1. Introduction
Nano-biotechnology represents one of the most promising technological domains, encompassing contemporary nanoscience and nanotechnology, which integrates various scientific disciplines, including biology, chemistry, physics, and material science as highlighted by [
1,
2]. Any particle falling within the size range of 1 to 100 nanometers is considered a viable nanoparticle that can be synthesized under specific controlled conditions [
3,
4]. Nanoscale metals exhibit enhanced electrical, optical, mechanical, unique chemical, and physical properties, rendering them highly versatile in numerous applications as observed in early studies [
5].
Nanoparticles are typically produced through either a top-down or bottom-up approaches. In the top-down method, large-scale substances are progressively reduced to nanometer-sized materials, while the bottom-up system entails the assembly of molecules to achieve nanometer-sized structures. The bottom-up method is commonly employed in the processes of biology and chemistry production of NPs [
6]. Traditional methods for creating metal nanoparticles rely on chemical and physical processes, but these approaches have several drawbacks, including high costs, the use of toxic and hazardous compounds for capping and reduction agents [
7], limited mono-dispersity, energy-intensive processes, heaviness, the generation of hazardous byproducts as noted in the study by Li et al. [
8].
Green chemistry finds diverse applications, including drug delivery, sterilization, DNA delivery, catalysis, antimicrobial agents, tissue repair, radiotherapy development, sensors, wound healing, and food preservation [
9]. Furthermore, silver nanoparticles (AgNPs) serve as natural antimicrobial agents and have a wide array of uses, including textile production and water purification methods. They are commonly found in air sterilizer sprays, socks, pillows, ventilators, detergents, moist towelettes, toothpaste, soaps, shampoos, refrigerator coatings, washing machines, food packaging, and even in phones [
10].
Given silver’s potent toxicity to various microbes, its utilization as an antimicrobial agent is widespread. Consequently, the advancement of silver nanoparticles opens up new avenues for disease treatment and prevention [
11]. The strong antibacterial effectiveness of silver NPs is attributed to their large surface area, allowing for more effective interaction with microbial cells compared to larger particles of the same material. Among metal nanoparticles, AgNPs are particularly renowned for their versatility, finding applications in biosensing, photonics, photocatalysis, pharmaceuticals, and microelectronics.
Moreover, AgNPs possess antimicrobial properties against fungi, viruses, and bacteria, while also demonstrating anti-inflammatory and anticancer activities, rendering them invaluable in fields like medicine and environmental applications, including clothing, cosmetics, and various consumer products [
12].
The biological synthesis of silver nanoparticles, particularly through algae like cyanobacteria, stands out as the most advantageous method. This approach offers safety, environmental friendliness, scalability, ease of production, low energy requirements, the convenience of operating at room temperature under normal conditions (rather than extreme conditions), and a rapid and stable process when compared to chemical or physical techniques [
13].
Cyanobacteria serve as an ideal biological model for nanoparticle biosynthesis due to their inherent characteristics, including accelerated growth rates, high productivity, and an efficient nitrate transport mechanism [
10,
14]. Biomolecules like lipids, carbohydrates, enzymes, and proteins are believed to play pivotal roles as capping agents in the process of reducing silver ions to silver NPs within the cell extract [
15,
16].
In this investigation, cyanobacterium Synechococcus sp. was employed to transform AgNO3 to AgNPs on a nanoscale. The biosynthesized AgNPs were evaluated using various machines, including UV-Vis spectroscopy, Zeta sizer and potential, SEM, and FTIR. Furthermore, the study explored the antibacterial properties of the prepared silver NPs.
2. Materials and Methods
2.1. Isolation, purification, and culture condition for Synechococcus sp.
Soil samples were collected and allowed to naturally air-dry at ambient temperature (25±2 °C) and subsequently underwent a meticulous sieving process to eliminate any impurities (see
Figure 1). Following this, one gram of the soil was mixed with 0.5 mL of water and introduced into sterilized Polytoma solid media [
17], which included 0.8 g/L agarose [
18], then placed in an incubator at a controlled temperature of 30±2 °C under a 14/10-hour day/night cycle with a light intensity of 35 µmol m
-2 s
-1. Weekly assessments were conducted to oversee and confirm the growth of cyanobacteria. The microalgal strain isolation was accomplished through the micromanipulation method and streak plates, and these procedures were iterated multiple times to attain purified cultures [
19]. Subsequently, the purified culture was cultured in BG11 medium [
20] to prepare working cultures. One hundred milliliters of stock culture were incubated in Erlenmeyer flasks containing sterilized medium at 30±2 °C at 14-10-hour day/night cycle under specific light conditions of 35 µmol m
-2 s
-1.
To confirm the axenic cultures in the isolated microalgal strain, its purity was tested using the following method: 1 mL of the purified Synechococcus sp. culture was blended with nutrient agar medium to encourage the growth of any potentially contaminating bacterial species. The mixture was then incubated at 37 °C for 24 hours.
2.2. Molecular identification
The isolated DNA by Qiagen DNeasy plant mini kit was sent to Novogene company UK to identify the purified isolate using 16S rRNA sequencing. The utilized specific primers (
Table 1) were used for PCR amplification of selected areas. The PCR outcomes were subjected to electrophoresis on a 2% agarose gel to identify the appropriately sized products. Subsequently, the PCR products from each sample were combined, and Illumina adapters were affixed to them. The library’s concentration was determined through Qubit and real-time PCR, while its size distribution was evaluated using a bioanalyzer. Following this, the sequence was cross-referenced with the NCBI database via BLAST, and the phylogenetic tree was crafted utilizing MEGA 11 software.
