1. Introduction
In recent years, low temperature disasters caused by global climate change have occurred frequently, seriously affecting the normal growth of crops. Low temperature stress causes a large accumulation of reactive oxygen species in plants, and then causes membrane lipid peroxidation, forming a large number of harmful substances malondialdehyde (Yan et al., 2016). A large number of studies have shown that as a micronutrient, selenium, especially as a component of the antioxidant glutathione peroxidase (GSH-Px), exogenous application of appropriate concentration of selenium fertilizer can participate in the removal of lipid hydrogen, hydrogen peroxide and other free radicals, alleviate the peroxidation damage caused by free radicals, and thus mitigate the adverse effects of low temperature on plant growth and development. Huang et al. (2018) studied the physiological indexes of strawberries under low temperature stress and found that selenium affected the ascorbic acid (ASA) -glutathione (GSH) cycle under low temperature environment to combat low temperature stress. Liu et al. (2021) found that selenium could not only enhance the cold resistance of tea plants, but also improve tea quality by promoting the accumulation of tea polyphenols, amino acids and sugars.
Selenium fertilizer mainly includes inorganic selenium, organic selenium and nano-selenium. Inorganic selenium fertilizers, such as sodium selenate or sodium selenite, exhibit low absorption and utilization conversion at low concentrations and high toxicity at high concentrations when applied through foliage or soil. As a nutrient element, the safe dose range of selenium is relatively narrow, and it is difficult to control its effective dosage (Wang et al., 2019). At present, organic selenium and nano-selenium fertilizers are mainly used. Nano-selenium refers to protein-dispersed selenium nanoparticles with particle size less than 60 nm, which have large specific surface area and high catalytic activity, biological function, antioxidant property and absorptivity. Research has indicated that inorganic selenium can not only enhance the effects of low temperature stress but also increase selenium content on plants; however, there is a scarcity of studies investigating the impacts of organic selenium and nano-selenium on crops.
The Suzhouqing variety is pak choi (Brassica chinensis var. pekinensis), belonging to the ordinary cabbage type. It possesses high nutritional value and ranks among the vegetables with high content of various vitamins and mineral elements. Moreover, it can effectively maintain blood vessel elasticity over an extended period of time. Additionally, this variety is rich in fiber, which aids in alleviating constipation and provides essential nutrients for normal physiological requirements while enhancing the body's innate immunity. In order to investigate whether organic selenium and nano-selenium can enhance low-temperature tolerance and increase selenium content in pak choi (Brassica chinensis var. pekinensis. cv. ‘Suzhouqing’), different concentrations (0, 5, 10, 20 mg L-1) of exogenous selenium were applied in this experiment to assess their effects on plant growth, nutritional quality and antioxidant properties.
In recent years, there has been an increasing number of studies utilizing RNA sequencing technology to investigate the effects of selenium on plants. Rao et al. (2020) integrated the sequencing results from PacBio SMRT and Illumina RNA-SEQ platforms, analyzing a total of 948 differential genes, out of which 11 were found to be associated with selenium metabolism in the selenium-accumulating plant Cardamine violifolia. They revealed candidate genes and pathways involved in selenium metabolism. Zhou et al. (2018) sequenced the RNA of Cardamine hupingshanensis seedlings after applying selenium and identified 25 genes that significantly responded to selenium stress, showing significant enrichment in four pathways. Guo et al. (2020) discovered structural genes, phosphate transporters, and sulfate transporters that may play a role in selenium metabolism when studying Pueraria lobata (Willd.) Ohwi's response to molecular-level selenium stimulation. Cao et al. (2018) observed that differential genes in tea trees responding to selenium stimulation were significantly enriched in four pathways.
Therefore, transcriptome sequencing analysis was conducted to identify differentially expressed genes and analyze enriched metabolic pathways associated with these genes. Ultimately, this study aims to reveal both physiological and biochemical mechanisms as well as molecular regulatory mechanisms underlying leaf responses to cold stress when applying selenium on leaf surfaces. These findings will provide a theoretical basis for future production strategies involving Suzhouqing.
2. Materials and methods
2.1. Plant materials and experimental conditions
The experiment was conducted on November 18, 2022, at the laboratory of Jiangxi Agricultural University in Nanchang, Jiangxi Province. The seeds were sown in a culture dish and allowed to grow for one week. Once the seedlings developed three leaves and a heart-shaped structure, they were transplanted into pot (33×24.4×13.5 cm) containing 320 g of soil for cultivation. Two seedlings were cultivated in each pot, and after 33 days (November 19, 2022) of growth, they were transferred to an artificial climate chamber for two weeks under low temperature stress conditions. The culture environment in the artificial climate chamber was set as follows: light intensity of 2000 lx with a photoperiod cycle of 12 hours light/12 hours dark, temperature maintained at 2℃, and humidity at 75%. For the experiment, organic selenium and nano-selenium fertilizers were used with varying concentrations: LO5 (organic selenium concentration of 5 mg L-1), LO10 (organic selenium concentration of 10 mg L-1), LO20 (organic selenium concentration of 20 mg L-1), LN5 (nano-selenium concentration of 5 mg L-1), LN10 (nano-selenium concentration of 10 mg L-1), LN20 (nano-selenium concentration of 20 mg L-1), and LCK (water, 5 mg L-1). The organic selenium fertilizer and nano-selenium fertilizer were applied three times throughout the entire growth period, specifically on December 9th, 16th, and 27th.
2.2. Determination of physiological and biochemical indexes
The chlorophyll a and b content, carotenoid content, superoxide dismutase (SOD) activity, peroxidase (POD) activity, malondialdehyde (MDA) content, reduced glutathione (GSH), soluble protein content and amino acid content were determined following Wang and Huang.'s study in 2015.
2.3. Transcriptomesequencing.
The total RNA was extracted using a plant total RNA extraction kit, and subsequently analyzed for RNA degradation and contamination through agarose-gel electrophoresis. The purity of the RNA was assessed using Nanodrop, while the concentration was accurately quantified with Qubit. Furthermore, the integrity of the RNA was precisely determined using Agilent 2100. Once qualified, the samples were subjected to Illumina sequencing, encompassing all eukaryotic RNAs. High-quality data facilitated splicing by Trinity, resulting in the longest transcript unigene that served as a reference sequence for subsequent analysis involving expression quantification and functional annotation.
3. Results
3.1. Photosynthetic pigments
After spraying different concentrations of organic selenium and nano-selenium on Suzhouqing leaves, the chlorophyll content of the leaves was measured and recorded in
Table 1. Compared to the control group, the treatments with LO5, LO10, and LO20 showed an increase in chlorophyll a content by 16.85%, 6.74%, and 71.91% respectively, with LO20 treatment exhibiting the most significant effect. Similarly, LN5, LN10, and LN20 treatments resulted in an increase in chlorophyll a content by 17.98%, 15.73%, and 11.24% respectively; among these treatments, LN10 had the highest chlorophyll a content observed. The application of LO5 led to a 4.44% increase in chlorophyll b content while LO20 exhibited a significant increase of 40%. Furthermore, carotenoid content increased significantly for all three treatments: LO5 (43.89%), LO10 (31.43%), and especially for LO20 (131%). Notably though there was no significant difference between carotenoid contents at concentrations LN10 and LN20 indicating that effective increases can be achieved even at lower concentrations.
3.2. Antioxidant properties of leaves
As illustrated in
Table 2, compared to the control group, Suzhouqing leaves treated with LO5, LO10, and LO20 exhibited a remarkable increase in SOD activity by 73.92%, 38.72%, and 341.16% respectively, with statistically significant differences observed; notably, the highest level was achieved under LO20 treatment conditions. In contrast, SOD activity displayed a trend of initial decline followed by subsequent elevation within LN5, LN10, and LN20 treatment groups. While LN5 treatment resulted in a significant decrease of 39.87% in SOD content compared to the control group, both LN10 and LN20 treatments led to substantial increases of 31.39% and 38.80% respectively; however, these differences were not statistically significant.
The POD activity of Suzuqingqing leaves treated with different concentrations of organic selenium is presented in
Table 2. In the LO5, LO10, and LO20 treatment groups, there was a significant increase in POD activity by 177.53%, 26.60%, and 100.06% respectively. In conclusion, appropriate concentration of organic selenium spray treatment can activate cellular antioxidant enzyme systems, enhance POD activity, improve stress resistance capabilities, and delay plant senescence. The table below illustrates the POD activity of leaves treated with different concentrations of nano-selenium. In LN5, LN10, and LN20 treatment groups, there was an initial decrease followed by an increase in POD activity trend observed. Compared to the control group, leaves treated with LN5 exhibited a significant decrease in POD content by 20.01%. However, leaves treated with LN10 and LN20 showed a significant increase in POD content by 24% and 31.69% respectively; among them, LN20 reached its maximum value.
As depicted in
Table 2, LO20 treatment demonstrated the most significant enhancement of CAT activity, exhibiting an increase of 85.96% compared to the control group. LO5 and LO10 treatments resulted in respective increases of 6.65% and 16.27% in CAT activity, with no significant difference observed for LO5 treatment. These findings indicate that under appropriate concentrations of organic selenium, CAT can effectively inhibit lipid peroxidation and stimulate antioxidant responses. In the LN5, LN10, and LN20 treatment groups, CAT activity exhibited a pattern of initial decrease followed by an increase before decreasing again. Compared to the control group, leaves treated with LN5 showed a notable decrease in CAT content by 30.40%, reaching statistical significance. Conversely, leaves treated with LN10 and LN20 displayed significant increases in CAT content by 69.11% and 27.77%, respectively; among them, LN10 achieved the highest value.
After the application of nano-selenium, the content of reduced glutathione (GSH) increased to 23.82%, 13.28%, and 4.98% respectively (
Table 2). In comparison with the control group, the GSH content sprayed with organic selenium was found to be 34.89%, 12.77%, and 9.55% respectively. Reduced glutathione serves as a crucial indicator for characterizing cellular antioxidant capacity, while its oxidation product is known as oxidized glutathione (GSSG). The increase in GSSG content indirectly suggests the presence of oxidative stress.
Table 2.
Antioxidant properties of green Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
Table 2.
Antioxidant properties of green Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
| Treatment |
LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| SOD activity (U g-1 fresh weight) |
286.76±
21.55d |
498.74±
16.02b |
397.78±
14.34c |
1265.06±
81.01a |
172.44±
14.78e |
376.78±
9.21c |
398.03±
22.41c |
| POD activity (U g-1 fresh weight) |
49.63±
3.16d |
137.74±
6.40a |
62.83±
3.78c |
99.29±
4.19b |
39.7±2.04e |
61.54±
2.09c |
65.36±
1.91c |
| CAT content (μmol min-1g-1 fresh weight) |
122.68±
7.78e |
130.84±
4.58de |
142.64±
4.32d |
228.14±
10.99a |
85.38±
4.22f |
207.46±
11.89b |
156.75±
8.74c |
| GSH content (μmol g-1 fresh weight) |
0.29±
0.01c |
0.33±
0.01b |
0.28±
0.02c |
0.46±
0.01a |
0.33±
0.01b |
0.35±
0.01b |
0.28±
0.01c |
3.4. Leaf weight and total selenium content
The leaf quality of individual plants is a crucial criterion for assessing the yield of green leaves in leaves. As depicted in
Table 3, in this experiment, the LO5, LO10, and LO20 treatments exhibited respective increases of 30.26%, 38.00%, and 0.95%. Similarly, the LN5, LN10, and LN20 treatments demonstrated increases of 16.19%, 28.66%, and 36.89% respectively. The total selenium content is presented in
Table 3, showing that spraying organic selenium and nano-selenium led to an increase in total selenium content. Specifically, the LO5, LO10, and LO20 treatments resulted in increases 80.00%, 190.00%, and 140.00% respectively; while the LN5 treatment showed an increase of 90.00%, followed by LN10 with an increase of 220.00% ,and finally LN20 with an increase of 160 .00%.
