 [
14], cytokines [
15,
16], HLA [
17,
18,
19], PON1 [
20], and the Y chromosome [
21]. None of these investigations revealed a significant divergence in the frequency of these variants from that of the younger control population. Gradually, the idea came up that the population of centenarians from the whole island could be still too heterogeneous to be informative. In addition, another source of confounding may be that a sizable proportion of centenarians who have lived for most of their existence in a rural environment often spend the final part of their existence in an urban environment, e.g. at their children's home, adopting habits that may somewhat influence the phenotype derived from their genetic endowment. As a matter of fact, the ideal approach would be to study only a carefully selected population from a geographically restricted area where longevity can manifest in a context of homogenous traditional living conditions. In 1999 a population living in “Ogliastra”, a rural subregion of the Mediterranean island of Sardinia, Italy ‒ whose genetic homogeneity is even higher than that of the population residing in the rest of the island ‒ was reported as one with the highest proportion of people living into their nineties, and centenarians were not rare [
22,
23]. This area was later dubbed Longevity Blue Zone (LBZ) due to the color used in the early geographical maps of longevity [
23]. Moreover, the centenarians in Sardinia, unlike most western populations, include a significantly greater proportion of men [
23]. The community living in this area exhibits both extreme genetic and cultural homogeneity providing a natural model to study the genetic factors of longevity, minimizing the effects of confounding.
Common Genetic Variants Associated with Human Longevity
Genetic variants involved in several metabolic pathways, and already reported to be associated with human longevity are listed in
Table 1 followed by a short background in the domain of longevity research.
Apolipoprotein E (APOE)
This 3 kb‒long gene is located in chromosome 19q13.32 and the exon 4 harbors two missense variants,
rs429358 (chr19:44908684 T>C, p.C112R) and
rs7412 (chr19:44908822 C>T, p.R158C). Different combinations of these SNPs define three major haplotypes, i.e. APOE ε3 (T at
rs429358 and C at
rs7412), APOE ε2 (T and T, respectively), and APOE ε4 (C and C, respectively), corresponding to isoform proteins differing from each other in amino acid substitutions at residues 112 and 158. In 1995 Schächter et al. reported a very low frequency of the ε4 allele in French centenarians [
24] prompting replication studies in several centenarians’ populations (reviewed by [
26]).
Angiotensin I Converting Enzyme (ACE1)
ACE1 is a 21 kb‒long gene, mapping in chromosome 17q23.3, contains the intron variant:
rs1799752 (chr17:63488530–63488543) i.e. an insertion/deletion (Ins/Del) polymorphism in intron 16 due to an Alu repetitive sequence. The Del/Del genotype is associated with increased activity of the ACE1 compared to the Ins/Ins genotype [
28]. The ACE Del-allele frequency was reported to increase in centenarians by Schächter [
24].
Interleukin 6 (IL-6)
The 6 kb‒long gene maps on chromosome 7p15.3 and codes for the pleiotropic cytokine IL-6, a mediator in the inflammatory response [
29]. The ‒174C allele of the functional
rs1800796 SNP within the IL-6 promoter, a minor allele in the Western population but a major allele in the Asian population according to the HapMap database, is associated with lower IL-6 plasma concentration compared with allele ‒174G carriers and was reported to be significantly increased in Italian centenarians [
30].
Forkhead Box O3A (FOXO3A)
FOXO3A, encoded by a gene mapping on chromosome 6q21, is a variant of the family of forkhead transcription factors, characterized by the presence of a fork-shaped molecular domain, which allows these proteins to interact with DNA [
38]. At the cellular level, it plays an important role in the regulation of gluconeogenesis and glycogenolysis, a protective role against oxidative stress; it is involved in cell proliferation, differentiation, cell cycle survival and quiescence, apoptosis, DNA repair, inhibition and promotion of differentiation, immune cell regulation, carcinogenesis, and stem cell maintenance [
31]. FOXO3A, in particular, has been shown to be associated with longevity in humans, in various populations [
39,
40] similar to the homologous gene Daf-16 in the nematode
Caenorhabditis elegans and dFOXO in the fruit fly
Drosophila melanogaster.
KLOTHO
Klotho is a transmembrane protein coded by the KL gene in chromosome 13q13.1, which, among various functions, controls the sensitivity of tissues to insulin [
33]. In humans, the protein is especially expressed at the level of the choroidal plexuses of the brain and in the distal tubule of the kidney. Some genetic variants of this protein appear to be involved in aging and loss of bone tissue; in fact, transgenic mice overexpressing the protein live longer than control mice [
34]. Conversely, mice with Klotho deficiency exhibit a syndrome that simulates accelerated human aging and develops rapidly progressive arteriosclerosis [
35]. Human heterozygous carriers for the KL ‒ VS polymorphism of the Klotho gene have an increased probability of survival into old age [
41] and a meta-analysis confirmed the increased longevity, albeit modest, in male carriers of the KL-VS variant [
42].