2.3. Silver nanoparticles biosynthesis
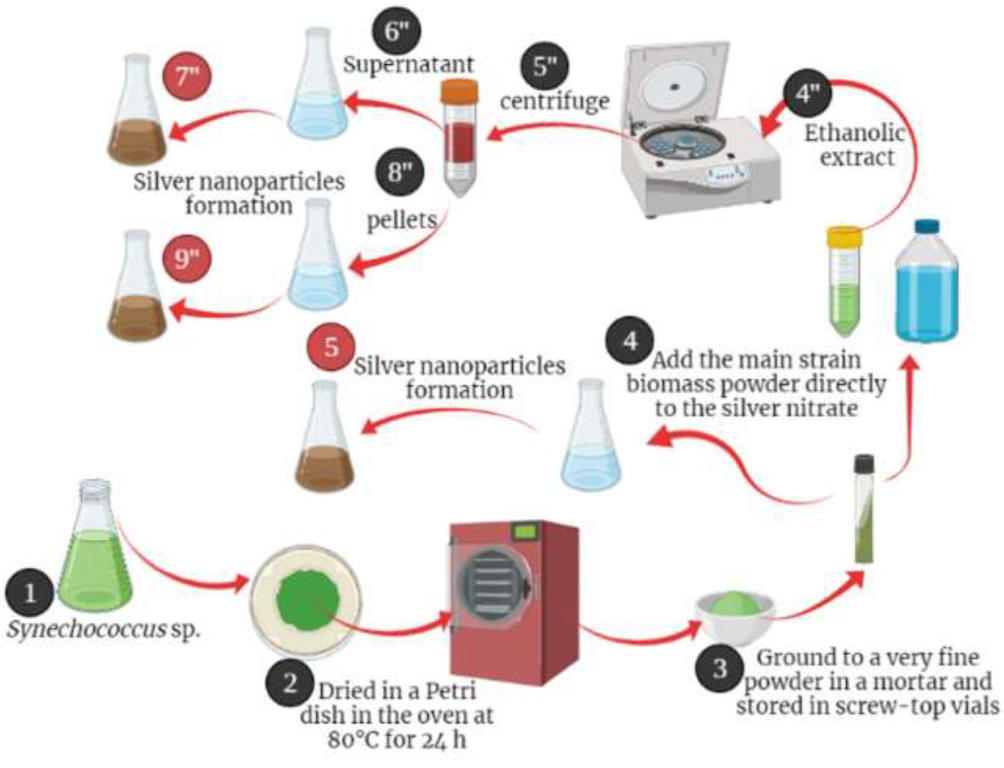
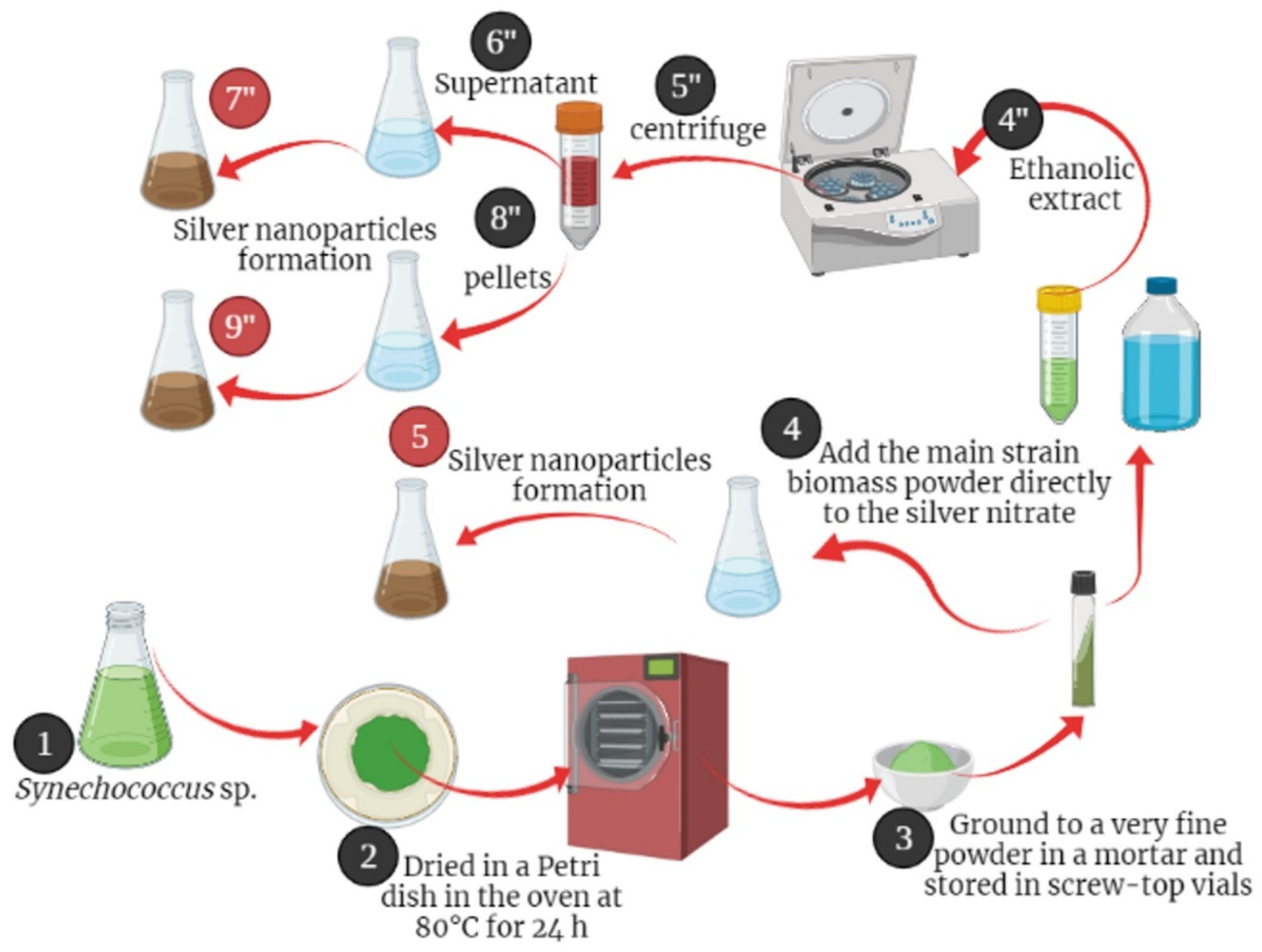
The culture of Cyanobacterium Synechococcus sp. was obtained through centrifugation at 4000 rpm for five minutes, and the resulting pellets were gathered and then dried in a Petri dish for 24 hours in an 80 °C oven. Subsequently, the biomass was finely pulverized into powder using a mortar and stored in sealed containers until required.
Figure 1 outlines the process of nanoparticle synthesis. In the initial experiment, we directly employed the primary microalgal biomass in its powder state as reducing agent for silver nitrate. This was achieved by adding 1g of the stock powder directly to 999 mL of AgNO3 solution (0.001M) under room temperature conditions with exposure to light at 25±2 °C. Another experiment involved using the absolute ethanol extract, which was obtained by immersing 2g of the
Synechococcus sp. biomass in 20 mL of ethanol for a duration of 24 hours, maintaining a uniform concentration of 100 mg/mL. Subsequently, the mixture was subjected to centrifugation to separate the pellets from the extract. The synthesis of silver NPs involved adding 1g of the air-dried extract granules to 1L of AgNO3 solution (0.001M).
Finally, the synthesized nanoparticles underwent two rinses with sterilized distilled water and were subsequently subjected to drying in a 70 °C oven for 24 hours to remove any remaining moisture. Subsequently, the dried AgNPs were finely powdered using a mortar and pestle.
2.4. Silver nanoparticles characterization
A Shimadzu UV-3600 UV spectrophotometer from Japan was employed to measure the diffuse reflectance absorption spectra of AgNPs across the UV-visible range, spanning from 250 to 800 nm. Furthermore, an FTIR-8400S Shimadzu spectrometer, also from Japan, was employed to investigate the chemical bond vibrations of silver NPs through FT-IR spectra covering the range of 400 to 4000 cm-1.
Furthermore, Zeta size and potential detection were conducted at 25 °C utilizing a Zeta sizer nano-ZS 90 equipped with transparent disposable Zeta cells manufactured by Malvern Instruments in the UK. The assessment of silver nanoparticle shape and size was carried out using scanning electron microscopy performed with a Zeiss Sigma 500 VP Analytical FE-SEM from Germany. The silver nanoparticle sample was meticulously prepared, loaded into the SEM chamber, and subjected to examination at various high magnifications. Images of the silver NPs were captured, archived, and quantified using computational software.
2.5. Antibacterial estimation
The antibacterial effectiveness of the biologically generated silver NPs and the cyanobacterium extract was assessed using the disc diffusion method at 2 distinct concentrations (0.5 and 1 mg/mL), with a 1 mg/mL Ampicillin solution as a positive control. This method was utilized to gauge the susceptibility of two G+ve bacteria, namely Staphylococcus aureus and Streptococcus sp., as well as three G-ve bacteria: Salmonella sp., Escherichia coli, and Pseudomonas sp.