Table 3.
Weight and total selenium content of Suzhouqing leaves.
Table 3.
Weight and total selenium content of Suzhouqing leaves.
| Treatment |
LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| Weight (g fresh weight) |
46.27±
0.32c |
60.27±
0.27b |
65.09±
0.18b |
46.71±
0.24c |
53.76±
0.73a |
59.53±
0.82c |
63.34±
0.69b |
Total selenium content
(mg kg-1) |
0.001±
0.001b |
0.0018±
0.0003a |
0.0029±
0.0002b |
0.0024±
0.0003c |
0.0019±
0.0003b |
0.0032±
0.0001b |
0.0026±
0.0002a |
3.4. Nutritional quality of leaves
Soluble amino acids are the fundamental constituents of an organism's protein. As depicted in Table 4, compared to the control group, leaves treated with LO10 exhibited a significant decrease in amino acid content by 11.82%, while those treated with LO5 and LO20 showed significant increases by 19.09% and 32.27% respectively, with LO20 reaching the highest value. In the LN5, LN10, and LN20 treatment groups, soluble sugar displayed an initial increase followed by a subsequent decrease trend. LN5 treatment led to a significant increase in leaf amino acid content by 9.09%, whereas leaves treated with LN10 and LN20 experienced decreases of 7.73% and 4.55% respectively, both reaching significance levels as well. The findings indicate that high concentrations of nano-selenium inhibit amino acid content.
The soluble protein content in plants is often considered as a crucial indicator of plant oxidative senescence. A significant portion of the soluble protein found in mature plant leaves is ribulose diphosphate carboxylase (RuBPCase), which indirectly regulates RuBPCase levels, thereby enhancing photosynthetic capacity during later stages of growth and effectively delaying crop senescence. As shown in Table 4, treatment with LO5, LO10, and LO20 resulted in an increase of 17.51%, 22.46%, and 60.93% respectively in soluble protein content, with LO20 exhibiting the highest value at a statistically significant level. The increased soluble protein content also provided ample nitrogen sources for antioxidant enzyme synthesis, thereby enhancing the plant's ability to scavenge reactive oxygen species and delay leaf senescence while increasing yield potential. In the LN5, LN10, and LN20 treatment groups, there was an initial increase followed by a subsequent decrease observed in soluble protein levels over time. Leaves treated with LN5 showed an increase of 11.06% in soluble protein content while those treated with LN10 exhibited a substantial increase of 49.39%. Additionally, LN10 treatment demonstrated the highest level of soluble protein content among all treatments with significant differences.
Table 3.
Nutritional quality of Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
Table 3.
Nutritional quality of Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
| Treatment |
Amino acid content (mg g-1 fresh weight) |
Soluble protein (mg g-1 fresh weight) |
| LCK |
2.2±0.02d |
28.49±1.14f |
| LO5 |
2.62±0.01b |
33.48±0.54cd |
| LO10 |
1.94±0.03g |
34.89±0.44c |
| LO20 |
2.91±0.01a |
45.85±0.99a |
| LN5 |
2.4±0.03c |
31.64±1.07e |
| LN10 |
2.03±0.03f |
42.56±1.74b |
| LN20 |
2.1±0.02e |
32.97±1.23de |
3.5. Leaf gene expression
3.5.1. Sequencing results and quality analysis
The image data of sequenced fragments were converted into sequence data (reads) using CASAVA base recognition. Transcriptomic libraries were obtained for the control group (LCK), nano-selenium treatment groups (LN5, LN10, and LN20), and organic selenium treatment groups (LO5, LO10, and LO20). Trimmomatic software was utilized to filter the sequencing data, ensuring that over 99% of bases had a Q30 quality score. The sequencing error rate for all samples was less than 0.1%, indicating high quality results (
Table 5). These findings demonstrate that RNA-Seq sequencing exhibited excellent quality with randomized and evenly distributed clean reads. By comparing reads that matched unique transcripts, differential genes could be identified.
Table 5.
Sequencing data statistics.
Table 5.
Sequencing data statistics.
| Treatment |
Raw base(G) |
Raw sequences |
Clean bases(G) |
Clean sequences |
Q30(%) |
GC(%) |
Error
Rate (%) |
| LCK |
8.13 |
54217790 |
7.5 |
50702038 |
99.85 |
47.0(%) |
0.03 |
| LO5 |
7.74 |
51575524 |
7.2 |
48567534 |
99.86 |
47.0(%) |
0.03 |
| LN5 |
7.95 |
52979962 |
7.39 |
49869758 |
99.86 |
46.0(%) |
0.03 |
| LO10 |
6.47 |
43115830 |
5.99 |
40471030 |
99.85 |
47.0(%) |
0.03 |
| LN10 |
6.25 |
41655152 |
5.77 |
38959214 |
99.85 |
47.0(%) |
0.03 |
| LO20 |
6.71 |
44757082 |
6.25 |
42197232 |
99.87 |
46.0(%) |
0.03 |
| LN20 |
6.06 |
40381854 |
5.6 |
37912748 |
99.86 |
47.0(%) |
0.03 |
3.5.2. Analysis of differentially expressed genes
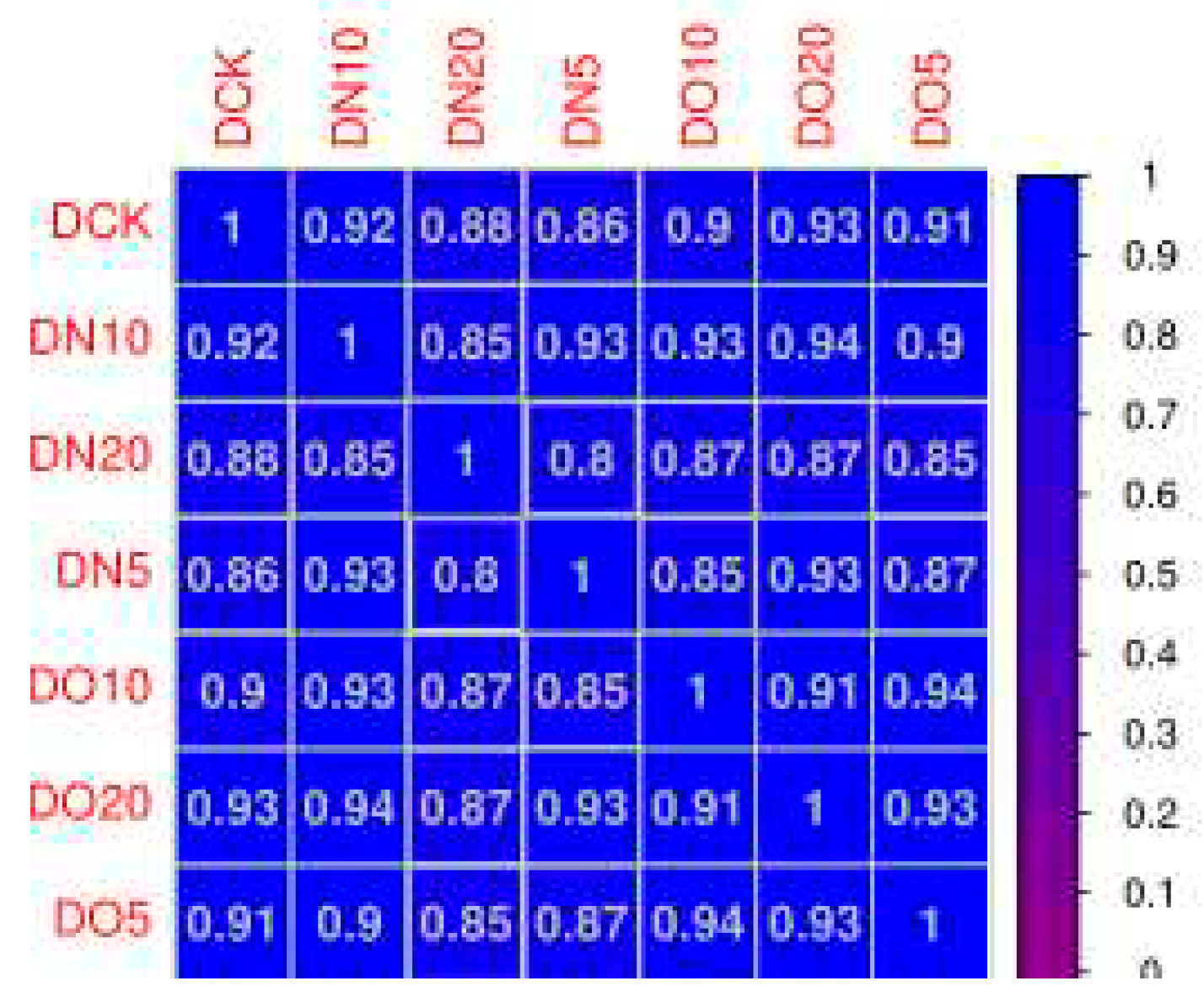
Correlation coefficients between groups were computed based on the TPM (Transcripts Per Kilobase Million) values of all genes in the sample. The results revealed that most Pearson correlation coefficients had an R
2 value greater than 0.8, indicating a strong correlation between gene expression levels across samples (
Figure 1).
The volcano plot of differentially expressed genes (
Figure 2) provides a visual representation of the overall distribution of these genes. The x-axis represents log
2(FoldChange), which indicates changes in gene expression multiples across different samples, while the y-axis represents log
10(padj), which reflects significance levels of expression differences.
3.5.3. Functional analysis of differentially expressed gene Gene Ontology (GO)
The functional significance enrichment analysis of GO identifies the significantly enriched GO functional items in the differentially expressed genes compared to the genomic background, thereby determining which biological functions are significantly correlated with these genes. Firstly, all differentially expressed genes were mapped to each term in the Gene Ontology database, and the gene count for each term was calculated. Subsequently, it was observed that the differentially expressed genes exhibited significant enrichment compared to the entire genome background. The enricher function of clusterProfiler utilizes a hypergeometric distribution test as part of its GO enrichment analysis method to identify significantly enriched GO Terms (P
adj≤0.05). Based on the results of this analysis, three categories of GO (biological process, cellular component and molecular function) were used to calculate separately the number of up-regulated genes, down-regulated genes, and total genes within each category (as shown in
Figure 3). In
Figure 3(a), metabolic process, plastid nucleoid, and alcohol binding had the highest number of up-regulated genes. In
Figure 3(b), chlorophyll metabolic process and regulation of dephosphorylation had a higher number of up-regulated genes along with tubulin complex and alcohol binding. In
Figure 3(c), regulation of dephosphorylation, cajal body, tubulin complex along with alcohol binding showed a higher count for up-regulated genes. Similarly in
Figure 3(d), metabolic process along with tubulin complex and alcohol binding displayed a higher count for up-regulated genes. In
Figure 3(e), metabolic process together with plastid nucleoid and phosphatase regulator activity exhibited a higher number for up-regulated genes. Lastly in
Figure 3(f), sesquiterpenoid metabolic process demonstrated the largest count for up-regulated genes followed by protein phosphatase type 2A complex and protein histidine kinase binding.
3.5.4. KEGG annotation and enrichment analysis of differentially expressed genes
(1) KEGG annotation of differentially expressed genes
Pathway enrichment analysis enables the identification of crucial biochemical metabolic pathways and signal transduction pathways associated with differentially expressed genes. KEGG (Kyoto Encyclopedia of Genes and Genomes) serves as the primary public database for Pathway information, integrating genome chemistry and system function data. The unit of analysis in pathway significant enrichment analysis is KEGG Pathway. A hypergeometric test was employed to identify pathways where differential genes were significantly enriched compared to all annotated genes. Similarly, a padj threshold less than 0.05 was used to determine significant enrichment in KEGG pathway analysis.