Glucose-6-Phosphate Dehydrogenase (G6PD)
The glucose-6-phosphate dehydrogenase (G6PD) is a “housekeeping” enzyme of the pentose phosphate pathway, which provides the coenzyme nicotinamide-adenine dinucleotide phosphate (NADPH) and ribose needed to synthesize DNA [
43]. Loss-of-function mutations in the G6PD gene, encoded on the Xq28 chromosome, lead to glutathione depletion and impaired anti-oxidant defense. Although G6PD deficiency has been reported in association with increased cardiovascular disease [
44,
45,
46] it is believed to reduce the risk of developing cancer both in vitro [
47] and in vivo [
48]. The frequency of G6PD deficiency was reported to be increased in Sardinian centenarians, especially in hemizygous males [
36,
37].
The purpose of the present study was to investigate the contribution of common gene variants previously associated to longevity with the exceptional longevity of Sardinian LBZ nonagenarians living in this area.
2. Materials and Methods
Study Population
The study participants were residents in 6 mountain villages (average altitude 649 mt a.s.l.; total population approximately 12,000) comprising a total area of 888 sq. km. Overall, they form a community isolated for centuries, living from seasonally-mobile pastoralism and subsistence farming until the post-WWII period. Many of the oldest inhabitants still live traditionally including food choices and physical activity. The six villages are those with the highest longevity rate within the Sardinian LBZ. From the six villages, a total of 331 subjects aged 90 years or older were considered eligible and 150 of them (45%) accepted to participate in the study. Eighty-nine were females and 61 were males. The response rate was relatively low as most nonagenarians lived with their children who were sometimes reluctant to grant an interview perceived as stressful for their relatives. The clinical and socio-demographic characteristics of the study participants have been previously reported [
49].
At the end of a home interview, about 10 mL of venous blood was collected in ethylenediamine tetraacetic acid (EDTA) for the extraction of genomic DNA. Routine hematological and clinical chemistry testing to ascertain the state of health were also performed. DNA was extracted from leukocytes by a salting-out procedure within 24 hours from the blood sample collection and resuspended in TE buffer. The control group consisted of an equal number of DNA samples from sex-matched subjects retrieved out of 633 unrelated subjects living in the four provinces of the island and assessed in the hospital reference Laboratory. More specifically, after stratifying the samples according to the province of residence, those belonging to the province of Nuoro (which comprises the LBZ) were chosen, excluding those older than 65 years or resident in any of the 6 municipalities of the LBZ to avoid a consanguinity bias with the long-lived involved in the study.
Genotyping
Selected markers are reported in
Table 1: APOE, including the three promoter polymorphisms APOE ‒491 (
rs449647), APOE ‒427 (
rs769446), and APOE ‒219 (
rs405509), the ACE1 Ins/Del gene polymorphism at intron 16, IL-6 –174 C/G polymorphism and one SNP within the FOXO3A gene, namely
rs2802292. DNA amplification by PCR was performed in a volume of 25μl containing 1μl of genomic DNA, 0.25μl of mix dNTPs, 3.75μl ddH
2O, and 5μl TaqMan Universal PCR Master Mix (Termo Fisher Scientific
®, Monza, Italy). The PCR cycle profile differed according to each genetic marker; APOE ε2/ε3/ε4 genotype was determined on DNA samples by a PCR-based method [
50]. ACE Ins/Del polymorphism was genotyped by direct PCR amplification of intron 16 [
28]. To avoid misclassification of Ins/Del and Del/Del genotypes a second amplification with the insertion-specific primer was performed in all Del/Del samples of the first amplification [
51]. The IL-6 -174G/C genotype was determined with the method of Olomolaiye et al. [
52]. The FOXO3A SNP has been genotyped by DNA sequencing methods using VIC and FAM as fluorescent tags using the Applied Biosystems 310 genetic analyzer.
Statistical Analysis
The LBZ samples were compared with the control group, to test for any difference in genotype/allele distribution. The statistical analysis was performed by the Pearson χ² test to evaluate differences between genotype and allele frequencies as well as to test for agreement of data with Hardy-Weinberg equilibrium (HWE), by using SPSS 16.0 (Chicago, IL, USA). P-values were corrected for multiple comparisons by Bonferroni analysis (P=0.05/number of comparisons), yielding a new P-value (P<0.007, number of comparisons =10). To estimate the strength of the longevity association, the P-value and the odds ratios (ORs) with 95% confidence interval (CI) were estimated. Haplotype analysis of the APOE locus was performed using Haploview 4.2 software.