To carry out this assessment, the pathogenic strains were first cultured freshly in nutrient broth for 24 hours. Following this, they were inoculated onto nutrient agar plates. Discs crafted from 5 mm Whatman filter paper were impregnated with 5 µL of each AgNPs concentration using a micropipette under sterile conditions. The dishes were subsequently kept at ambient temperature for 20 minutes before being placed in an incubator at 37 °C for a duration of 24 hours. The size of the inhibitory region was then determined using a ruler and presented in millimeters [
21].
2.6. Statistical analysis
Every experiment was executed following a design based on complete randomization and was replicated. The results are reported as (means ± standard error) and visualized utilizing GraphPad Prism software version 8.4.2.
3. Results and Discussion
3.1. Identification at the molecular level
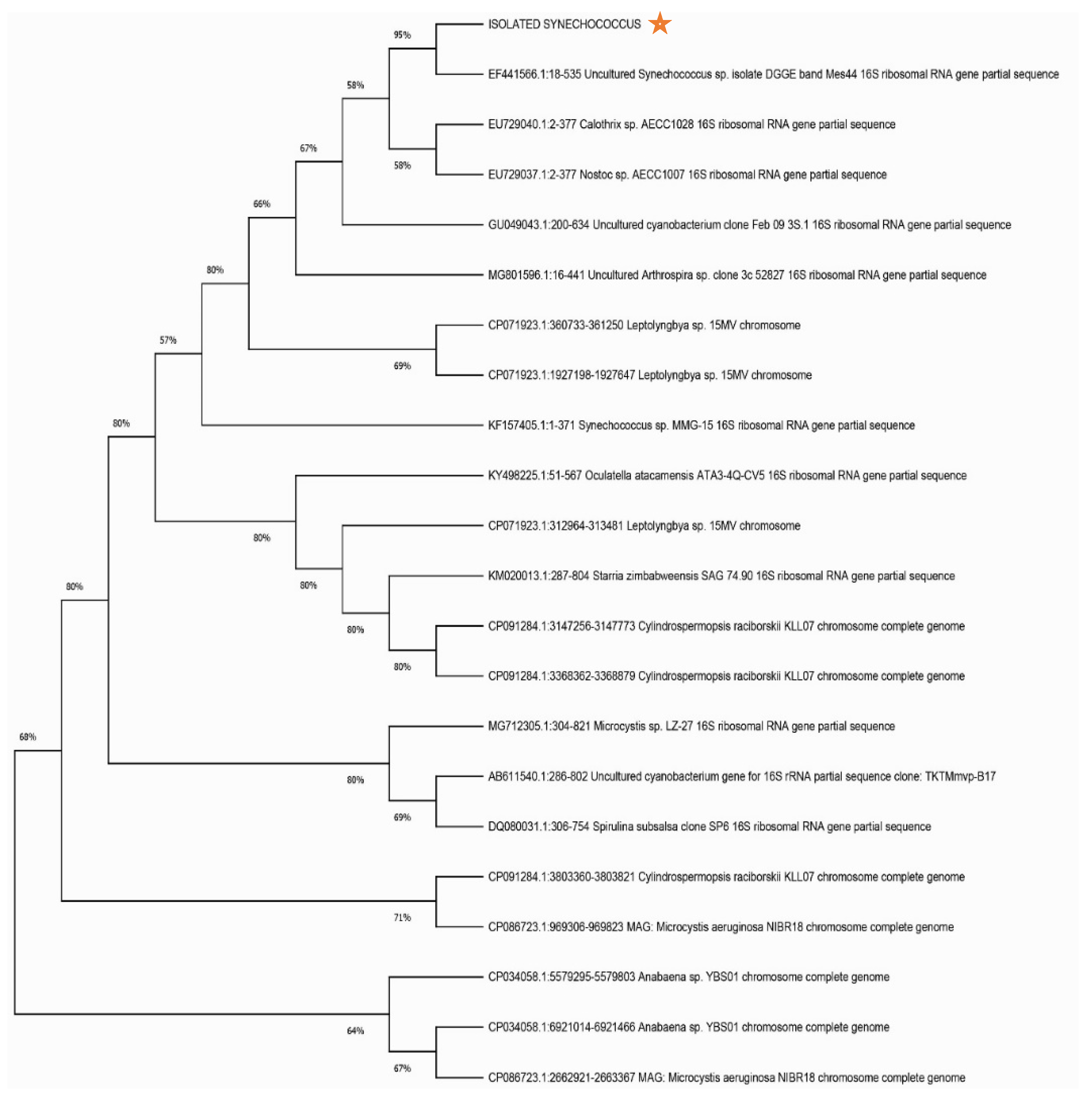
Cyanobacterial strain isolated from the natural environment was molecularly detected as
Synechococcus DGGE with accession number EF441566.1. This determination was established by constructing a phylogenetic tree with the assistance of the MEGA software, utilizing data acquired from the NCBI BLAST tool (
Figure 2).
3.2. Rapid indication for AgNPs preparation
Characterization based on visual observation involved noting the transformation of the aqueous solution to a brown hue, triggered by surface plasmon resonance activation when AgNO3 was combined with cyanobacterium powder, pellets, and ethanol extract, as compared to the control [
3,
14]. The duration of this bio-reduction process was documented, as illustrated in
Figure 3.
Upon direct addition of the Synechococcus sp. powder and ethanol extract pellets, the AgNO3 aqueous solution changed its color to a shade of yellowish-green, whereas the ethanol extract exhibited a subtle greenish tint. After 30 minutes, the powder method shifted to a faint gray color, the pellets method assumed a mild brown tint, and the extract method turned black-green. After 60 minutes, the powder method transformed into a dark gray shade, the pellets method became brown-gray, and the extract method shifted to brown. After 90 minutes, both the powder and pellets turned into a darker gray color, and the extract method produced a deeper brown color, signifying the completion of the bio-reduction process and the formation of AgNPs. Additionally, after 24 hours, the black-brown color was examined to highly study bio-reduced AgNPs generated using the powder and pellets techniques, while the extract method remained unchanged.