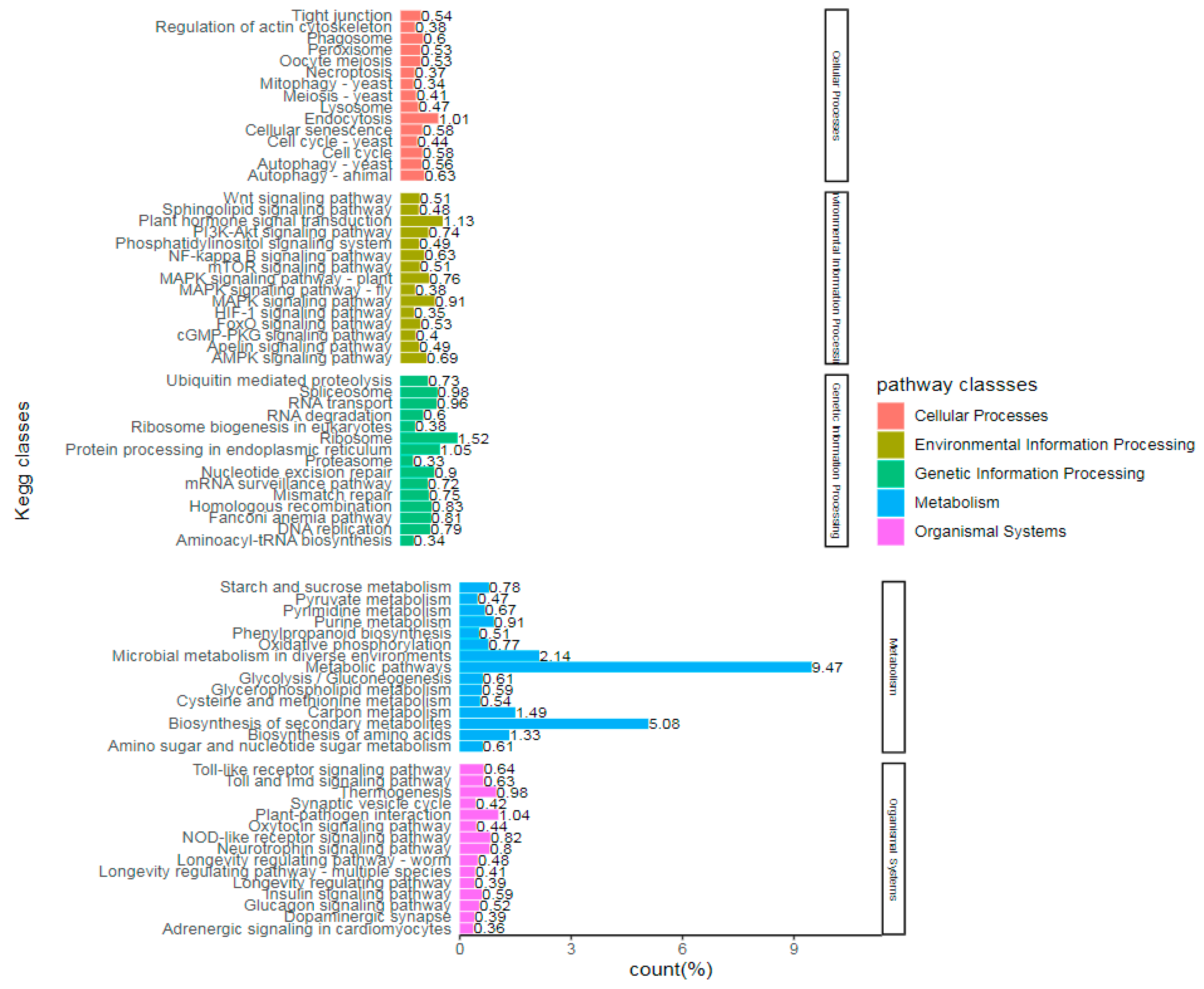
Figure 4 illustrates the proportion of annotated genes and included pathways by comparing unigene with the KEGG database.
(2) Enrichment of KEGG pathway diagram
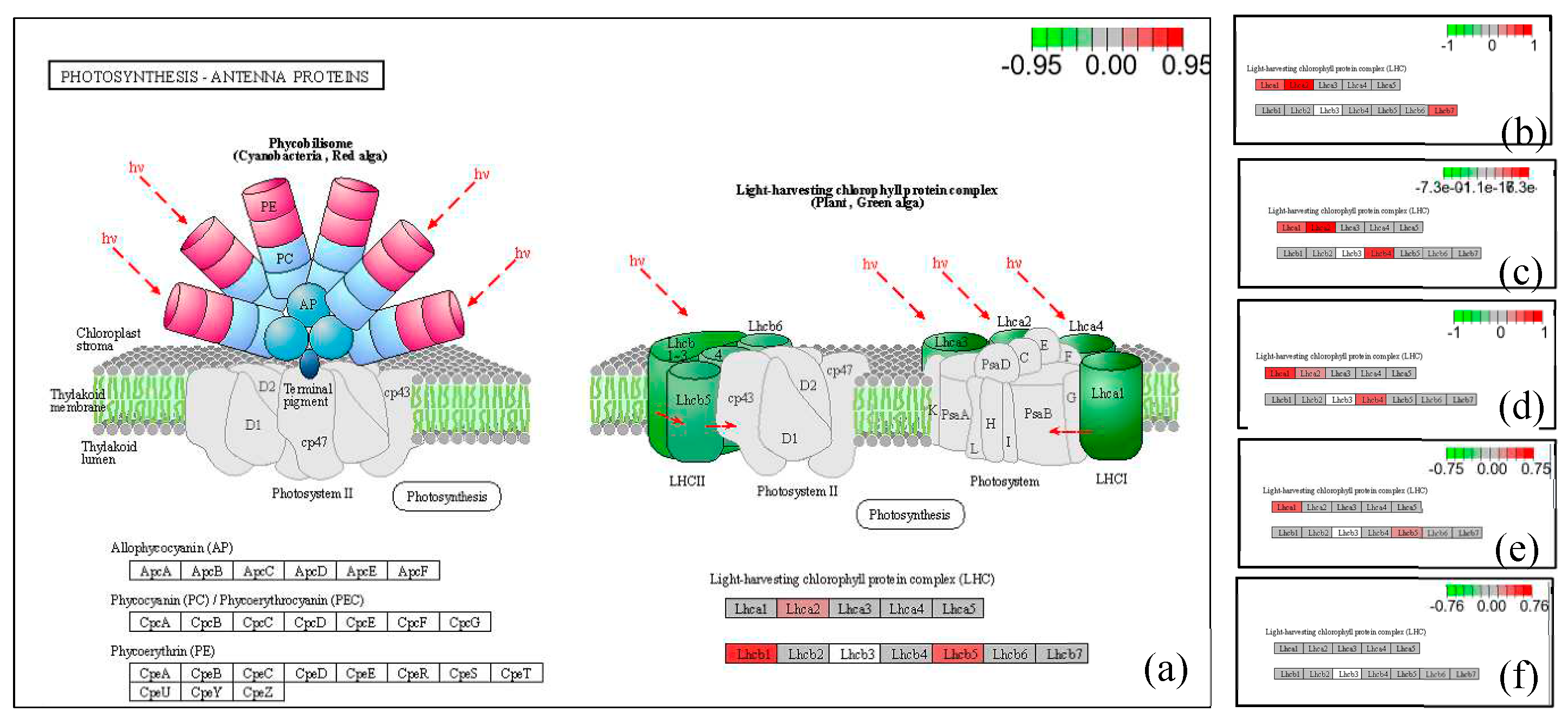
The pathway map illustrating the enrichment of differentially expressed genes is presented in
Figure 5. To visualize the distribution of these genes within the pathway map, differential genes were annotated, with red indicating up-regulated genes and green representing down-regulated genes.
The light-harvesting protein complex in the pathway of light-antenna proteins comprises both up-regulated and down-regulated genes. The chlorophyll-binding subunits of photosystems I and II serve as internal antenna light-harvesting proteins for oxygenic photosynthesis. In cyanobacteria, phycobilisomes contain antenna proteins, while green plants possess light-harvesting chlorophyll protein complexes acting as peripheral antenna systems to enhance the efficiency of light energy absorption.
The expression of LHca2 (light-harvesting complex I chlorophyll a/b binding protein 2) was up-regulated in LN5 compared to LCK. Additionally, LHcb1 (light-harvesting complex II chlorophyll a/b binding protein 1) showed an increase in expression, as well as LHcb5 (light-harvesting complex II chlorophyll a/b binding protein 5). In LN10 vs LCK, there was an up-regulation of LHca1 (light-harvesting complex I chlorophyll a/b binding protein 1), along with the up-regulation of LHca2 and LHcb7 (light-harvesting complex II chlorophyll a/b binding protein 7). Similarly, in LN20 vs LCK, there was an up-regulation of LHca1, along with the up-regulation of LHca2 and LHcb4 (light-trapping complex II chlorophyll a/b binding protein). Furthermore, in LO5 vs LCK, there was an up-regulation of LHca1. Specifically, there was an increase in the expression of chlorophyll a/b binding protein 2 or namely known as LHca2. Moreover, both LHcb1 and Lhcb2 were found to be up-regulated. Lastly, both LHcb4 and LHcb6 exhibited increased expression levels. In LO10 vs LCK specifically showed an increase in the expression level for LHca1 while also showing increased levels for LHcb5.