3. Results
Table 2 displays the genotype and allele frequencies of the analyzed markers. All genotypes were within the limits of the Hardy-Weinberg equilibrium (P>0.05). None of the polymorphisms showed a statistically significant difference between long-livers and controls except for IL-6 –174GC which showed in LBZ agers a slightly increased frequency of the CC genotype (14.7 vs 8.7, n.s.), associated with a blunted inflammatory response. Of note, the frequency of the APOE ε4 allele among LBZ older individuals was similar to controls although no ε4/4 homozygotes were detected.
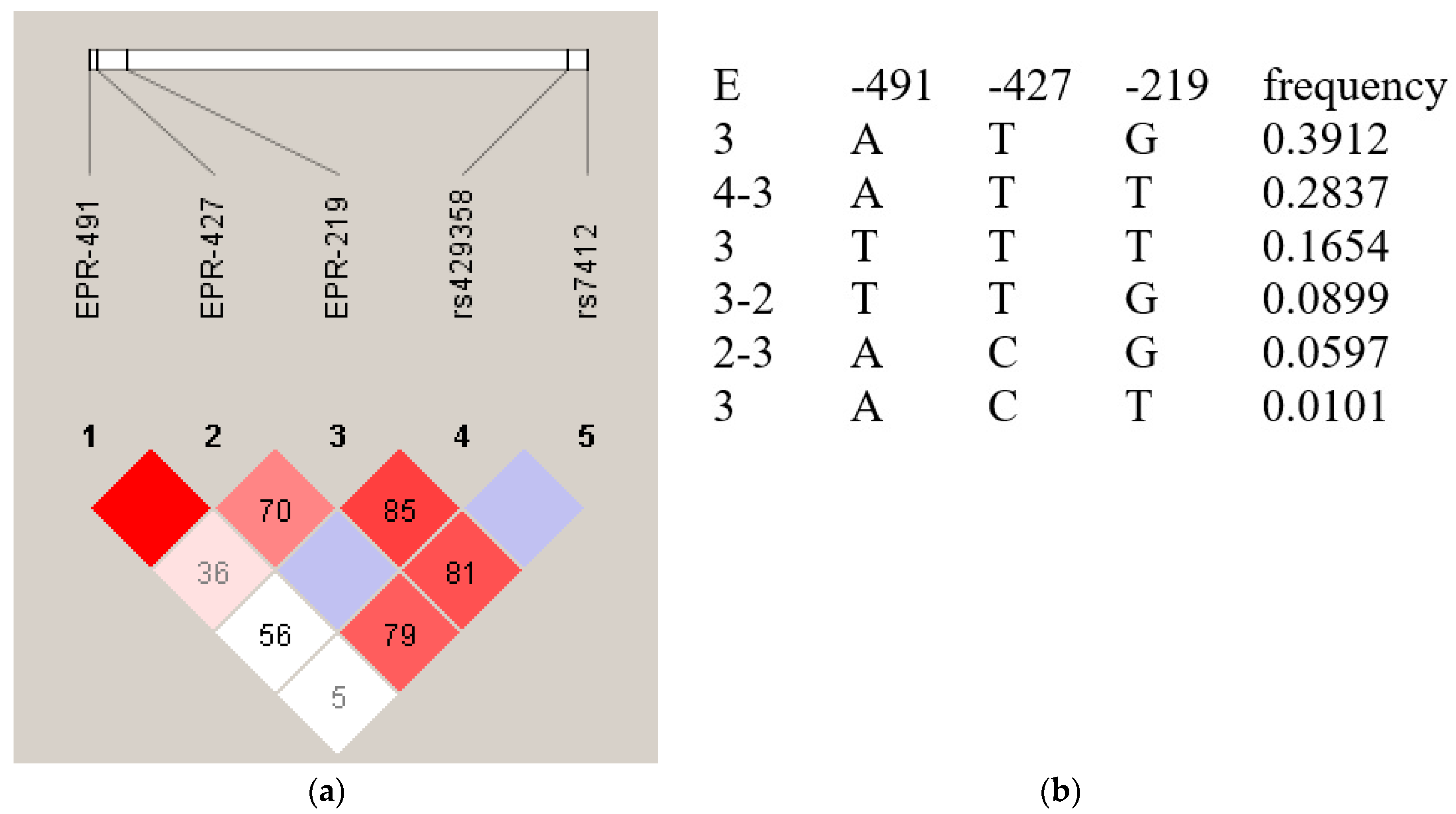
The distribution of genotypes and alleles in the promoter region of the APOE gene did not reach any significant statistical difference between the long-lived compared to the control group. Based on the frequencies of the haplotypes (reported in
Figure 1) it appears that the most frequent haplotype (0.3912) in positions ‒491, ‒427, and ‒219, was A‒T‒G, in linkage with the ε3 allele at the coding region, followed by the haplotype A‒T‒T (0.2837) in linkage with the ε4 allele. Notably, the frequencies of the two haplotypes did not differ in the two groups analyzed, therefore a significant role in the longevity of the Sardinian LBZ population cannot be attributed to the APOE locus as previously reported in other populations [
32] and even in Sardinia in a numerically smaller sample [
20].