3.3. Characterization of the produced silver nanoparticles
3.3.1. UV spectrum
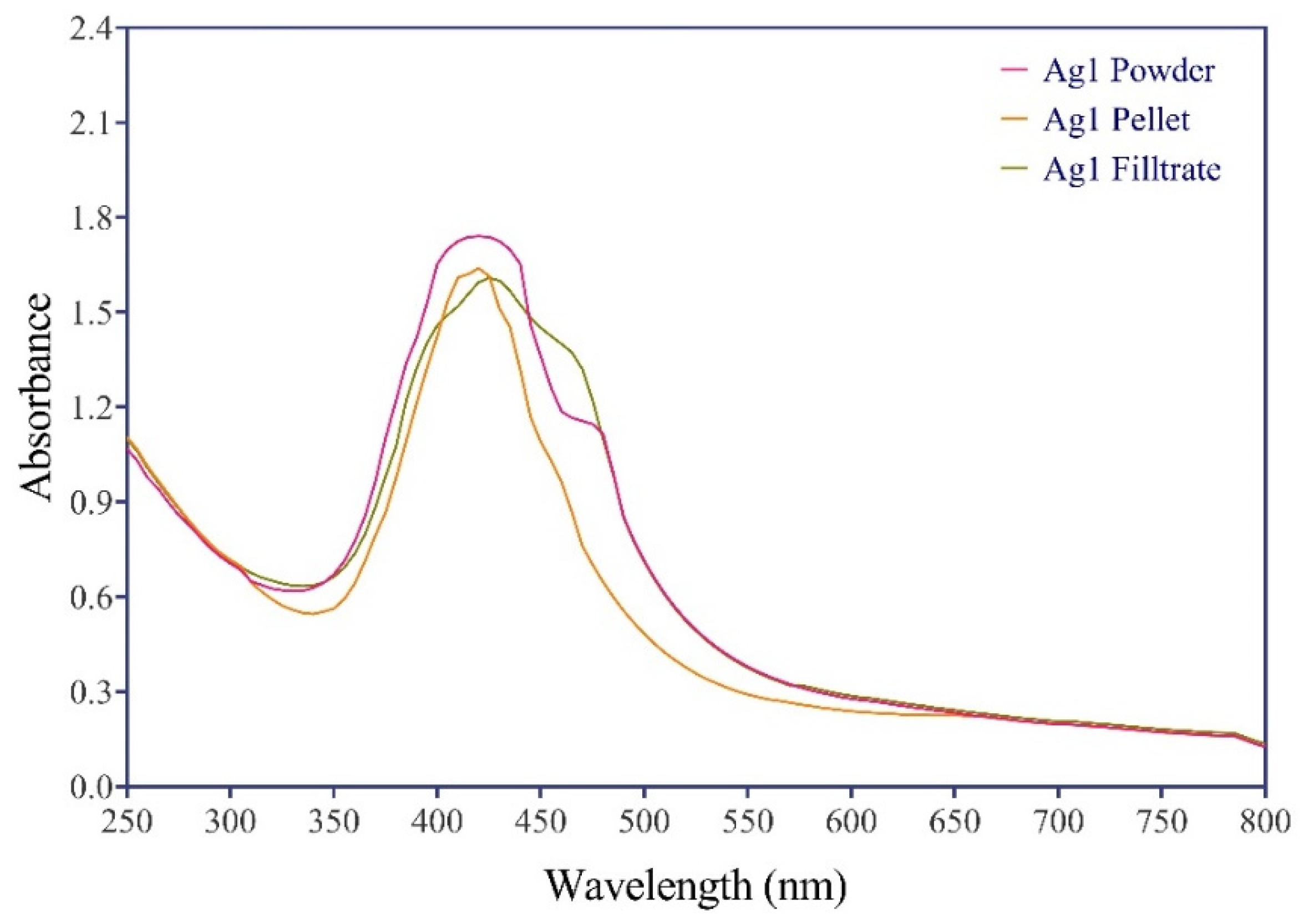
UV-Vis spectroscopy is employed as a method for examining the indirect bio-reduction of silver NPs from an aqueous AgNO
3 solution. To confirm the reduction of Ag+, the UV-Vis spectra of the reaction mixture were compared to a blank containing distilled water. The emergence of a dark brown color, induced by surface plasmon resonance, is considered a significant early indicator of the presence of AgNPs [
22]. The emergence of surface plasmon resonance bands at 420 nm signified the generation of AgNPs. Although the peak intensities of both methods were not identical, the formation of AgNPs using the pellet-based method was notably faster, resulting in a broadening and sharpening of the 420 nm peak as AgNPs were generated. These results also revealed that AgNPs in aqueous solutions were polydisperse and did not exhibit signs of aggregation. The UV-Vis spectral analysis (
Figure 4) demonstrates a transition to a dark brownish color with distinct bands around 420 nm, confirming the formation of bio-capped AgNPs.
Several previous studies have documented a similar UV spectrum of AgNPs bio-synthesized from green sources, featuring a peak around the 400 nm region [
23,
24]. In the AgNPs spectra, surface plasmon resonance bands were detected, aligning with the wavelength at which both the real and imaginary components of silver’s dielectric function practically disappeared. Multiple factors, such as the medium’s dielectric constant and particle volume, influence the position of absorbance, as depicted in
Figure 4. The strength of the silver NPs peak produced by the powder method was 1.7, subsequently by the pellet method at 1.6, and lastly, the ethanol extract approach also at 1.6. All of these methods exhibited a slight shift in the band position, suggesting the presence of polydisperse particles of varying sizes. Significantly, the plasmon band displayed broadening and shifting, indicating particle agglomeration and an increase in volume [
24].
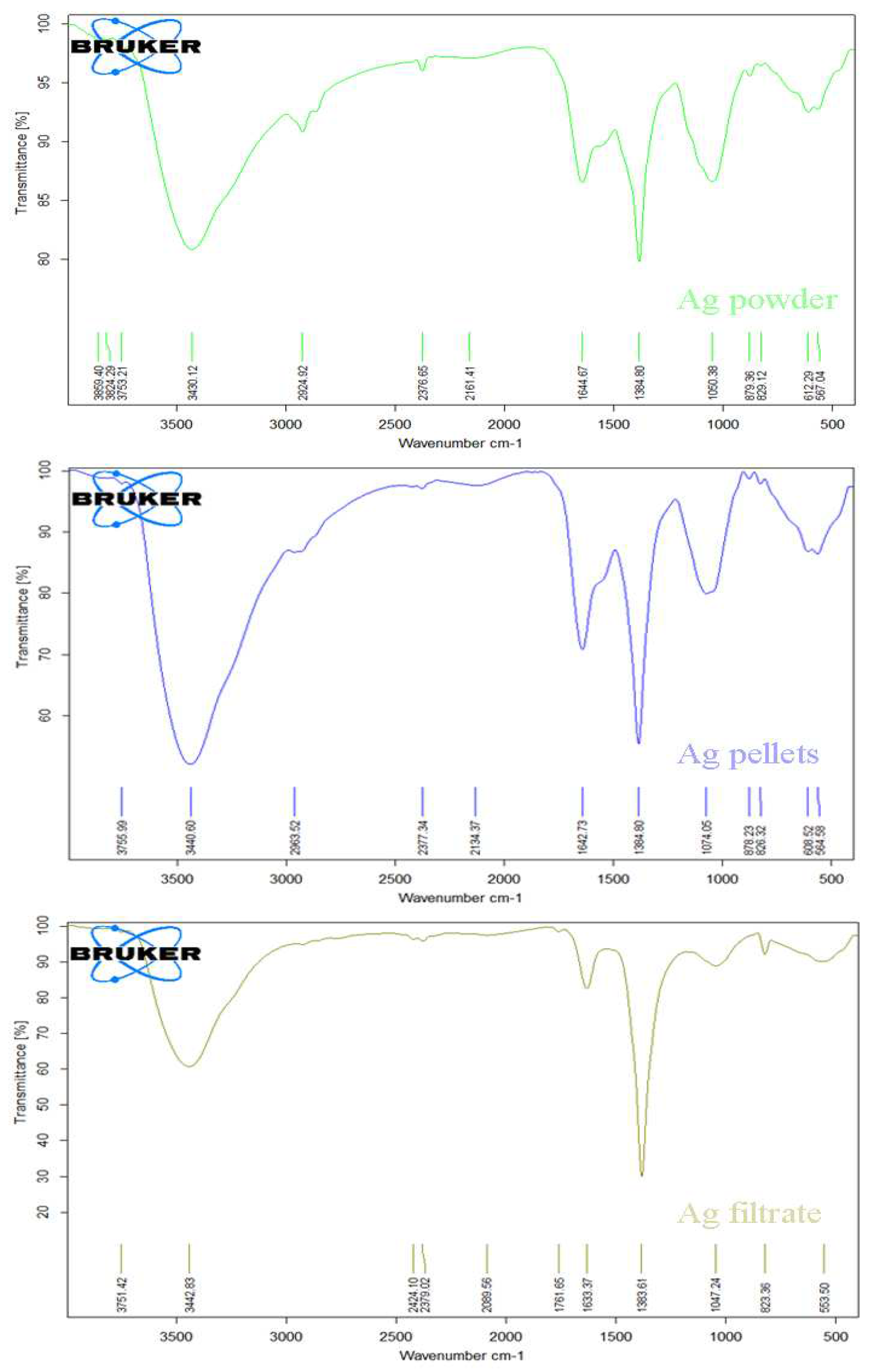
3.3.2. FT-IR spectrum
FTIR analysis was utilized to detect potential interfacial groups that may exist between the capping agents and silver nanoparticles. This analytical technique was also utilized to pinpoint the primary functional groups accountable for the conversion of Ag
+ to Ag0 in those produced by the cyanobacterium (see the FT-IR spectrum in
Figure 5). The spectra exhibit peaks at 3430, 2924, 2376, 1644, 1384, 1050, 879, and 612 cm
−1 in the AgNPs’ spectra synthesized using the powder method. For AgNPs synthesized by pellets, the FT-IR spectrum displays prominent bands at 3440, 2963, 2856, 2377, 2134, 1642, 1384, 1074, 878, and 608 cm
−1. Moreover, noticeable differences in the spectrum of AgNPs from the filtrate include peaks at 3442, 2379, 1761, 1383, 1047, 823, and 553 cm
−1.