3.5.5. Weighted geneco-expression network analysis (WGCNA)
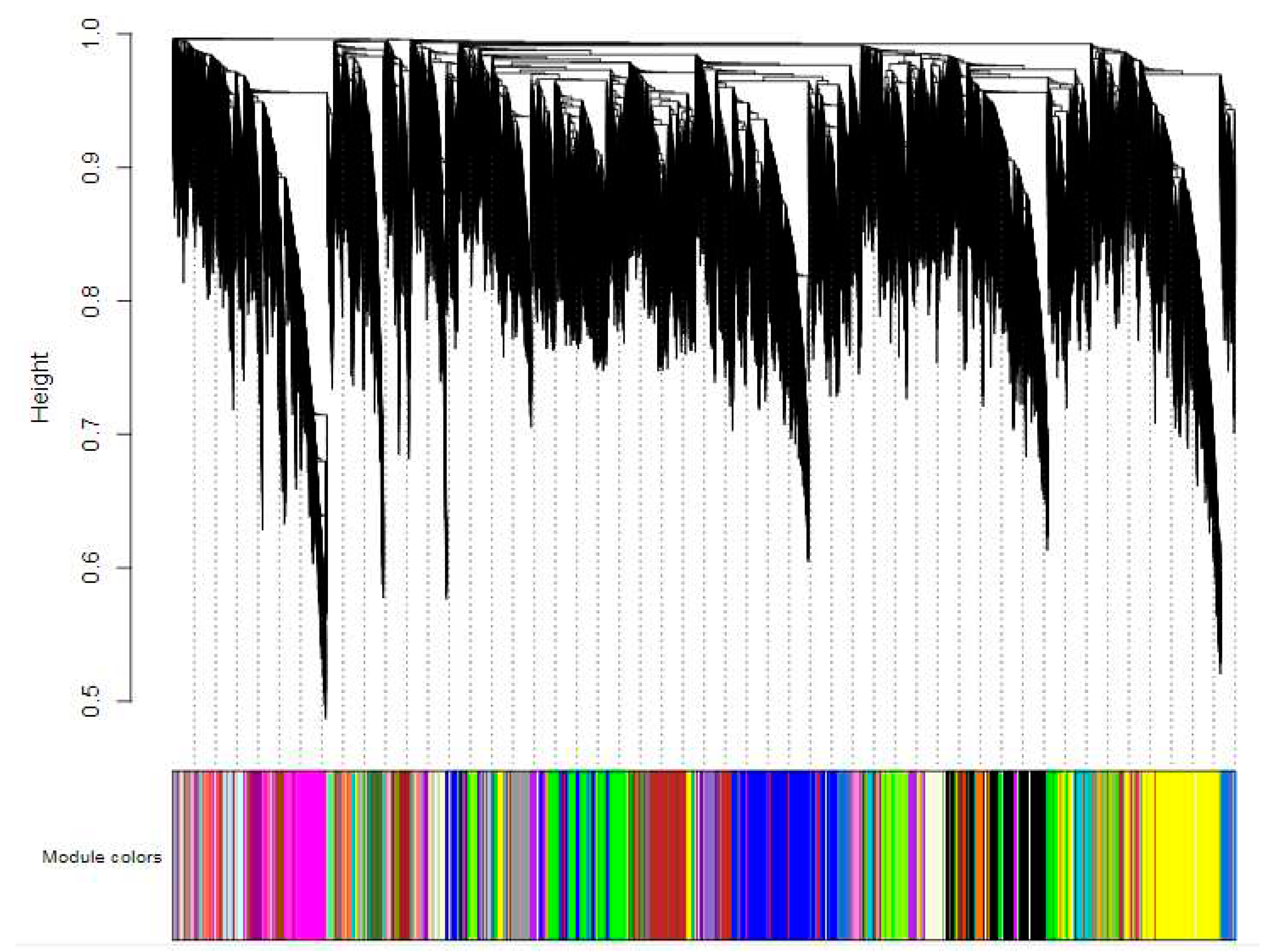
In order to identify gene co-expression modules, a weighted gene co-expression network analysis (WGCNA) was conducted for all expressed genes. Firstly, the coexpression network was constructed based on the transcriptome sequencing results using a defined fitting index R
2 = 0.8. Low expression genes were excluded and the cluster of coexpressed genes was dynamically cut. A cluster tree was then generated according to the correlation between gene expression levels (
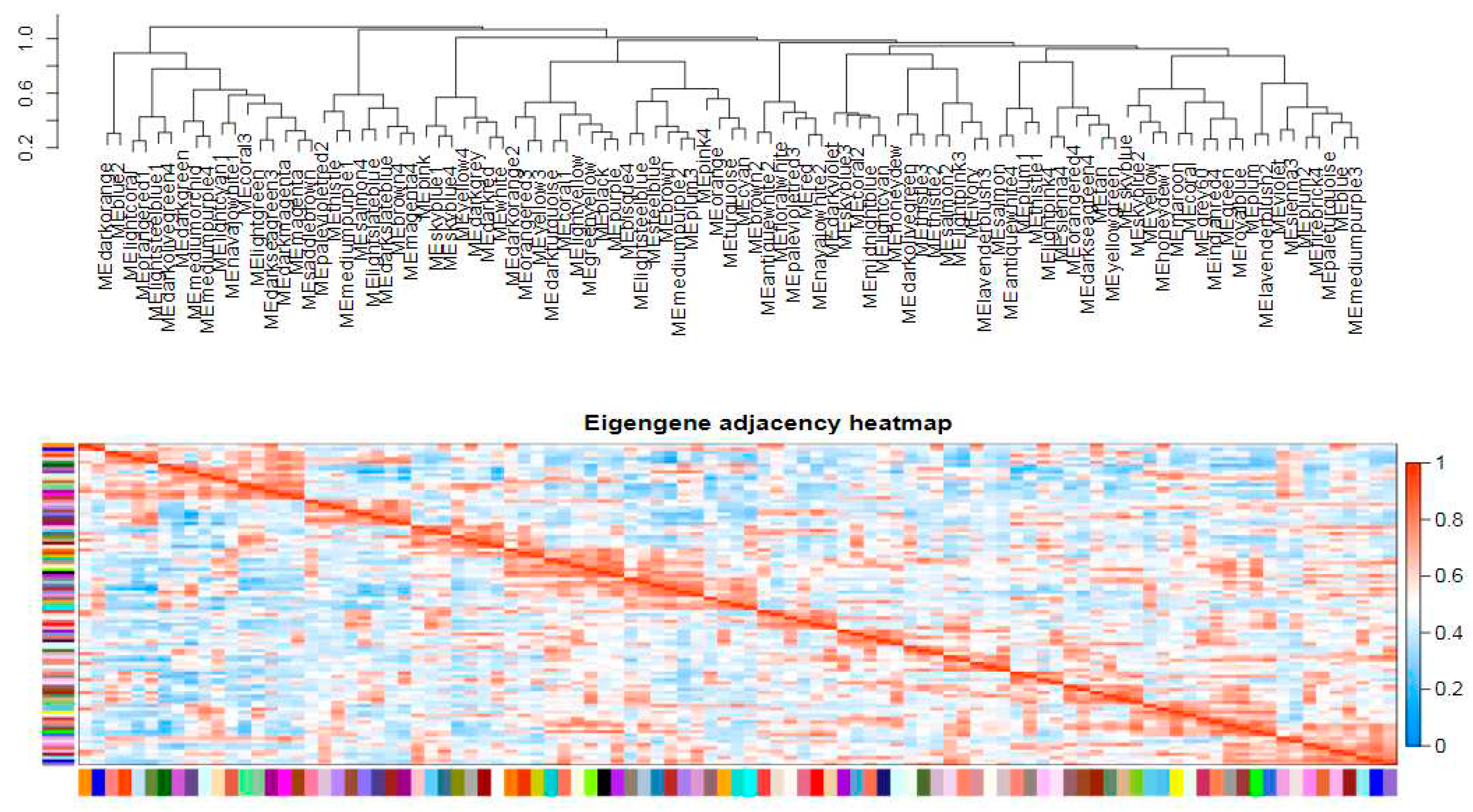
Figure 6), resulting in module divisions. Each color in the figure represents a specific module that includes genes belonging to the same cluster in the clustering tree. If certain genes consistently exhibit similar changes in expression during a physiological process or across different tissues, it suggests their functional relationship and they can be defined as a module. The vertical distance reflects the dissimilarity between two nodes (genes). The correlation heat map between modules (
Figure 7) can be divided into two parts: the upper part clusters modules based on their eigengene values, while each row and column in the bottom half represents an individual module. Blue indicates negative correlation with a coefficient ranging from 0 to 0.5; closer values to zero indicate stronger negative correlations. Red indicates positive correlation with coefficients ranging from 0.5 to 1; closer values to one indicate stronger positive correlations. The results demonstrated a significant correlation of 0.749 between the Coral module and Darkturquoise module, thereby establishing these two modules as potential target gene modules. Subsequently, an independent analysis was conducted on the expression levels of all genes and eigenvector genes within the two modules across all samples. It was observed that the gene expression levels within each module exhibited a high degree of correlation, while the expression levels of eigenvector genes were also strongly correlated with the overall expression level of their respective modules. This finding suggests that eigenvector genes in target modules can effectively represent the overall gene expressions within their corresponding modules (
Figure 8). Finally, hub genes from the two modules were selected based on their connectivity ranking with other genes; specifically, those with higher kWithin rankings indicating central hub positions were chosen. The Darkturquoise module yielded five core genes, while the Coral module identified four core genes, resulting in a total of hub core genes (
LOC103857346, Actin-related protein 2/3 complex subunit 1A;
LOC103828797, Actin-related protein 2/3 complex subunit 2A;
LOC103874199, Superoxide dismutase [Fe] 3; LOC103838242, Monodehydroascorbate reductase; LOC103846588, Thioredoxin M2; LOC103873206, ABC transporter A family member 7;
LOC106353797, ABC transporter B family member 26;
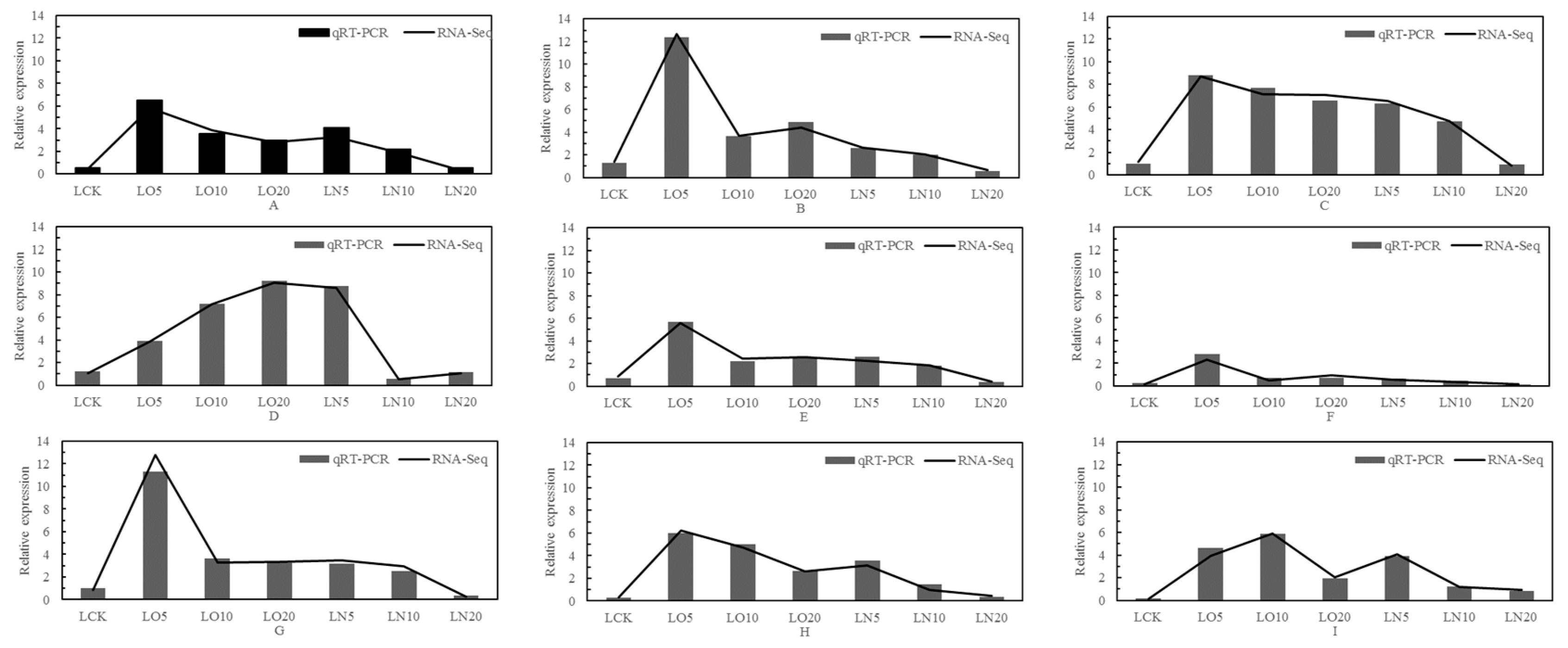
LOC103859979, ATP sulfurylase 1; LOC103859070, Cysteine synthase D1) associated with the antioxidant stress, absorption, transport, and metabolism of selenium. Real-time fluorescence quantitative PCR was conducted to validate these nine hub genes, as depicted in
Figure 9.
3.5.6. Fluorescence quantitative PCR analysis
The differential genes were validated using qRT-PCR, and the mapping analysis yielded an high R
2 value calculated based on the 2
-△△Ct method. The concordance between the qRT-PCR validation and RNA-Seq results demonstrated the reliability of the experimental findings (
Figure 9).