Similarly, the frequency of the G allele of the FOXO3A gene did not show any difference in long-livers and younger controls.
Finally, in the G6PD locus mapping to the X chromosome, only 4 carriers of G6PD mutations were found (three mutations c.563C→T defining the
Mediterranean variant, and one mutation c.1342 A→G named
Sant’Antioco) out of a total of 150 informative DNA samples, likely associated with a reduction in enzymatic activity. This at most corresponds to a slight reduction in the frequency of mutated alleles in the long-lived group, consistent with epidemiological data which did not suggest a selection of the mutation in the older age groups [
53] compatible with the recently ascertained role of functional mutations in the G6PD locus as cardiovascular risk factors [
45].
4. Discussion
The population living in the inner region of Sardinia is known both for its exceptional longevity and for the long period of isolation, leading to a divergence of many gene variants from the European average frequencies. This distinct genetic background may suggest that such genetic structure could favor longevity. In the present work, the association of Sardinian LBZ longevity with known variants previously studied in other long-lived populations was investigated through 11 genetic markers. For most of the markers, the frequencies of the genotypes and alleles in the control group did not differ significantly from those recorded in other Western populations, including mainland Italy, avoiding the risk of result misinterpretation due to Sardinian peculiarity. In addition, the homogeneity of this population is very high, minimizing the weight of confounding effects given by socioeconomic or environmental factors. The sample was sized enough reducing type two error bias.
No significant differences in the distribution of genetic variants emerged between LBZ and the general population of Sardinia that may account for the increased survival in the LBZ. In particular, no significant role of APOE was found. The APOE has been investigated in several populations of long-livers showing variable and sometimes conflicting results [
25]. For example, the frequency of APOE ε4 (which is the ancestral haplotype) is the lowest among Sardinians (5%) and the highest among African Pygmies (41%) [
54]. Similarly to our study, [
55] reported no significant effect of APOE ε2ε2 on human longevity in Spanish subjects analyzed. On the contrary a meta-analysis by Sebastiani et al. including seven case-control studies (28,297 participants) reported a marginally significant association of ε2ε2 with human longevity (OR=2.39, 95% CI: 0.99, 5.76) [
56]. Since the promoter region of the APOE gene contains functional polymorphisms, in the present study we explored their association with LBZ longevity. The polymorphisms are
rs449647 (-491A/T),
rs769446 (-427T/C), and
rs405509 (-219T/G) which are in linkage disequilibrium with the polymorphisms of the coding region rs429358 and rs7412: the frequency of each of them has been determined as well as that of haplotypes. Again, the analysis of APOE promoter polymorphisms variability did not reveal any association with LBZ longevity.
The FOXO3A variant rs2802292 has been reported in association with longevity in the Okinawa population [
39]. However, in a previous study conducted on a small number of LBZ subjects [
20], no association was detected between FOXO3A polymorphisms and longevity. The present study confirmed these results. It is noteworthy that neither of the two markers is consistently associated with longevity in replication studies, i.e. APOE and FOXO3A display differences in the Sardinian population, despite the probability of finding a difference of at least eight percentage points with a statistical power higher than 80% and around proportions of about 10%.
Regarding the ACE1 locus, a meta-analysis by Garatchea et al. examined 12 studies and reported a modest, albeit significant, positive association of the ACE1 Del allele and Del/Del genotype with exceptional longevity [
57], an apparently counterintuitive finding considering that the ACE1 Del-allele is usually associated with higher circulating levels of the potent vasoconstrictor angiotensin II [
58].
Regarding the inflammation genes, a meta-analysis of three Italian studies did not find a significant difference in the IL-6 genotype between the oldest old and controls, although a frequency of the IL-6 -174 GG genotype significantly lower than the other genotypes was detected, supporting a negative association between the GG genotype and longevity [
16]. A second meta-analysis of four studies covering a total of 2945 long-lived individuals and 2992 controls did not find evidence that the IL-6 G174C SNPs affect the probability of reaching an advanced age in Caucasians [
59]. In our study, we observed a weakly significant difference, i.e. a higher frequency of the homozygous ‒174CC genotype in LBZ nonagenarians, although, at the allelic level, the frequency of the C allele was not significantly higher than the G allele. These data suggest a modest role of the inflammatory genes in the Sardinia LBZ longevity, as described in centenarians of mainland Italy [
30] and in contrast to findings reported in populations with different genetic structures such as Finnish [
60]. Interestingly, a previous study that compared centenarians from all over the island with young controls living in the four provinces of the island did not reveal a significant association [
15]. This suggests that the genetic makeup of the oldest LBZ members may be partially different from that of the Sardinian long-lived persons as a whole, because of the different selection rates operating in the evolutionary history of the Sardinians by environmental factors. On a speculative level, because the Sardinian population was characterized until 1950 by a strong prevalence of malaria [
61], this infectious disease probably was able to select inflammatory alleles, and in particular, to induce an increased C-allele frequency at the position ‒174 in the IL-6 gene promoter. The selective pressure towards alleles able to induce a weaker inflammatory response protecting against severe forms of malaria may have contributed, through different pathways, to longevity avoiding chronic inflammatory disorders.