The bands detected in the FTIR spectrum can be elucidated in the following manner: The wide peak at around 3400 cm
−1 is attributed to the stretching and vibrations of hydroxyl groups present in phenolic, flavonoid, and triterpenoid compounds derived from the cyanobacterium
Synechococcus sp. biomass [
25]. Peaks in the vicinity of 2900 cm
−1 correspond to the broadened (C–H) bands, suggesting the existence of aliphatic compounds contributing to the bio-reduction process [
26]. Moreover, the bands approximately at 2200 cm
−1 may be associated with the protein extension vibrations within the (N-H) amide group [
14]. The band at 1600 cm
−1, attributed to the (C=O) stretching, implies that proteins can potentially bind to Ag
+ through either (RCOO) or free (NH
2) groups [
25]. Additionally, the band at approximately 1300 cm
−1 can be associated with the (O-H) bending characteristic of phenolic compounds or the presence of the (C-H) functional group observed in bio-capped AgNPs [
16]. The bands around 1100 cm
−1 are attributed to the broadening vibrations of (C-O) bonds present in proteins, carboxylic acids, esters, and polysaccharides, which are associated with the C-Alkyl halide stretch group. As indicated by the FTIR spectrum, these naturally occurring organic compounds play crucial roles in both reducing Ag+ ions and stabilizing the biologically generated AgNPs.
3.3.3. Evaluation through Zeta Sizer and Potential analysis:
The particle size of biogenically produced silver NPs is illustrated in
Figure 6. As depicted in the figure, the obtained AgNPs exhibit polydispersity. The mean size of AgNPs synthesized using the powder technique is 189 nm, with a polydispersity index (PDI) of 0.456. AgNPs produced through the pellet method exhibit an approximately size of 124.2 nm, accompanied by a PDI of 0.371. Lastly, AgNPs produced using the ethanol extract method have an approximately size of 158.9 nm, with a PDI of 0.396.
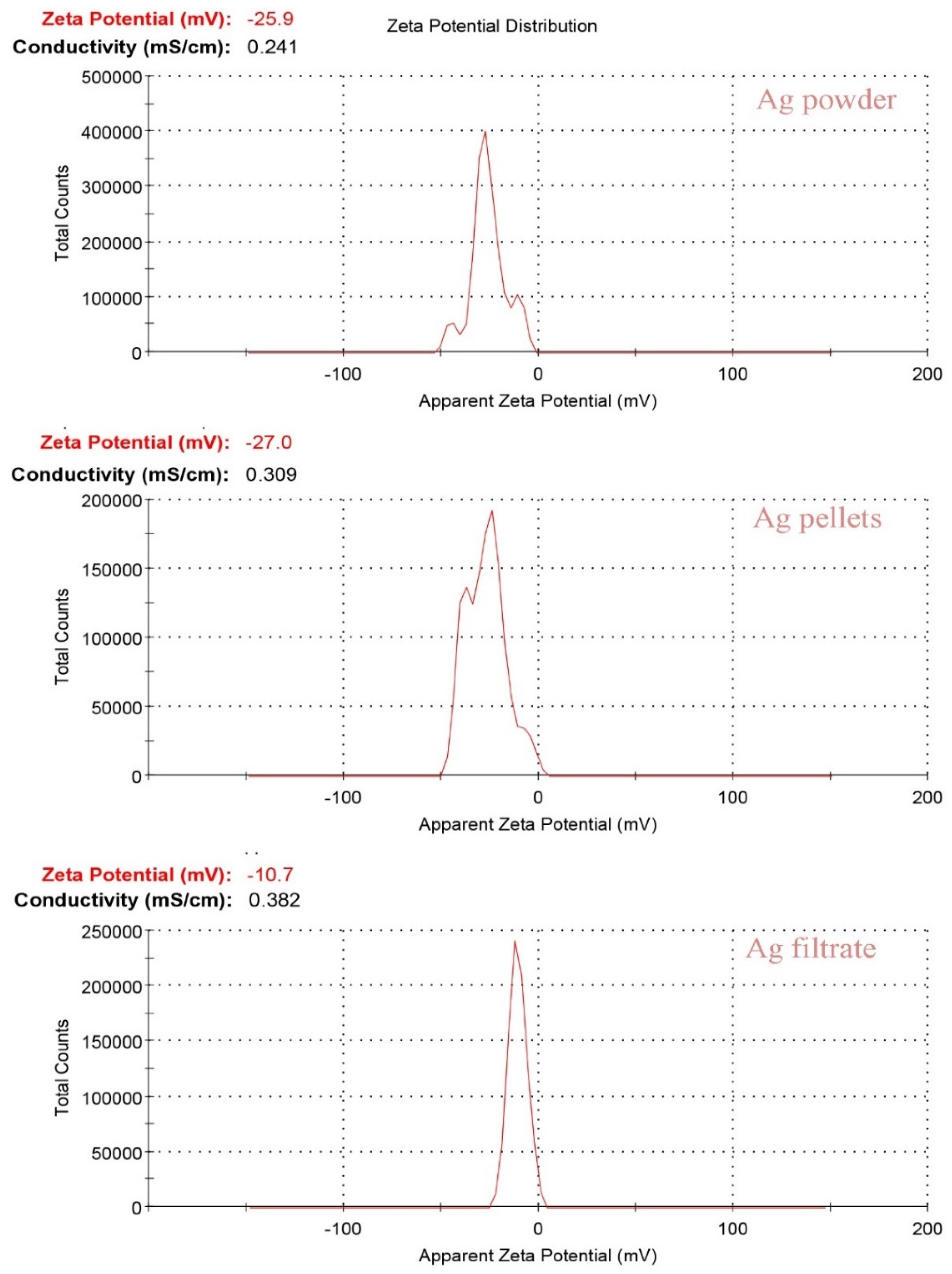
Zeta potential, a physical property of particles in suspension that predicts their long-term stability, was investigated. It represents the surface potential of molecules and is influenced by changes in the interface within the dispersion due to the presence of functional groups on the molecule’s surface or the adsorption of ionic species in the liquid dispersion medium [
27]. The stability of the biologically synthesized AgNPs was verified by conducting Zeta potential assessments. The surface charge of AgNPs produced from
Synechococcus sp. is depicted in
Figure 7. For the powder method, the Zeta potential was determined to be -25.9 mV, with a conductivity of 0.241 mS/cm, while in the pellet method, it registered -27 mV, with a conductivity of 0.309 mS/cm. In the ethanolic extract method, the average Zeta potential was found to be -10.7 mV, with a conductivity of 0.382 mS/cm.