3.5.6. Differential gene function analysis
(1) Genes related to selenium absorption - endocytosis
Endocytosis is a cellular mechanism employed for the acquisition of extracellular nutrients. Further analysis showed that endocytosis included not only the hub genes (LOC103857346, Actin-related protein 2/3 complex subunit 1A;
LOC103828797, Actin-related protein 2/3 complex subunit 2A), but also other genes as shown in
Table 5. Transcriptome analysis revealed 32 differentially expressed genes associated with endocytic proteins, as presented in
Table 5. Notably, Actin-related protein 2/3 complex subunit 1A, nuclear-pore anchor, and vacuolar protein sorting-associated protein exhibited enhanced expression upon treatment with organic selenium and nano-selenium 2 homolog 3, along with upregulation observed in vacuolar protein sorting-associated protein 28 homolog 2 and Vacuolar protein sorting-associated protein 60. This suggests that endocytosis may facilitate the absorption of selenium.
Table 5.
The related gene in endocytosis.
Table 5.
The related gene in endocytosis.
| Gene name |
Transcripts Per Kilobase Million (TPM) |
Function |
| LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| LOC103857346 |
0 |
43.1 |
72.6 |
15.4 |
75 |
121 |
55 |
Actin-related protein 2/3 complex subunit 1A |
| LOC103828797 |
25 |
24.8 |
19.5 |
11.2 |
21.8 |
0 |
17.7 |
Actin-related protein 2/3 complex subunit 2A |
| LOC103838512 |
187 |
62.1 |
97.2 |
146 |
191 |
229 |
70.8 |
Actin-related protein 2/3 complex subunit 3 |
| LOC103863562 |
18.5 |
9.9 |
5.04 |
0 |
11.5 |
0 |
19.8 |
Actin-related protein 2/3 complex subunit 4 |
| LOC103829270 |
723 |
667 |
236 |
242 |
432 |
360 |
389 |
ADP-ribosylation factor 2-B |
| LOC103834200 |
223 |
230 |
138 |
47.7 |
152 |
246 |
167 |
ADP-ribosylation factor GTPase-activating protein AGD12 |
| LOC103838481 |
4.56 |
0 |
2 |
0 |
0 |
1.37 |
3.9 |
ADP-ribosylation factor GTPase-activating protein AGD2 |
| LOC103845528 |
67.72 |
112.4 |
44.77 |
40.66 |
49.35 |
51.99 |
19.62 |
AP-2 complex subunit alpha-1 |
| LOC103853730 |
393 |
273 |
278 |
246 |
350 |
337 |
264 |
AP-2 complex subunit mu |
| LOC103859264 |
15 |
16 |
11 |
2 |
16 |
23 |
12 |
Clathrin heavy chain 2 |
| LOC103839878 |
262 |
61.5 |
0 |
114 |
123 |
101 |
5.6 |
E3 ubiquitin-protein ligase UPL5 |
| LOC103836651 |
95.1 |
35.6 |
50.4 |
53.4 |
0 |
35.3 |
60.6 |
EH domain-containing protein 1 |
| LOC103831794 |
318 |
211 |
169 |
729 |
288 |
154 |
217 |
ESCRT-related protein CHMP1B |
| LOC103859096 |
390 |
425 |
369 |
149 |
405 |
360 |
365 |
F-actin-capping protein subunit alpha |
| LOC103830490 |
0 |
3 |
1 |
18 |
0 |
1 |
0 |
Nuclear-pore anchor |
| LOC103848932 |
7.79 |
0 |
12.1 |
0 |
4 |
0 |
3.39 |
Phosphatidylinositol 4-phosphate 5-kinase 6 |
| LOC103831688 |
38.4 |
0 |
33.7 |
0 |
1.08 |
0 |
0 |
Probable F-actin-capping protein subunit beta |
| LOC103840633 |
79.5 |
4.75 |
21.9 |
11.3 |
20.2 |
39.7 |
50.4 |
Probable mediator of RNA polymerase II transcription subunit 37e |
| LOC103866553 |
730 |
42.7 |
0 |
684 |
363 |
46 |
39.1 |
Protein homolog of mammalian lyst-interacting protein 5 |
| LOC103827815 |
42 |
39 |
32 |
31 |
34 |
28 |
19 |
Ras-related protein RABF2a |
| LOC125608966 |
154 |
0 |
148 |
0 |
49.8 |
108 |
142 |
Ras-related protein RABG3f |
| LOC103827890 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
Vacuolar protein sorting-associated protein 2 homolog 2 |
| LOC103844298 |
0 |
16.3 |
6.77 |
0 |
15.9 |
17.3 |
9.31 |
Vacuolar protein sorting-associated protein 2 homolog 3 |
| LOC103847195 |
130 |
271 |
106 |
106 |
258 |
107 |
137 |
Vacuolar protein sorting-associated protein 20 homolog 2 |
| LOC103861671 |
0 |
1.25 |
0 |
0 |
0 |
0 |
0 |
Vacuolar protein sorting-associated protein 22 homolog 1 |
| LOC103857037 |
13.9 |
7.68 |
18.1 |
39.2 |
13.8 |
3.77 |
22.8 |
Vacuolar protein sorting-associated protein 25 |
| LOC103839783 |
22 |
25 |
39 |
102 |
38 |
18 |
44 |
Vacuolar protein sorting-associated protein 28 homolog 2 |
| LOC103853808 |
806 |
406 |
455 |
701 |
1167 |
590 |
468 |
Vacuolar protein sorting-associated protein 32 homolog 2 |
| LOC103847243 |
365 |
329 |
418 |
858 |
239 |
278 |
456 |
Vacuolar protein sorting-associated protein 36 |
| LOC103853060 |
312 |
196 |
501 |
171 |
240 |
267 |
217 |
Vacuolar protein sorting-associated protein 45 homolog |
| LOC103859354 |
29.8 |
55.7 |
51.5 |
33.7 |
97.1 |
62.8 |
62.2 |
Vacuolar protein sorting-associated protein 60.1 |
| LOC103863440 |
0 |
1.25 |
0 |
0 |
0 |
0 |
0 |
Vacuolar protein-sorting-associated protein 37 homolog 1 |
(2) Genes involved in antioxidant stress
Low temperature can induce the generation of Reactive Oxygen Species (ROS) in plants, and excessive ROS accumulation can trigger cell apoptosis. To enhance the ROS response, plants up-regulate the expression of antioxidant coding genes, promote antioxidants biosynthesis, and thereby improve their antioxidant capacity. In addition to the hub genes mentioned above, there are other genes involved in antioxidant activity. In both organic selenium and nano-selenium treatment groups, superoxide dismutase (SOD), superoxide dismutase [Fe] 3, superoxide dismutase [Fe] 2, monodehydroascorbate reductase, and thioredoxin M2 were upregulated significantly, playing a crucial role in combating oxidative stress caused by low temperature in leaves (
Table 6).
(3) Selenium transport-related gene ----ABC transporter
The ABC transporter plays a crucial role in the cell membrane by facilitating the transport of substances across the cell or extracellular space through ATP. In addition to the aforementioned hub genes, there exist additional genes implicated in the transportation of selenium. Comparative analysis revealed differential expression of 23 ABC transporter genes, with upregulation observed for ABC transporter B family member 29, ABC transporter C family member 10, ABC transporter E family member 2, ABC transporter G family member 11, ABC transporter G family member 40, and ABC transporter I family member 1.
Table 7.
The genes of ABC transporter gene family.
Table 7.
The genes of ABC transporter gene family.
| Gene name |
Transcripts Per Kilobase Million (TPM) |
Function |
| LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| LOC103873206 |
824.4 |
630.78 |
496.96 |
128.14 |
582.37 |
345.51 |
431.95 |
ABC transporter A family member 7 |
| LOC106353797 |
2169.8 |
2721 |
1746.08 |
1866 |
2128.89 |
1854 |
1048.58 |
ABC transporter B family member 26 |
| LOC103847350 |
15.2 |
30.03 |
0 |
40.27 |
14.82 |
18.59 |
24.13 |
ABC transporter B family member 29 |
| LOC103866318 |
296.88 |
203.33 |
710.22 |
204.74 |
559.52 |
133.95 |
153.34 |
ABC transporter B family member 4 |
| LOC103867037 |
266.9 |
216.14 |
115.52 |
311.32 |
258.27 |
197.28 |
109.8 |
ABC transporter B family member 6 |
| LOC103841771 |
182.07 |
436 |
721 |
291.67 |
1832 |
488.35 |
665.6 |
ABC transporter C family member 10 |
| LOC103868683 |
328.47 |
155.67 |
263.61 |
375.64 |
412.3 |
209.69 |
223.52 |
ABC transporter C family member 4 |
| LOC103838010 |
394.11 |
449.39 |
79.4 |
308.17 |
497.77 |
513.89 |
135.88 |
ABC transporter C family member 5 |
| LOC103869976 |
980.68 |
1062.82 |
450.92 |
526.92 |
857.17 |
772.79 |
0 |
ABC transporter D family member 2 |
| LOC103869976 |
0 |
5 |
1 |
0 |
0 |
0 |
1.02 |
ABC transporter D family member 2 |
| LOC103865536 |
0 |
0 |
0 |
34.46 |
0 |
31.82 |
6.76 |
ABC transporter E family member 2 |
| LOC103857266 |
0 |
5.19 |
306 |
0 |
628.18 |
324.33 |
0 |
ABC transporter E family member 2 |
| LOC103851055 |
0 |
0 |
0 |
456.64 |
314.58 |
47.79 |
0 |
ABC transporter F family member 1 |
| LOC103867761 |
10.94 |
0 |
1.95 |
30.92 |
0 |
20.46 |
7.7 |
ABC transporter G family member 1 |
| LOC103835938 |
674.37 |
334.08 |
1137.08 |
801.13 |
939.33 |
1435.65 |
800.87 |
ABC transporter G family member 11 |
| LOC103867209 |
0 |
0 |
0 |
11.45 |
0 |
0 |
76.09 |
ABC transporter G family member 33 |
| LOC103872310 |
4.97 |
0 |
0 |
0 |
0 |
1.65 |
0 |
ABC transporter G family member 35 |
| LOC103872346 |
120 |
632 |
245 |
368 |
32 |
402 |
212 |
ABC transporter G family member 40 |
| LOC103872346 |
44.06 |
247.04 |
33.05 |
111.63 |
0 |
141.49 |
45.11 |
ABC transporter G family member 40 |
| LOC103832795 |
5693.61 |
1452.2 |
4726 |
640.2 |
0 |
3356.75 |
2220.56 |
ABC transporter G family member 7 |
| LOC103838079 |
25.13 |
0 |
66 |
53.12 |
42.03 |
73 |
0 |
ABC transporter I family member 1 |
| LOC103847034 |
126 |
0 |
0 |
78.17 |
0 |
0 |
170.22 |
ABC transporter I family member 10 |
| LOC103859358 |
7 |
3 |
6 |
0 |
5 |
0 |
1 |
ABC transporter I family member 6 |
(4) Genes related to selenium metabolism
The metabolic pathways of cysteine and methionine, sulfur, selenide, and glutathione, which are closely associated with selenium metabolism, were analyzed. The subsequent analysis revealed that selenium metabolism involved not only the hub genes but also other genes, as indicated in
Table 8. After selenium absorption, the expression of genes related to the sulfur metabolic pathway was either up-regulated or down-regulated. The expression of chloroplastic ATP-ylase 1 genes was up-regulated following treatment with organic selenium and nano-selenium. Key genes controlling the expression of Cysteine synthase showed varying degrees of up-regulation, with LN5 Transcripts TPM exhibiting the highest value. Furthermore, it was observed that genes involved in selenide metabolism-related enzymes were up-regulated. These included glutathione S-transferase U12 and S-sulfo-L-cysteine synthase (O-acetyl-L-serine-dependent). On the other hand, glutathionyl-hydroquinone reductase YqjG and probable glutathione peroxidase 8 were down-regulated. In summary, there was a higher overall abundance of up-regulated gene expression compared to down-regulated gene expression, and significant differences in gene expression levels were observed between organic selenium and nano-selenium treatments. Notably, under nano-selenium treatment conditions, certain key genes exhibited significantly higher expression levels than under organic selenium treatment conditions, indicating a superior absorption effect for nano-selenium.