A trend of decrease in G6PD mutated allele frequency was also observed, with borderline significance. This finding is intriguing since this allele has been involved in cardiovascular disease [
45,
62], although some literature data have raised the possibility that G6PD deficiency could be favorable to longevity owing to the potential lifetime protection against certain types of cancers [
36,
37]. The findings of the present work do not seem to support a definite role of this marker in LBZ longevity.
5. Conclusions
In conclusion, the findings of the present work do not seem to support a definite role of the selected genetic markers with longevity in the Sardinian LBZ. The lack of a positive association, with the exception of IL-6, should be interpreted as a relatively low weight of genetic factors in the exceptional longevity of this region, not merely reflecting the particular choice of candidate genes.
Author Contributions
Conceptualization, A.E. and G.M.P.; methodology, G.M.P.; software, G.M.P.; validation, A.E.., G.M.P. and M.P.D.; formal analysis, G.M.P.; investigation, A.E..; resources, M.P.D.; data curation, G.M.P.; writing—original draft preparation, A.E., G.M.P. and M.P.D.; writing—review and editing, A.E., G.M.P. and M.P.D.; visualization, G.M.P..; supervision, M.P.D..; project administration, M.P.D.; funding acquisition, M.P.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethics Committee of “Azienda Ospedaliero-Universitaria di Sassari” (Prot. N. 136/CE, 9/2/2012).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data will be available upon specific request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Langner, G. Schätzung von Säuglingssterblichkeit und Lebenserwartung im Zeitalter des Imperium Romanum: methodenkritische Untersuchung. Historical Social Research 1998, 23, 299-326. [CrossRef]
- Floris, P.; Dore, M.P.; Pes, G.M. Does the longevity of the Sardinian population date back to Roman times? A comprehensive review of the available evidence. PLoS One 2021, 16, e0245006. [CrossRef]
- Stumpferl, S.W.; Brand, S.E.; Jiang, J.C.; Korona, B.; Tiwari, A.; Dai, J.; Seo, J.G.; Jazwinski, S.M. Natural genetic variation in yeast longevity. Genome Res 2012, 22, 1963-1973. [CrossRef]
- Finch, C.E.; Tanzi, R.E. Genetics of aging. Science 1997, 278, 407-411. [CrossRef]
- Giuliani, C.; Garagnani, P.; Franceschi, C. Genetics of Human Longevity Within an Eco-Evolutionary Nature-Nurture Framework. Circ Res 2018, 123, 745-772. [CrossRef]
- McGue, M.; Vaupel, J.W.; Holm, N.; Harvald, B. Longevity is moderately heritable in a sample of Danish twins born 1870-1880. J Gerontol 1993, 48, B237-244. [CrossRef]
- Herskind, A.M.; McGue, M.; Holm, N.V.; Sorensen, T.I.; Harvald, B.; Vaupel, J.W. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870-1900. Hum Genet 1996, 97, 319-323. [CrossRef]
- Pilling, L.C.; Atkins, J.L.; Bowman, K.; Jones, S.E.; Tyrrell, J.; Beaumont, R.N.; Ruth, K.S.; Tuke, M.A.; Yaghootkar, H.; Wood, A.R.; et al. Human longevity is influenced by many genetic variants: evidence from 75,000 UK Biobank participants. Aging (Albany NY) 2016, 8, 547-560. [CrossRef]
- Puca, A.A.; Daly, M.J.; Brewster, S.J.; Matise, T.C.; Barrett, J.; Shea-Drinkwater, M.; Kang, S.; Joyce, E.; Nicoli, J.; Benson, E.; et al. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci U S A 2001, 98, 10505-10508. [CrossRef]
- Skytthe, A.; Valensin, S.; Jeune, B.; Cevenini, E.; Balard, F.; Beekman, M.; Bezrukov, V.; Blanche, H.; Bolund, L.; Broczek, K.; et al. Design, recruitment, logistics, and data management of the GEHA (Genetics of Healthy Ageing) project. Exp Gerontol 2011, 46, 934-945. [CrossRef]
- Herr, M.; Jeune, B.; Fors, S.; Andersen-Ranberg, K.; Ankri, J.; Arai, Y.; Cubaynes, S.; Santos-Eggimann, B.; Zekry, D.; Parker, M.; et al. Frailty and Associated Factors among Centenarians in the 5-COOP Countries. Gerontology 2018, 64, 521-531. [CrossRef]
- Tan, Q.; Zhao, J.H.; Li, S.; Kruse, T.A.; Christensen, K. Power assessment for genetic association study of human longevity using offspring of long-lived subjects. Eur J Epidemiol 2010, 25, 501-506. [CrossRef]