These results suggest that the biogenetically produced AgNPs exhibit polydispersity and long-term stability, as indicated by the negatively charged surface of the nanoparticles they generated. Furthermore, it’s worth mentioning that nanoparticles generally need a Zeta potential value exceeding +30 mV or falling below -30 mV to maintain adequate electrostatic repulsion, which guarantees their stability in a solution [
28]. In this context, it’s worth noting that all the AgNPs we biogenically synthesized are prominently anionic, a significant characteristic for their biological applications [
29]. Additionally, a relatively elevated Zeta potential value is necessary for colloidal suspensions to remain chemically and physically stable since substantial repulsive forces prevent aggregation resulting from inadvertent interactions among neighboring nanoparticles [
30].
3.3.4. Scanning electron microscope
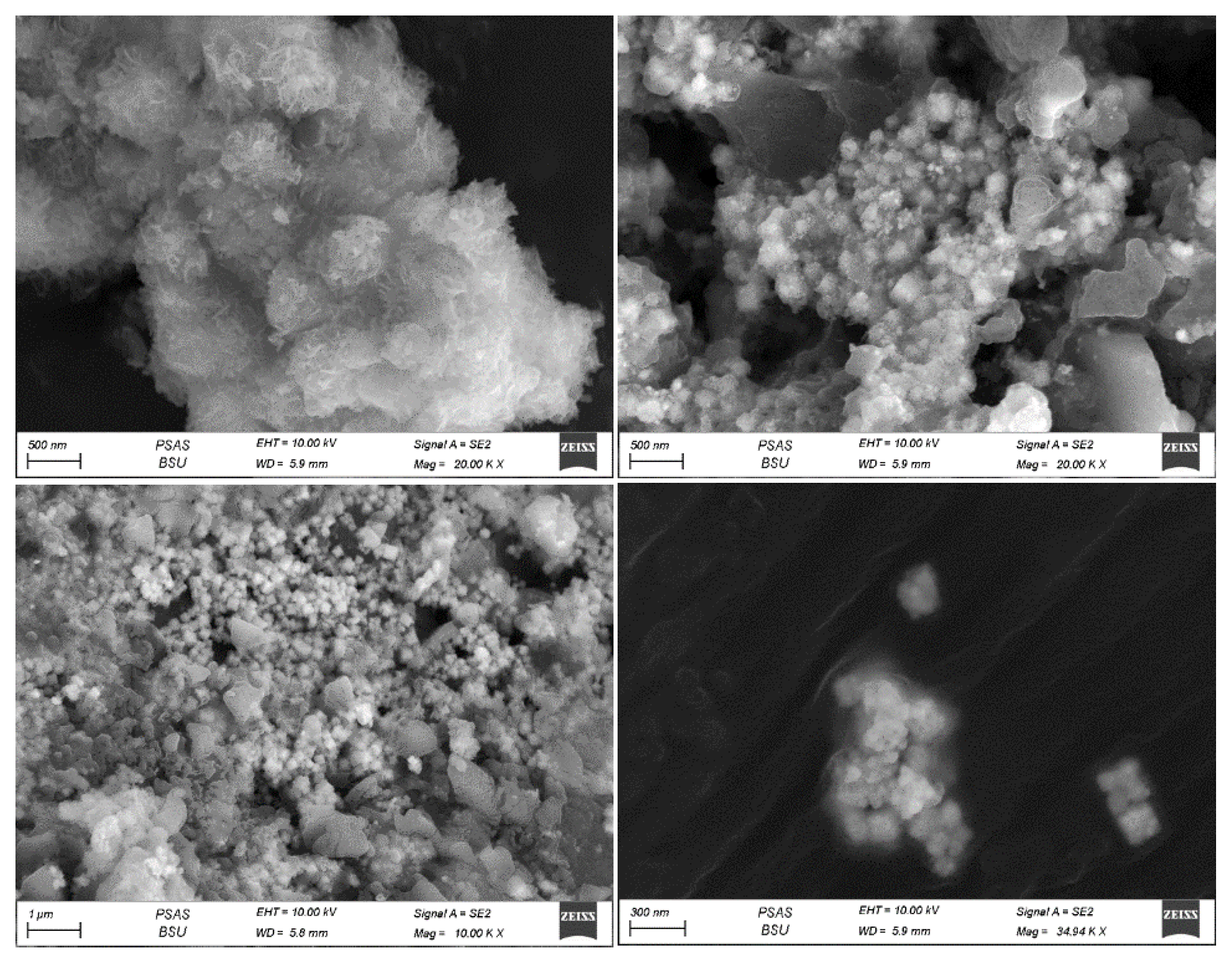
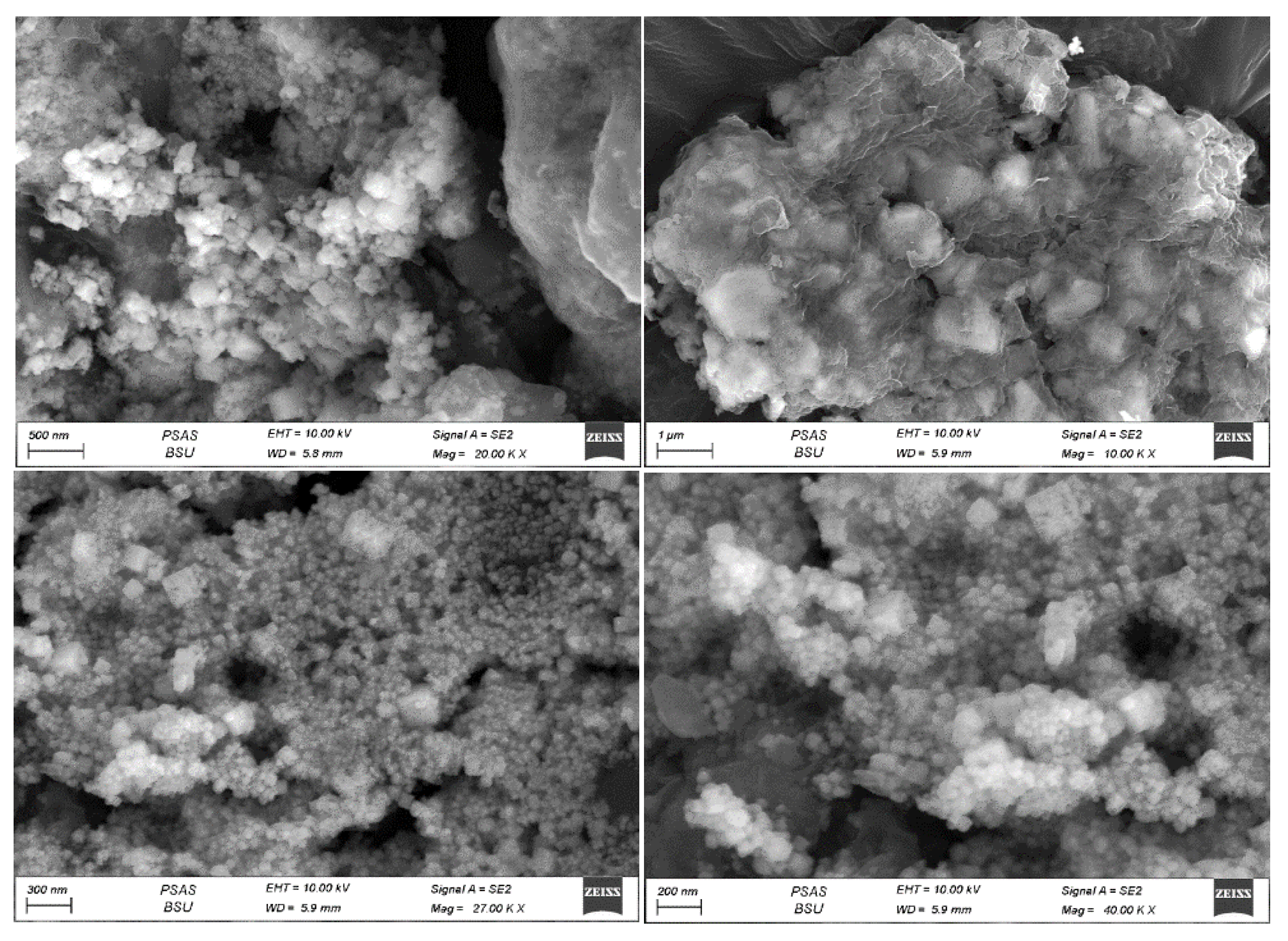
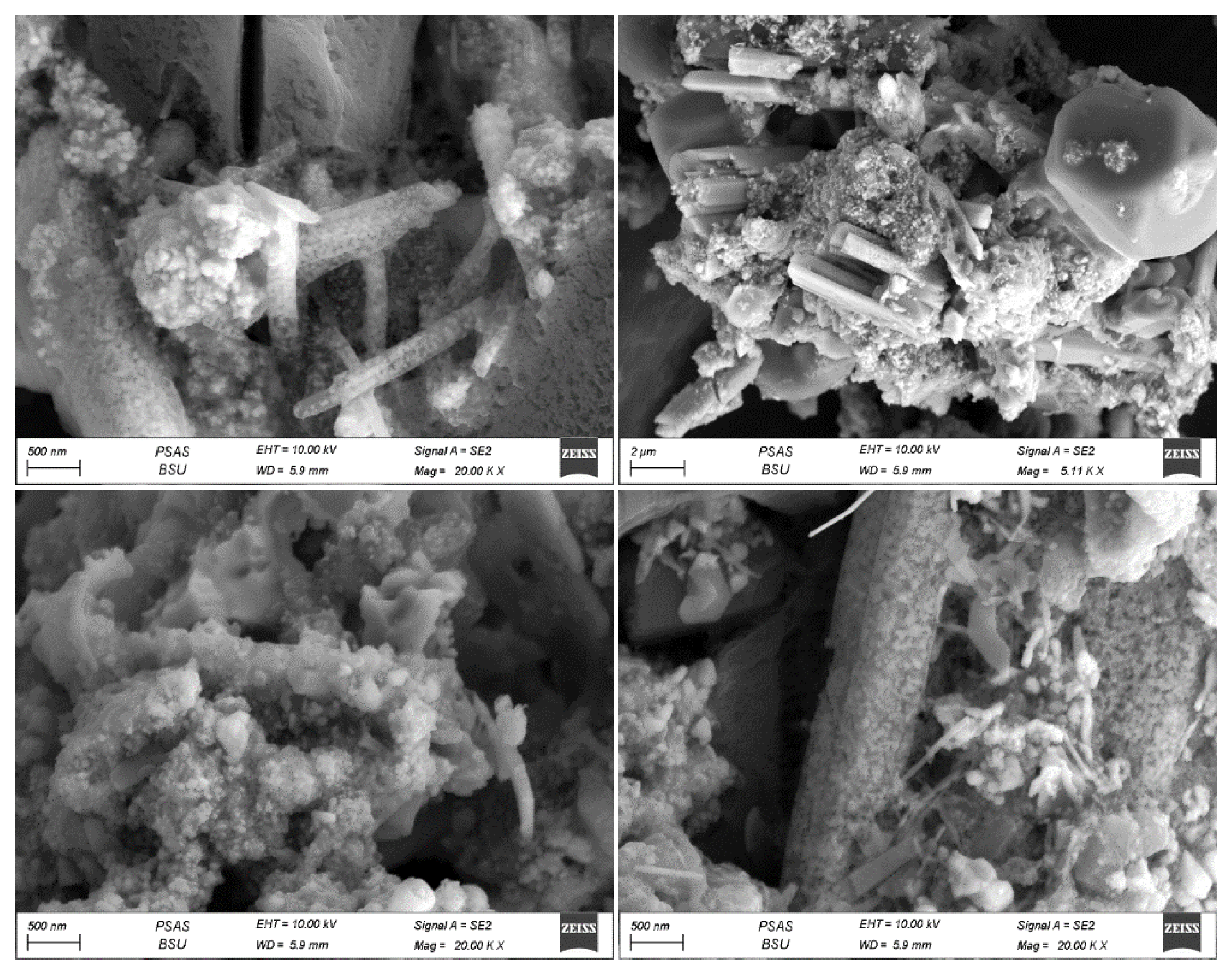
SEM analysis was employed to characterize the morphological features of the silver nanoparticles, a standard method for discerning the shape and size of biogenically synthesized nanoparticles. SEM images obtained through the three techniques revealed that the particles displayed polydispersity and exhibited various forms, including cubic, spherical, rod-shaped, and irregular nanoparticles.
For the powder technique, the particle sizes of individual sliver nanoparticles ranged from 20 to 80 nm, displaying plate-like, spherical, and cubic shapes (refer to