4. Discussion
4.1. Effects of spraying organic selenium and nano-selenium fertilizer on leaf photosynthesis under low temperature stress
Studies have indicated that the photosynthetic process of plants is negatively impacted by low temperature stress. The original chlorophyll undergoes destruction due to low temperatures, leading to a decrease in chlorophyll content (Yang et al., 2022). Consequently, the photosynthetic capacity of plants weakens, hindering carbon assimilation and retarding plant growth rate (Tang, 2011). Application of an appropriate concentration of selenium can regulate porphyrin biosynthesis in plants, thereby influencing chlorophyll metabolism (Zhong et al., 2017). Moreover, an optimal selenium concentration facilitates the absorption of P, K, Ca, Mg and other elements by crops. Most of these elements actively participate in or promote the synthesis process of chlorophyll (Han and Wei, 2015) while regulating photosynthesis. Results demonstrate that under low temperature stress conditions, foliar application with suitable concentrations of organic selenium (20 mg L-1) and nano-selenium (10 mg L-1) significantly enhances leaf chlorophyll content. Furthermore, this study reveals that spraying organic selenium (20 mg L-1) and nano-selenium (10 mg L-1) substantially increases the chlorophyll a/b ratio-a finding consistent with Zhang et al.'s research on grapes in 2018 and Hu et al.'s investigation on purple potatoes in 2019. The increase in the chlorophyll a/b ratio indicates an enhancement in the stacking degree of thylakoid membranes within chloroplasts, thereby improving the efficiency of light energy conversion through enhanced absorption, transfer, and utilization of photons by plants. Additionally, it was observed that lower concentrations led to a significant increase in organic selenium carotenoid content, while carotenoid content initially increased and then decreased with increasing nano-selenium concentration. This suggests that an appropriate selenium concentration can promote carotenoid synthesis to some extent and protect plant chlorophyll molecules from photooxidation damage, which aligns with Wang 's (2019) research on cress and mustard.
4.2. Effects of spraying organic selenium and nano-selenium fertilizer on the regulation of antioxidant system of leaves under low temperature stress
The regulation of selenium on the plant antioxidant system: Secondary metabolites, such as ascorbic acid (ASA) and plant phenolic compounds (TP), are commonly recognized as non-enzymatic substances possessing potent antioxidant capacity. The ascorbate-glutathione cycle (ASA-GSH cycle) represents a highly efficient detoxification pathway in both the cytoplasm and chloroplasts of plants. ASA exhibits the capability to donate electrons for numerous antioxidant reactions within the organism. Superoxide dismutase (SOD) serves as the primary defense against superoxide anions by catalyzing their dismutation into oxygen and hydrogen peroxide. Additionally, catalase (CAT) and peroxidase (POD) further decompose H2O2 to prevent oxidative damage to plant cells. Song et al. (2019) demonstrated that lettuce seedlings could mitigate membrane lipid peroxidation damage under low temperature stress by increasing the activities of superoxide dismutase (SOD) and peroxidase (POD). Similarly, Wang et al. (2019) revealed that suitable spraying concentrations of nano-selenium fertilizer enhanced SOD and POD activities. This study reveals that different concentrations of organic selenium spray improved the activities of POD, SOD, and catalase (CAT) in leaves to varying degrees under low temperature stress, with the optimal concentration being 20 mg L-1. Conversely, lower concentrations of nano-selenium fertilizer decreased the activity of these three enzymes in leaves under low temperature stress; however, their activity increased with higher concentrations. Additionally, this study found that ascorbate peroxidase content increased to varying degrees with increasing organic selenium concentration during low temperature stress, reaching its peak at a concentration of 5 mg L-1 and again at 20 mg L-1. As for nano-selenium concentration, ascorbate peroxidase content also increased to different extents and reached its maximum at a concentration of 20 mg L-1.
4.3. Effects of spraying organic selenium and nano-selenium fertilizer on leaf nutritional quality under low temperature stress
Under low temperature stress, the levels of amino acids, soluble sugars, and flavonoids in plants are known to increase (Zhou et al., 2021). Huang et al. (2023) and Yuan et al. (2008) reported that under low temperature stress, the levels of soluble sugars and soluble proteins showed varying degrees of increase in small-seed coffee seedlings and spirulina. Adverse conditions weaken plant metabolic reactions while enhancing hydrolysis and weakening synthesis, leading to degradation of macromolecular compounds such as soluble proteins into amino acids, soluble sugars, peptides, etc. Additionally, metabolic disorders under adverse conditions result in an accumulation of excess oxygen free radicals within the plant body. Consequently, plants synthesize a large number of flavonoid substances to enhance free radical scavenging and antioxidant activity. This study demonstrated that leaves exhibited increased levels of soluble sugar, soluble protein, amino acid, and flavonoid content under low temperature stress with increasing concentrations of organic selenium up to 20 mg L-1. The high concentration of organic selenium treatment may have affected the respiratory electron transport chain and tolerance to organic selenium stress in leaves, leading to the use of their own physiological metabolism to alleviate exogenous selenium stress (Liu et al., 2019). However, spraying with lower concentrations of nano-selenium (5 mg L-1) significantly increased soluble sugar and amino acid contents, while spraying with 10 mg L-1 of nano-selenium significantly increased soluble protein and flavonoid contents. The increase in soluble protein content from nano-selenium spray was due to its direct participation in protein synthesis through selenocysteine (Se-Cys) and selenomethionine (Se-Met), which reduced cysteine (Cys) and methionine (Met) levels in free amino acids. Additionally, as a component of tRNA ribonucleic acid chains in plants that transport amino acids, selenium also influenced other free amino acids. Excessive concentrations of nano-selenium decreased free amino acid content; however, under optimal concentration treatment at 10 mg L-1, the ability for nano-selenium to convert free amino acids into soluble proteins likely weakened but still resulted in an increase in soluble protein content. Further research is needed on this specific mechanism.
4.4. Absorption of organic selenium and nano-selenium at different concentrations in leaves
Currently, research on the absorption of inorganic selenium in plants primarily focuses on the mechanism of root absorption, while there is limited reporting on the absorption of organic selenium and nano-selenium in leaves. It has been speculated that transporters associated with plant cysteine methionine may be involved in certain processes of organic selenide absorption. Kong (2021) discovered significant differences in genes related to the endocytosis process after treatment with nano-selenium compared to the control group. Therefore, it is hypothesized that millet's absorption of nano-selenium is linked to the endocytosis process, and further tests are needed to verify whether genes related to PIP5K, Hsc70, PLD, and ArfGAP are involved in millet's selenium uptake and how they participate in this process. This study concludes absorption of nano-selenium and organic selenium is associated with endocytosis, aligning with previous findings.
4.5. Transport of different concentrations of organic selenium and nano-selenium in leaves
The study revealed that ABC transporters play a crucial role in promoting plant growth by facilitating the transportation of nutrients, hormones, lipids, secondary metabolites, metal ions, and foreign substances. Additionally, they contribute to disease resistance and response to stress in plants. Moreover, these transporters are involved in regulating ion channels. The ABCB subgroup transporters primarily function in transporting heavy metals and secondary metabolites for resistance purposes. On the other hand, the ABCC subfamily is responsible for transporting glutathione compounds. Byrne SL et al. (2010) reported that 28 ABC transporters in ryegrass may be associated with selenide accumulation. In the investigation of genes related to selenium absorption and metabolism in tea tree roots conducted by Hu et al. (2016), 16 key genes of ABC transporter were identified suggesting their potential involvement in selenate transport within roots. However, Kong et al. (2021) examined selenium response genes in millet leaves and found that out of 7 differential genes belonging to ABC transporter family, 5 were up-regulated while 2 were down-regulated contradicting previous studies' findings. These discrepancies might arise from variations observed among different species regarding the expression patterns of ABC transporters.
4.6. Metabolism of different concentrations of organic selenium and nano-selenium in leaves
Usually, selenite is reduced to selenide by sulfite reductase or glutathione, and selenide can also be produced through various mechanisms such as methionol in the action of selenium-binding proteins. Subsequently, cysteine synthase catalyzes the conversion of selenide into selenocysteine (SeCys) (Terry et al., 2000). Kong (2021) discovered that key genes regulating the expression of Se-binding protein 1 and cysteine synthase were upregulated to varying degrees during this process. The key genes associated with cysteine synthase in this study exhibit similarities to those identified in previous research. Following the biosynthesis of selenocysteine, a portion of it undergoes further conversion into selenocysta by cystathionine-gamma-synthetase (CγS), which is subsequently decomposed into se-homocysteine (SeHCys) by cystathionine-β-lyase (CBL). Finally, under the influence of cysteine methyltransferase (MET), it is transformed into selenomethionine (SeMet). Some synthetic SeCys and SeMet can substitute Cys and Met within plant protein chains, thereby influencing their structure and function (Hoewyk et al., 2008). Other non-protein selenium amino acids like Methylselenol and its derivatives, as well as non-amino acid selenides, can be generated through the action of methionin-gamma-lyase to mitigate the toxic effects exerted by SeMet on plants' growth processes (Whanger,2002,2004). Inserting SMT gene in Arabidopsis Thaliana resulted in an increased content of SeMSeCys along with enhanced tolerance towards selenium (Hugh et al., 2004). Hu did not detect any SMT genes in tea tree roots following selenium treatment, indicating that SMT is not implicated in selenium regulation (discovery and analysis of key genes for selenium uptake and metabolism in tea tree roots). Similarly, Tan (2009) found no differential expression of SMT due to selenium induction. In this study, while no SMT was identified, the crucial gene homocysteine S-methyltransferase was discovered, and it was observed that the selenocysteine S-methyltransferase gene CsSMT mRNA could interact with the homocysteine S-methyltransferase gene of Arabidopsis thaliana (2016). Furthermore, the upregulation of glutaredoxin-C1, glutaredoxin-C6 and glutathione S-transferase U12 suggests that selenium plays a role in regulating the synthesis of glutathione, cysteine and methionine which ultimately affects its accumulation.
5. Conclusions
In conclusion, this study concludes that spraying with appropriate concentrations of organic selenium (20 mg L-1) or nano-selenium (10 mg L-1) is a useful strategy for improving “Suzhouqing” tolerance to low temperature, and subsequently yield.
Moreover, the gene expression related to selenium absorption, transport and metabolism of Suzhouqing was systematically screened and analyzed by RNA-Seq. Among the gene ontology (GO) terms, Chlorophyll metabolic process and plastid nucleoid were prominent mainly in biological process and cellular components. KEGG showed different pathways such as endocytosis, plant hormone signal transduction, metabolic pathways, biosynthesis of secondary metabolites under comparative treatments. Based on the transcriptome data mentioned above, a Weighted Gene Co-expression Network Analysis (WGCNA) was employed to construct a network that associates with physiological traits related to stress resistance. Two highly correlated gene co-expression modules were identified, and within them, nine hub genes associated with endocytosis, antioxidant stress, absorption, transport, and metabolism of selenium were discovered. Functional annotation revealed their predominant involvement in the regulatory pathway of hypothermia stress. Subsequently, the nine hub genes were subjected to fluorescence quantitative PCR verification, and the results obtained were consistent with those from transcriptome sequencing. The study revealed that, in addition to the aforementioned hub genes enhancing the response of plants to low temperature stress, 32 differential genes related to endocytosis, 23 differentially expressed ABC transporter genes, also exerted an influence on crop growth. Furthermore, the upregulation of Superoxide dismutase, Monodehydroascorbate playing a crucial role in combating oxidative stress caused by low temperature. Moreover, the regulation of glutaredoxin-C1, glutaredoxin-C6 and glutathione S-transferase U12 suggests that selenium plays a role in regulating the synthesis of glutathione, cysteine and methionine which ultimately affects its accumulation.