- Bucci, L.; Ostan, R.; Cevenini, E.; Pini, E.; Scurti, M.; Vitale, G.; Mari, D.; Caruso, C.; Sansoni, P.; Fanelli, F.; et al. Centenarians' offspring as a model of healthy aging: a reappraisal of the data on Italian subjects and a comprehensive overview. Aging (Albany NY) 2016, 8, 510-519. [CrossRef]
- Bonafe, M.; Olivieri, F.; Mari, D.; Baggio, G.; Mattace, R.; Berardelli, M.; Sansoni, P.; De Benedictis, G.; De Luca, M.; Marchegiani, F.; et al. P53 codon 72 polymorphism and longevity: additional data on centenarians from continental Italy and Sardinia. Am J Hum Genet 1999, 65, 1782-1785. [CrossRef]
- Pes, G.M.; Lio, D.; Carru, C.; Deiana, L.; Baggio, G.; Franceschi, C.; Ferrucci, L.; Oliveri, F.; Scola, L.; Crivello, A.; et al. Association between longevity and cytokine gene polymorphisms. A study in Sardinian centenarians. Aging Clin Exp Res 2004, 16, 244-248. [CrossRef]
- Di Bona, D.; Vasto, S.; Capurso, C.; Christiansen, L.; Deiana, L.; Franceschi, C.; Hurme, M.; Mocchegiani, E.; Rea, M.; Lio, D.; et al. Effect of interleukin-6 polymorphisms on human longevity: a systematic review and meta-analysis. Ageing Res Rev 2009, 8, 36-42. [CrossRef]
- Lio, D.; Pes, G.M.; Carru, C.; Listi, F.; Ferlazzo, V.; Candore, G.; Colonna-Romano, G.; Ferrucci, L.; Deiana, L.; Baggio, G.; et al. Association between the HLA-DR alleles and longevity: a study in Sardinian population. Exp Gerontol 2003, 38, 313-317. [CrossRef]
- Scola, L.; Lio, D.; Candore, G.; Forte, G.I.; Crivello, A.; Colonna-Romano, G.; Pes, M.G.; Carru, C.; Ferrucci, L.; Deiana, L.; et al. Analysis of HLA-DRB1, DQA1, DQB1 haplotypes in Sardinian centenarians. Exp Gerontol 2008, 43, 114-118. [CrossRef]
- Scola, L.; Lio, D.; Crivello, A.; Candore, G.; Forte, G.I.; Colonna-Romano, G.; Pes, M.G.; Carru, C.; Ferrucci, L.; Deiana, L.; et al. Analysis of HLA-DQA, HLA-DQB frequencies in a group of Sardinian centenarians. Rejuvenation Res 2006, 9, 157-160. [CrossRef]
- Poulain, M.; Herm, A.; Errigo, A.; Chrysohoou, C.; Legrand, R.; Passarino, G.; Stazi, M.A.; Voutekatis, K.G.; Gonos, E.S.; Franceschi, C.; et al. Specific features of the oldest old from the Longevity Blue Zones in Ikaria and Sardinia. Mech Ageing Dev 2021, 198, 111543. [CrossRef]
- Passarino, G.; Underhill, P.A.; Cavalli-Sforza, L.L.; Semino, O.; Pes, G.M.; Carru, C.; Ferrucci, L.; Bonafe, M.; Franceschi, C.; Deiana, L.; et al. Y chromosome binary markers to study the high prevalence of males in Sardinian centenarians and the genetic structure of the Sardinian population. Hum Hered 2001, 52, 136-139. [CrossRef]
- Pes, G.M. The Sardinian Centenarian Study. In Proceedings of the Research Workshop on “Genes, Genealogy and Longevity”, Montpellier (France), 1999.
- Poulain, M.; Pes, G.M.; Grasland, C.; Carru, C.; Ferrucci, L.; Baggio, G.; Franceschi, C.; Deiana, L. Identification of a geographic area characterized by extreme longevity in the Sardinia island: the AKEA study. Exp Gerontol 2004, 39, 1423-1429. [CrossRef]
- Schächter, F.; Faure-Delanef, L.; Guenot, F.; Rouger, H.; Froguel, P.; Lesueur-Ginot, L.; Cohen, D. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet 1994, 6, 29-32. [CrossRef]
- Gerdes, L.U.; Jeune, B.; Ranberg, K.A.; Nybo, H.; Vaupel, J.W. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a "frailty gene," not a "longevity gene". Genet Epidemiol 2000, 19, 202-210. [CrossRef]
- Garatachea, N.; Marin, P.J.; Santos-Lozano, A.; Sanchis-Gomar, F.; Emanuele, E.; Lucia, A. The ApoE gene is related with exceptional longevity: a systematic review and meta-analysis. Rejuvenation Res 2015, 18, 3-13. [CrossRef]
- Feng, J.; Xiang, L.; Wan, G.; Qi, K.; Sun, L.; Huang, Z.; Zheng, C.; Lv, Z.; Hu, C.; Yang, Z. Is APOE epsilon3 a favourable factor for the longevity: an association study in Chinese population. J Genet 2011, 90, 343-347. [CrossRef]
- Tiret, L.; Rigat, B.; Visvikis, S.; Breda, C.; Corvol, P.; Cambien, F.; Soubrier, F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet 1992, 51, 197-205.