Figure 8). Meanwhile, the pellet approach yielded particles with sizes ranging from 42 to 89 nm, also appearing in plate-like, spherical, and cubic shapes (see
Figure 9). Additionally, the particle size distribution of sliver nanoparticles produced using the ethanolic extract method (depicted in
Figure 10) ranged from 40 to 140 nm, with spherical forms spanning from 40 to 90 nm.
Images acquired from all three techniques explained that the particles exhibited a tendency to aggregate within substantial clusters, enveloped by a bio-capped layer. The existence of varied polyphenolic elements, encompassing flavonoids and terpenoids, aided in the reduction of Ag+ ions, conferred stability to the surface of the capped AgNPs, and resulted in in vivo reduction and capping [
31]. Many of these bio-capped particles exhibited various sizes, encompassing cubic, spherical, irregular plate-like, and rod-shaped structures.
SEM images clearly reveal multiple aggregations, encased by a layer of biomolecules that actively participated in both the bio-reduction of Ag
+ and the stabilization of the resulting AgNPs. This observation lends support to the presence of distinctive peaks and shoulders in the UV absorption spectra, as well as the notable Zeta sizer values. It is worth noting that the impact of these nanoparticles, particularly in terms of cellular activation, cellular uptake, and intercellular distribution, is primarily governed by factors like shape, size, charge, functionalization, and core structure [
32]. Consequently, this study represents a promising exploration into the antimicrobial properties of silver nanoparticles in various shapes and sizes.