The beneficial effects on yield and total selenium content under low temperature were attributed to (1) protection of photosynthetic pigments for enhancing photosynthetic capacity by the up-regulation of LHca2, LHcb1, LHca1, LHcb4 in KEGG pathway: photosynthesis-antenna proteins; (2) activation of antioxidant system for efficient ROS homeostasis such as SOD, POD and CAT by the genes such as Superoxide dismutase, Monodehydroascorbate and (3) selenium absorption by endocytosis, seleninum transportation by ABC transporter gene family and selenium metabolism related genes such as Cysteine synthase, Glutaredoxin.
Therefore, these findings provide a foundation for further investigation into the molecular mechanisms underlying the effects of organic selenium and nano-selenium on cyanochloride production at low temperatures.
Funding
This study was supported by the Fund project of Natural Science Foundation of Jiangxi Province (No. 20224BAB215033), Key R&D Project in Jiangxi Province (20223BBF61016) and the Natural Science Foundation of China (No. 42167045), and the Natural Science Foundation of China (No. 31960331).
Acknowledgments
The authors would like to thank all the reviewers who participated in the review.
References
- Byrne, S.L.; Durandeau, K.; Nagy, I.; Barth, S. Identification of ABC transporters from Lolium perenne L. that are regulated by toxic levels of selenium. Planta 2010, 231, 901–911. [Google Scholar] [PubMed]
- Campion, E.M.; Loughran, S.T.; Walls, D. Protein quantitation and analysis of purity. Methods in Molecular Biology 2011, 681, 229–258. [Google Scholar] [PubMed]
- Cao, D.; Liu, Y.; Ma, L.; et al. Transcriptome analysis of differentially expressed genes involved in selenium accumulation in tea plant (Camellia sinensis). PloS One 2018, 13, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Yao, Y.; Yang, M.; et al. Transcriptome sequencing and analysis reveals the molecular response to selenium stimuli in Pueraria lobata (willd. ) Ohwi. PeerJ 2020, 8, 1–18. [Google Scholar] [CrossRef]
- Han, X.X.; Wei, H.Y. Research progress in nutritional biology of selenium. Journal of Southern Agriculture 2015, 46, 1798–1804. [Google Scholar]
- Hoewyk, D.V.; Pilon, M.; Pilon-Smits, E.A.H. The functions of NifS-like proteins in plant sulfur and selenium metabolism. Plant Science 2008, 174, 120–123. [Google Scholar]
- Huang, C.; Qin, N.; Sun, L.; et al. Selenium improves physiological parameters and alleviates oxidative stress in strawberry seedlings under low-temperature stress. International journal of molecular sciences 2018, 19, 1913. [Google Scholar] [CrossRef]
- Huang, L.F.; Long, Y.Z.; Li, J.Q.; et al. Physiological and biochemical characteristics of Coffea Arabica seedling under low temperature stress. Journal of Agricultural Science and Technology 2023, 25, 1–8. [Google Scholar]
- Hugh, H.; Wood, K.V.; Brett, L.; et al. Production of Se-methylselenocysteine in transgenic plants expressing selenocysteine methyltransferase. BMC Plant Biology 2004, 4, 1–11. [Google Scholar]
- Hu, W.X.; Zhao, B.S.; Shi, Y.; et al. Effects of ecological nano-selenium on the growth and photosynthetic characteristics of purple potatoes. Journal of Northern Agriculture 2019, 47, 64–69. [Google Scholar]
- Hu, Y.R. 2016. Identification and analysis of genes related to selenium assimilation and metabolism in tea plant roots. Chinese Academy of Agricultural Sciences.
- Kong, Q.H.; Li, F.F.; Qin, L.; et al. Screening and analysis of Se responsive genes in leaves of foxtail millet. Molecular Plant Breeding 2021, 19, 2798–2810. [Google Scholar]
- Liu, J.X.; Li, X.F.; Shi, Y.; et al. Effects of foliar spraying ecological nano-selenium on quality of lettuce. Zhejiang Agricultural Sciences 2019, 60, 803–806. [Google Scholar]
- Liu, K.; Li, S.; Han, J.; et al. Effect of selenium on tea (Camellia sinensis) under low temperature: Changes in physiological and biochemical responses and quality. Environmental and Experimental Botany 2021, 188, 104475. [Google Scholar] [CrossRef]
- Rao, S.; Yu, T.; Cong, X.; et al. Integration analysis of PacBio SMRT-and Illumina RNA-seq reveals candidate genes and pathway involved in selenium metabolism in hyperaccumulator Cardamine violifolia. BMC Plant Biology 2020, 20, 492. [Google Scholar] [CrossRef]
- Song, Y.P.; Gong, F.R.; Yang, X.K.; et al. Comprehensive evaluation on cold tolerance of various lettuce seedlings. Acta Agriculturae Jiangxi 2019, 31, 21–25. [Google Scholar]
- Tan, Z. 2009. Studies on the transcription and expression levels of selenocysteine methyltransferase in the leaves of Camellia Sinensis. Anhui Agricultural University.
- Tang, Y.Y. 2011. Screening of peanut genotypes for low temperature tolerance and identification of low temperature responsive genes. Ocean University of China.
- Terry, N.; Zayed, A.M.; Souza, M.P.D.; et al. Selenium in higher plants. Annual Review of Plant Physiology & Plant Molecular Biology 2000, 51, 401. [Google Scholar]
- Wang, L.L. 2019. Effects of cadmium and selenium treatment on growth and physiological quality of Cardamine flexuosa and Brassica alboglabra. Zhejiang A&F University.
- Wang, X.K.; Huang, J.L. 2015. Principles and techniques of plant physiological biochemical experiment. Higher Education Press.
- Wang, Y.C.; Zhao, T.Y.; Chang, S.M.; et al. Effect of nano-selenium spraying on the growth characteristics, nutritional quality, phenolics content and antioxidant activity of mung bean sprouts. Journal of China Agricultural University 2019, 24, 39–46. [Google Scholar]
- Whanger, P.D. Selenocompounds in plants and animals and their biological significance. Journal of the American College of Nutrition 2002, 21, 223–232. [Google Scholar] [CrossRef]
- Whanger, P.D. Selenium and its relationship to cancer: an update. British Journal of Nutrition 2004, 91, 11–28. [Google Scholar] [CrossRef]
- Xu, L.L.; Xu, Y.M. Study on trace element selenium and human health. Agricultural Technology Service 2016, 33, 85–86. [Google Scholar]
- Yan, W.H.; Ma, Y.B.; Zhang, J.R.; et al. The influence of physiological characteristics and cold hardiness evaluation of 8 Agropyron Gaertn seedlings under low temperature stress. Grassland and Prataculture 2016, 28, 22–30. [Google Scholar]
- Yang, F.F.; Halike, G.L.Z.Y.; Maimaiti, A.L.J.; et al. Effects of exogenous trehalose on photosynthesis of Hami melon seedlings under chilling stress. Northern Horticulture 2023, 524, 27–30. [Google Scholar]
- Yuan, S.Z.; Li, S.Y.; Qiao, C. The effect of low temperature stress on content of soluble sugar in spirulina (Arthrospira). Chinese Agricultural Science Bulletin 2008, 167, 113–116. [Google Scholar]
- Zhang, X.; Li, X.Y.; Liu, J.; et al. The effect of foliar spraying selenium to fruit quality and chlorophyll contenton 87-1 grape. Journal of Inner Mongolia Agricultural University (Natural Science Edition) 2018, 39, 24–28. [Google Scholar]
- Zhong, S.Z.; Zhang, B.J.; Zhang, M.; et al. Effects of selenium on photosynthesis and antioxidation of rice. Soil and Fertilizer Sciences in China 2017, 4, 134–139. [Google Scholar]
- Zhou, P.; Lin, Z.K.; Guo, R.; et al. Effects of low temperature treatment on gene expression and flavonoids biosynthesis metabolism in peach (Prunus persica) leaves. Journal of Agricultural biotechnology 2021, 29, 1283–1294. [Google Scholar]
- Zhou, Y.; Tang, Q.; Wu, M.; et al. Comparative transcriptomics provides novel insights into the mechanisms of selenium tolerance in the hyperaccumulator plant Cardamine hupingshanensis. Scientific Reports 2018, 8, 2789. [Google Scholar] [CrossRef]
Figure 1.
Heat map of correlation coefficient between samples.
Figure 1.
Heat map of correlation coefficient between samples.
Figure 2.
Volcano map of differentially expressed genes. Note: The horizontal coordinate is log2(fold change), that is, the pair value of the fold change value; The ordinate is -log10(padj), which is the inverse of the logarithm of the corrected p-value.
Figure 2.
Volcano map of differentially expressed genes. Note: The horizontal coordinate is log2(fold change), that is, the pair value of the fold change value; The ordinate is -log10(padj), which is the inverse of the logarithm of the corrected p-value.
Figure 3.
GO enrichment histogram of differential genes. Note: B: Biological process; C: Cellular component; M: Molecular function; B1: Chlorophyll metabolic process; B2: Chloroplast fission; B3: Circadian regulation of gene expression; B4: Clathrin-dependent endocytosis; B5: Ethanolamine-containing compound metabolic process; B6: Maltose metabolic process; B7: Peptidyl-histidine modification; B8: Photosynthesis, light harvesting; B9: Plastid fission; B10: Positive regulation of histone H3-K27 methylation; B11: Positive regulation of protein import; B12: Positive regulation of protein import into nucleus; B13: Positive regulation of protein localization to nucleus; B14: Primary miRNA processing; B15: Regulation of dephosphorylation; B16: Regulation of histone H3-K27 methylation; B17: Regulation of phosphatase activity; B18: Regulation of phosphoprotein phosphatase activity; B19: Regulation of photosynthesis, light reaction; B20: Regulation of protein dephosphorylation; B21: Systemic acquired resistance, salicylic acid mediated signaling pathway; B22: Cell plate formation involved in plant-type cell wall biogenesis; B23: Cellular response to gravity; B24: Galacturonate biosynthetic process; B25: Photosynthetic electron transport chain; B26: Spindle assembly involved in meiosis; B27: Plastid transcription; B28: Plastid translation; B29: Regulation of protein localization to nucleus; B30: Regulation of transcription from RNA polymerase II promoter in response to stress; B31: Ribosomal protein import into nucleus; B32: Salicylic acid mediated signaling pathway; B33: Regulation of defense response to oomycetes; B34: Regulation of generation of precursor metabolites and energy; B35: Induced systemic resistance, jasmonic acid mediated signaling pathway; B36: Regulation of DNA methylation; B37: Regulation of miRNA metabolic process; B38: Cell plate assembly; B39: Cellular response to disaccharide stimulus; B40: Cellular response to sucrose stimulus; B41: Ciliary basal body-plasma membrane docking; B42: Lateral growth; B43: Negative regulation of iron ion transport; B44: Negative regulation of lipoprotein particle clearance; B45: Negative regulation of low-density lipoprotein particle clearance; B46: Negative regulation of photosynthesis; B47: Negative regulation of photosynthesis, light reaction; B48: Nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay; B49: Peptidyl-serine dephosphorylation; B50: Photoinhibition; B51: Photosynthesis, light harvesting in photosystem I; B52: Photosynthetic electron transport in photosystem II; B53: Photosystem II repair; B54: Positive regulation of histone H3-K27 methylation; B55: Positive regulation of protein import; B56: Protein repair; B57: Regulation of chlorophyll catabolic process; B58: Regulation of generation of precursor metabolites and energy; B59: Regulation of histone H3-K9 dimethylation; B60: Regulation of lipoprotein particle clearance; B61: Regulation of low-density lipoprotein particle clearance; B62: Regulation of protein import; B63: Regulation of tetrapyrrole catabolic process; B64: Secondary growth; B65: Sesquiterpenoid metabolic process; B66: Spindle assembly involved in meiosis; B67: Visual behavior; C1: Cajal body; C2: Chloroplast thylakoid membrane protein complex; C3: Chromatin silencing complex; C4: Cytoplasmic chromosome; C5: Plastid nucleoid; C6: Transcription repressor complex; C7: Tubulin complex; C8: Light-harvesting complex; C9: Thylakoid light-harvesting complex; C10: Protein phosphatase type 2A complex; C11: Ski complex; C12: Transcriptionally active chromatin; M1: Abscisic acid binding; M2: Alcohol binding; M3: Chlorophyll binding; M4: Isoprenoid binding; M5: Methionine adenosyltransferase activity; M6: Phosphatase regulator activity; M7: Protein phosphatase regulator activity; M8: Cytokinin receptor activity; M9: Pigment binding; M10: Protein C-terminal methylesterase activity; M11: Protein histidine kinase binding; M12: Pyrophosphate hydrolysis-driven proton transmembrane transporter activity.