- Ershler, W.B.; Keller, E.T. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu Rev Med 2000, 51, 245-270. [CrossRef]
- Bonafè, M.; Olivieri, F.; Cavallone, L.; Giovagnetti, S.; Mayegiani, F.; Cardelli, M.; Pieri, C.; Marra, M.; Antonicelli, R.; Lisa, R.; et al. A gender--dependent genetic predisposition to produce high levels of IL-6 is detrimental for longevity. Eur J Immunol 2001, 31, 2357-2361. [CrossRef]
- Barthel, A.; Schmoll, D.; Unterman, T.G. FoxO proteins in insulin action and metabolism. Trends Endocrinol Metab 2005, 16, 183-189. [CrossRef]
- Revelas, M.; Thalamuthu, A.; Oldmeadow, C.; Evans, T.J.; Armstrong, N.J.; Kwok, J.B.; Brodaty, H.; Schofield, P.R.; Scott, R.J.; Sachdev, P.S.; et al. Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity. Mech Ageing Dev 2018, 175, 24-34. [CrossRef]
- Chen, G.; Liu, Y.; Goetz, R.; Fu, L.; Jayaraman, S.; Hu, M.C.; Moe, O.W.; Liang, G.; Li, X.; Mohammadi, M. alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 2018, 553, 461-466. [CrossRef]
- Dubal, D.B.; Zhu, L.; Sanchez, P.E.; Worden, K.; Broestl, L.; Johnson, E.; Ho, K.; Yu, G.Q.; Kim, D.; Betourne, A.; et al. Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J Neurosci 2015, 35, 2358-2371. [CrossRef]
- Kurosu, H.; Yamamoto, M.; Clark, J.D.; Pastor, J.V.; Nandi, A.; Gurnani, P.; McGuinness, O.P.; Chikuda, H.; Yamaguchi, M.; Kawaguchi, H.; et al. Suppression of aging in mice by the hormone Klotho. Science 2005, 309, 1829-1833. [CrossRef]
- Manganelli, G.; Masullo, U.; Passarelli, S.; Filosa, S. Glucose-6-phosphate dehydrogenase deficiency: disadvantages and possible benefits. Cardiovasc Hematol Disord Drug Targets 2013, 13, 73-82. [CrossRef]
- Schwartz, A.G.; Pashko, L.L. Dehydroepiandrosterone, glucose-6-phosphate dehydrogenase, and longevity. Ageing Res Rev 2004, 3, 171-187. [CrossRef]
- Martins, R.; Lithgow, G.J.; Link, W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 2016, 15, 196-207. [CrossRef]
- Willcox, B.J.; Donlon, T.A.; He, Q.; Chen, R.; Grove, J.S.; Yano, K.; Masaki, K.H.; Willcox, D.C.; Rodriguez, B.; Curb, J.D. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A 2008, 105, 13987-13992. [CrossRef]
- Flachsbart, F.; Caliebe, A.; Kleindorp, R.; Blanche, H.; von Eller-Eberstein, H.; Nikolaus, S.; Schreiber, S.; Nebel, A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A 2009, 106, 2700-2705. [CrossRef]
- Di Bona, D.; Accardi, G.; Virruso, C.; Candore, G.; Caruso, C. Association of Klotho polymorphisms with healthy aging: a systematic review and meta-analysis. Rejuvenation Res 2014, 17, 212-216. [CrossRef]
- Zhu, Z.; Xia, W.; Cui, Y.; Zeng, F.; Li, Y.; Yang, Z.; Hequn, C. Klotho gene polymorphisms are associated with healthy aging and longevity: Evidence from a meta-analysis. Mech Ageing Dev 2019, 178, 33-40. [CrossRef]
- Luzzatto, L.; Arese, P. Favism and Glucose-6-Phosphate Dehydrogenase Deficiency. N Engl J Med 2018, 378, 1068-1069. [CrossRef]
- Thomas, J.E.; Kang, S.; Wyatt, C.J.; Kim, F.S.; Mangelsdorff, A.D.; Weigel, F.K. Glucose-6-Phosphate Dehydrogenase Deficiency is Associated with Cardiovascular Disease in U.S. Military Centers. Tex Heart Inst J 2018, 45, 144-150. [CrossRef]
- Pes, G.M.; Parodi, G.; Dore, M.P. Glucose-6-phosphate dehydrogenase deficiency and risk of cardiovascular disease: A propensity score-matched study. Atherosclerosis 2019, 282, 148-153. [CrossRef]
- Parsanathan, R.; Jain, S.K. Glucose-6-phosphate dehydrogenase (G6PD) deficiency is linked with cardiovascular disease. Hypertens Res 2020, 43, 582-584. [CrossRef]
- Zhang, C.; Zhang, Z.; Zhu, Y.; Qin, S. Glucose-6-phosphate dehydrogenase: a biomarker and potential therapeutic target for cancer. Anticancer Agents Med Chem 2014, 14, 280-289. [CrossRef]
- Pes, G.M.; Errigo, A.; Soro, S.; Longo, N.P.; Dore, M.P. Glucose-6-phosphate dehydrogenase deficiency reduces susceptibility to cancer of endodermal origin. Acta Oncol 2019, 58, 1205-1211. [CrossRef]
- Pes, G.M.; Errigo, A.; Tedde, P.; Dore, M.P. Sociodemographic, Clinical and Functional Profile of Nonagenarians from Two Areas of Sardinia Characterized by Distinct Longevity Levels. Rejuvenation Res 2020, 23, 341-348. [CrossRef]
- Hixson, J.E.; Vernier, D.T. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res 1990, 31, 545-548.
- Ueda, S.; Heeley, R.P.; Lees, K.R.; Elliott, H.L.; Connell, J.M. Mistyping of the human angiotensin-converting enzyme gene polymorphism: frequency, causes and possible methods to avoid errors in typing. J Mol Endocrinol 1996, 17, 27-30. [CrossRef]
- Olomolaiye, O.; Wood, N.A.; Bidwell, J.L. A novel NlaIII polymorphism in the human IL-6 promoter. Eur J Immunogenet. 1998, 25.
- Pes, G.M.; Errigo, A.; Bitti, A.; Dore, M.P. Effect of age, period and birth-cohort on the frequency of glucose-6-phosphate dehydrogenase deficiency in Sardinian adults. Ann Med 2018, 50, 68-73. [CrossRef]
- Corbo, R.M.; Scacchi, R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a 'thrifty' allele? Ann Hum Genet 1999, 63, 301-310. [CrossRef]
- Garatachea, N.; Emanuele, E.; Calero, M.; Fuku, N.; Arai, Y.; Abe, Y.; Murakami, H.; Miyachi, M.; Yvert, T.; Verde, Z.; et al. ApoE gene and exceptional longevity: Insights from three independent cohorts. Exp Gerontol 2014, 53, 16-23. [CrossRef]
- Sebastiani, P.; Gurinovich, A.; Nygaard, M.; Sasaki, T.; Sweigart, B.; Bae, H.; Andersen, S.L.; Villa, F.; Atzmon, G.; Christensen, K.; et al. APOE Alleles and Extreme Human Longevity. J Gerontol A Biol Sci Med Sci 2019, 74, 44-51. [CrossRef]
- Garatachea, N.; Marin, P.J.; Lucia, A. The ACE DD genotype and D-allele are associated with exceptional longevity: a meta-analysis. Ageing Res Rev 2013, 12, 1079-1087. [CrossRef]
- Rigat, B.; Hubert, C.; Alhenc-Gelas, F.; Cambien, F.; Corvol, P.; Soubrier, F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 1990, 86, 1343-1346. [CrossRef]
- Wei, G.Z.; Wang, F.; Zhao, Y.G.; Li, S.S.; Shi, M.L.; Gao, K.; Luo, Y.; Tang, W.R. Association of longevity with TNF-alpha G308A and IL-6 G174C polymorphic inflammatory biomarkers in Caucasians: a meta-analysis. Z Gerontol Geriatr 2016, 49, 706-713. [CrossRef]
- Wang, X.Y.; Hurme, M.; Jylha, M.; Hervonen, A. Lack of association between human longevity and polymorphisms of IL-1 cluster, IL-6, IL-10 and TNF-alpha genes in Finnish nonagenarians. Mech Ageing Dev 2001, 123, 29-38. [CrossRef]
- Tognotti, E. Program to eradicate malaria in Sardinia, 1946-1950. Emerg Infect Dis 2009, 15, 1460-1466. [CrossRef]
- Dore, M.P.; Parodi, G.; Portoghese, M.; Pes, G.M. The Controversial Role of Glucose-6-Phosphate Dehydrogenase Deficiency on Cardiovascular Disease: A Narrative Review. Oxid Med Cell Longev 2021, 2021, 5529256. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).