Figure 3.
GO enrichment histogram of differential genes. Note: B: Biological process; C: Cellular component; M: Molecular function; B1: Chlorophyll metabolic process; B2: Chloroplast fission; B3: Circadian regulation of gene expression; B4: Clathrin-dependent endocytosis; B5: Ethanolamine-containing compound metabolic process; B6: Maltose metabolic process; B7: Peptidyl-histidine modification; B8: Photosynthesis, light harvesting; B9: Plastid fission; B10: Positive regulation of histone H3-K27 methylation; B11: Positive regulation of protein import; B12: Positive regulation of protein import into nucleus; B13: Positive regulation of protein localization to nucleus; B14: Primary miRNA processing; B15: Regulation of dephosphorylation; B16: Regulation of histone H3-K27 methylation; B17: Regulation of phosphatase activity; B18: Regulation of phosphoprotein phosphatase activity; B19: Regulation of photosynthesis, light reaction; B20: Regulation of protein dephosphorylation; B21: Systemic acquired resistance, salicylic acid mediated signaling pathway; B22: Cell plate formation involved in plant-type cell wall biogenesis; B23: Cellular response to gravity; B24: Galacturonate biosynthetic process; B25: Photosynthetic electron transport chain; B26: Spindle assembly involved in meiosis; B27: Plastid transcription; B28: Plastid translation; B29: Regulation of protein localization to nucleus; B30: Regulation of transcription from RNA polymerase II promoter in response to stress; B31: Ribosomal protein import into nucleus; B32: Salicylic acid mediated signaling pathway; B33: Regulation of defense response to oomycetes; B34: Regulation of generation of precursor metabolites and energy; B35: Induced systemic resistance, jasmonic acid mediated signaling pathway; B36: Regulation of DNA methylation; B37: Regulation of miRNA metabolic process; B38: Cell plate assembly; B39: Cellular response to disaccharide stimulus; B40: Cellular response to sucrose stimulus; B41: Ciliary basal body-plasma membrane docking; B42: Lateral growth; B43: Negative regulation of iron ion transport; B44: Negative regulation of lipoprotein particle clearance; B45: Negative regulation of low-density lipoprotein particle clearance; B46: Negative regulation of photosynthesis; B47: Negative regulation of photosynthesis, light reaction; B48: Nuclear-transcribed mRNA catabolic process, 3'-5' exonucleolytic nonsense-mediated decay; B49: Peptidyl-serine dephosphorylation; B50: Photoinhibition; B51: Photosynthesis, light harvesting in photosystem I; B52: Photosynthetic electron transport in photosystem II; B53: Photosystem II repair; B54: Positive regulation of histone H3-K27 methylation; B55: Positive regulation of protein import; B56: Protein repair; B57: Regulation of chlorophyll catabolic process; B58: Regulation of generation of precursor metabolites and energy; B59: Regulation of histone H3-K9 dimethylation; B60: Regulation of lipoprotein particle clearance; B61: Regulation of low-density lipoprotein particle clearance; B62: Regulation of protein import; B63: Regulation of tetrapyrrole catabolic process; B64: Secondary growth; B65: Sesquiterpenoid metabolic process; B66: Spindle assembly involved in meiosis; B67: Visual behavior; C1: Cajal body; C2: Chloroplast thylakoid membrane protein complex; C3: Chromatin silencing complex; C4: Cytoplasmic chromosome; C5: Plastid nucleoid; C6: Transcription repressor complex; C7: Tubulin complex; C8: Light-harvesting complex; C9: Thylakoid light-harvesting complex; C10: Protein phosphatase type 2A complex; C11: Ski complex; C12: Transcriptionally active chromatin; M1: Abscisic acid binding; M2: Alcohol binding; M3: Chlorophyll binding; M4: Isoprenoid binding; M5: Methionine adenosyltransferase activity; M6: Phosphatase regulator activity; M7: Protein phosphatase regulator activity; M8: Cytokinin receptor activity; M9: Pigment binding; M10: Protein C-terminal methylesterase activity; M11: Protein histidine kinase binding; M12: Pyrophosphate hydrolysis-driven proton transmembrane transporter activity.
Figure 4.
KEGG metabolic pathway classification statistics. Note: The ordinate is the annotated KEGG pathway, and the abscissa is the proportion of genes annotated to the pathway.
Figure 4.
KEGG metabolic pathway classification statistics. Note: The ordinate is the annotated KEGG pathway, and the abscissa is the proportion of genes annotated to the pathway.
Figure 5.
Pathway diagram of KEGG enrichment analysis (a: LO5 vs LCK, b: LO10 vs LCK, c: LO20 vs LCK; d: LN5 vs LCK; e: LN10 vs LCK, f: LN20 vs LCK). Note: red indicates that the KO node contains up-regulated genes, and green indicates that the KO node contains down-regulated genes.
Figure 5.
Pathway diagram of KEGG enrichment analysis (a: LO5 vs LCK, b: LO10 vs LCK, c: LO20 vs LCK; d: LN5 vs LCK; e: LN10 vs LCK, f: LN20 vs LCK). Note: red indicates that the KO node contains up-regulated genes, and green indicates that the KO node contains down-regulated genes.
Figure 6.
Hierarchical clustertingtree.
Figure 6.
Hierarchical clustertingtree.
Figure 7.
Module eigenegene correlation between different modules of WGCNA.
Figure 7.
Module eigenegene correlation between different modules of WGCNA.
Figure 8.
Expression levels of all genes and corresponding module eigengene in MEdarkturquoise (left) module and Coral (right) module.
Figure 8.
Expression levels of all genes and corresponding module eigengene in MEdarkturquoise (left) module and Coral (right) module.
Figure 9.
qRT-PCR identification and RNA-Seq analysis. Note: A: LOC103857346, Actin-related protein 2/3 complex subunit 1A; B: LOC103828797, Actin-related protein 2/3 complex subunit 2A; C: LOC103874199, Superoxide dismutase [Fe] 3; D: LOC103838242, Monodehydroascorbate reductase; E: LOC103846588, Thioredoxin M2; F: LOC103873206, ABC transporter A family member 7; G: LOC106353797, ABC transporter B family member 26; H: LOC103859979, ATP sulfurylase 1; I: LOC103859070, Cysteine synthase D1.
Figure 9.
qRT-PCR identification and RNA-Seq analysis. Note: A: LOC103857346, Actin-related protein 2/3 complex subunit 1A; B: LOC103828797, Actin-related protein 2/3 complex subunit 2A; C: LOC103874199, Superoxide dismutase [Fe] 3; D: LOC103838242, Monodehydroascorbate reductase; E: LOC103846588, Thioredoxin M2; F: LOC103873206, ABC transporter A family member 7; G: LOC106353797, ABC transporter B family member 26; H: LOC103859979, ATP sulfurylase 1; I: LOC103859070, Cysteine synthase D1.
Table 1.
Photosynthetic pigment content of Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
Table 1.
Photosynthetic pigment content of Suzhouqing treated with different concentrations of organic selenium and nano-selenium.
| Treatment |
Chlorophyll a content (mg g-1 fresh weight) |
Chlorophyll b content (mg g-1 fresh weight) |
Total chlorophyll content (mg g-1 fresh weight) |
Chlorophyll a/b |
Carotenoid content (μg g-1) |
| LCK |
0.89±0.02e |
0.45±0.02bc |
1.34±0.01c |
1.96±0.15c |
107.85±4.39d |
| LO5 |
1.04±0.05bc |
0.47±0.02b |
1.5±0.04b |
2.23±0.16ab |
155.18±7.11bc |
| LO10 |
0.95±0.02de |
0.43±0.02c |
1.37±0.02c |
2.22±0.1b |
141.75±13.35c |
| LO20 |
1.53±0.03a |
0.63±0.02a |
2.16±0.03a |
2.44±0.1a |
249.83±16.58a |
| LN5 |
1.05±0.05b |
0.47±0.02b |
1.52±0.07b |
2.24±0.11ab |
165.49±13.51b |
| LN10 |
1.03±0.04bc |
0.45±0.04bc |
1.48±0.06b |
2.29±0.2ab |
145.46±7.23c |
| LN20 |
0.99±0.05cd |
0.46±0.01b |
1.45±0.05b |
2.13±0.09bc |
146.14±18.27bc |
Table 6.
Genes involved in antioxidative stress.
Table 6.
Genes involved in antioxidative stress.
| Gene name |
Transcripts Per Kilobase Million (TPM) |
Function |
| LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| LOC103874199 |
30.28 |
19.88 |
57.58 |
0 |
51.84 |
59.11 |
19.66 |
Superoxide dismutase [Fe] 3 |
| LOC103857203 |
386 |
441 |
791 |
251 |
798 |
332 |
458 |
Superoxide dismutase [Fe] 2 |
| LOC103838242 |
0 |
214.58 |
7.63 |
0 |
33.25 |
14.4 |
0 |
Monodehydroascorbate reductase |
| LOC103846588 |
4.08 |
9.16 |
6.09 |
0 |
20.4 |
3.19 |
4.52 |
Thioredoxin M2 |
Table 8.
Selenium metabolism related genes.
Table 8.
Selenium metabolism related genes.
| Gene name |
Transcripts Per Kilobase Million (TPM) |
Function |
| LCK |
LO5 |
LO10 |
LO20 |
LN5 |
LN10 |
LN20 |
| LOC103859979 |
385 |
820 |
667 |
601 |
2072 |
935 |
820 |
ATP sulfurylase 1 |
| LOC103859070 |
0 |
32.81 |
12.43 |
18.59 |
128.88 |
9.64 |
7.02 |
Cysteine synthase D1 |
| LOC103873728 |
53.22 |
12.86 |
20.8 |
0 |
0 |
0 |
0 |
Glutaredoxin-C1 |
| LOC103834501 |
1.48 |
0 |
3.32 |
0 |
34.39 |
9.1 |
5.9 |
Glutaredoxin-C6 |
| LOC103831537 |
1.09 |
406 |
76.93 |
52.03 |
0 |
90.99 |
221.85 |
Glutathione S-transferase U12 |
| LOC103848467 |
338.38 |
278.31 |
268.22 |
0 |
80.84 |
332.68 |
163.07 |
Glutathionyl-hydroquinone reductase YqjG |
| LOC103838277 |
245.06 |
71.83 |
35.63 |
14.66 |
160.63 |
93.06 |
51.59 |
Probable glutathione peroxidase 8 |
| LOC103828203 |
387 |
580 |
338 |
150 |
419 |
349 |
331 |
S-sulfo-L-cysteine synthase (O-acetyl-L-serine-dependent) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).