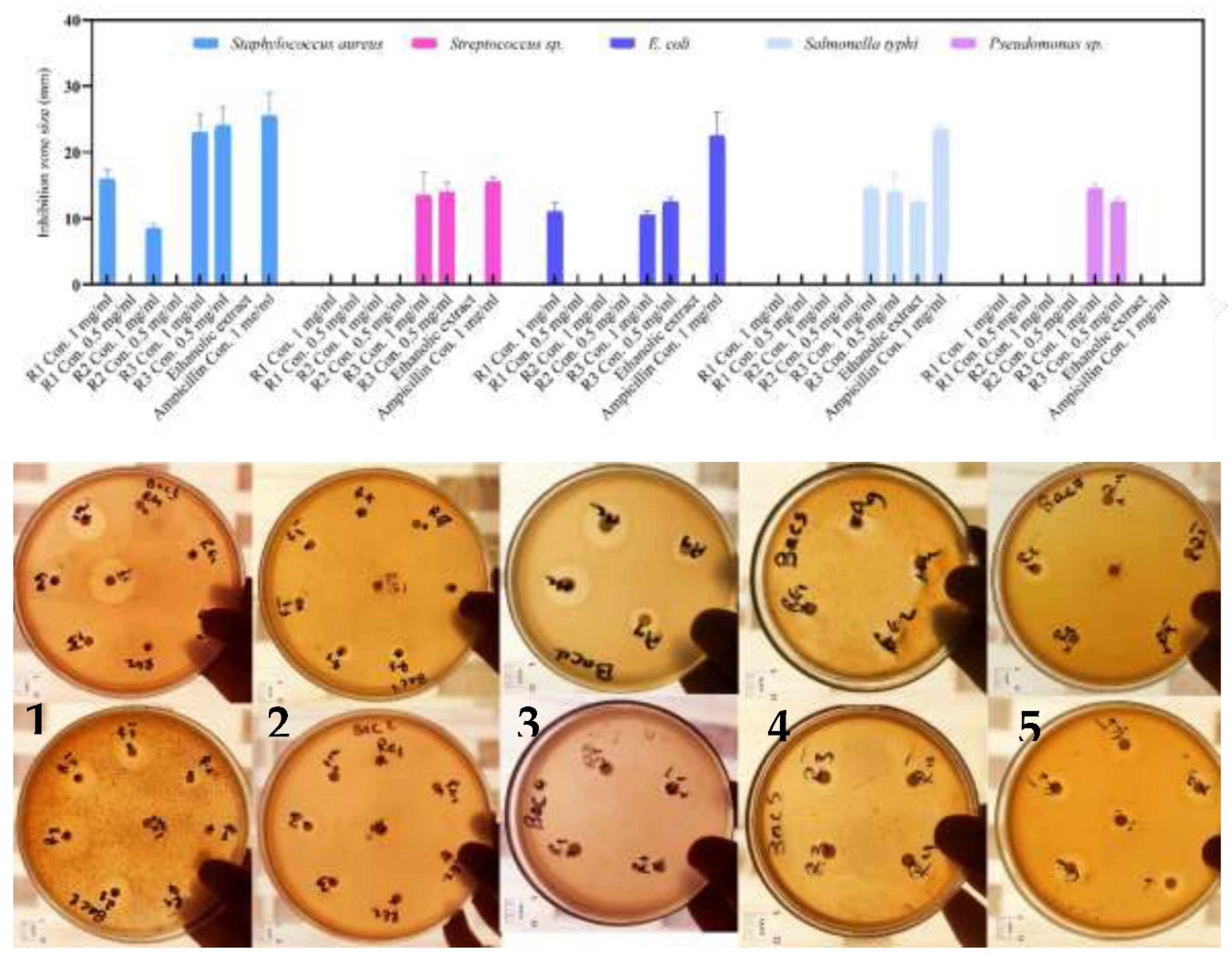
3.3.5. Antibacterial potentials
Silver nanoparticles have exhibited their efficacy against a wide range of pathogens, numbering over 650 species. Nevertheless, the exact mechanism responsible for their antibacterial activity remains enigmatic [
33]. We employed the agar disc diffusion technique to assess the antimicrobial efficacy of both the
Synechococcus extract and the silver NPs biologically synthesized against specific pathogenic bacteria, as illustrated in
Figure 11.
The Synechococcus extract had no inhibition zone with the bacterial strains subjected to the most extensive testing, however the biosynthesized NPs displayed antibacterial activity with some variation among pathogenic strains. In this respect, the greatest inhibition zones (24 mm and 23 mm) were achieved by application of 1 and 0.5 mg/ml, respectively by ethanolic extract AgNPs against Staphylococcus aureus. However, the low concentrations of the produced nanoparticles (0.5 mg/ml) generated by the powder and pellet techniques showed negative results. However, when using a concentration of 1 mg/ml of silver NPs, the powder method resulted in a distinct zone of 16 mm, while the pellet method yielded a clear zone measuring 9 mm.
On the other hand, when using the powder and pellet methods, AgNPs did not show any inhibitory effect against Streptococcus sp. However, the ethanolic extract method resulted in a clear zone of 14 mm at dosages of 1 and 0.5 mg/ml, which was similar to the result obtained by Ampicillin as a positive control, producing a clear zone of 15.5 mm. Conversely, when testing Gram-negative bacteria, the E. coli strain was inhibited by AgNPs prepared using the powder technique with a concentration of 1 mg/ml, resulting in a clear zone of 11 mm. AgNPs of the filtrate method with a low dose of 0.5 mg/ml also created a clear zone of 12.5 mm. Finally, when combined with Salmonella sp. and Pseudomonas sp., AgNPs of the filtrate produced a clear zone of 14.5 mm.
The obtained data aligns with earlier research [
15,
34], where nanoparticles were produced and led to a 22 mm suppression of
Staphylococcus growth. Furthermore, these studies demonstrated the efficacy of these nanoparticles against both pathogenic bacteria and the unicellular organism
Candida albicans.
When comparing the three methods employed this investigation to evaluate the antibacterial efficacy of the blue-green algae
Synechococcus, it became evident that silver NPs prepared by the ethanol method exhibited a significant impact on both G+ve and G-ve bacteria. This phenomenon can be attributed to the remarkably small size of these silver NPs. In this context, some workers discussed that the antibacterial activity of silver nanoparticles is associated with four distinct mechanisms: (1) The adherence of silver nanoparticles to the cell wall and the outer surface of the cell membrane, (2) The penetration of silver nanoparticles into the cell, causing damage to intracellular structures such as vacuoles, ribosomes, mitochondria, and biomolecules like lipids, proteins, and DNA. (3) The inhibition of cytotoxicity and oxidative stress induced by oxygen-active molecules and free radicals, (4) The modulation of signal transduction pathways [
35].
4. Conclusions
In the current investigations, Synechococcus nanoparticles were created using a non-harmful method. Powder, pellets, and ethanol extract of the cyanobacterium Synechococcus were utilized. UV spectroscopy indicated a peak with shoulders at 420 nm region, while SEM revealed different morphological forms of AgNPs as cubic, spherical, plates, and rod shapes seen in aggregation and single particles with diameters from 20 to 140 nm. Additionally, from the several possible reduction groups detected by FT-IR, these biologically manufactured AgNPs have highly unique features that traditional chemistry cannot create. To sum up, the antibacterial properties of the prepared Synechococcus AgNPs suggest that they can be used as antimicrobial agents to combat a broad range of bacteria. Our prepared AgNPs demonstrated broad antibacterial effects, particularly against Gram-positive bacteria, such as Staphylococcus. The ethanolic extract method resulted in a 24 mm clear zone, while the powder and pellet methods were less effective, indicating that larger particles may not be as useful as antibacterial agents but may have advantages in other areas such as wastewater treatment. Due to their antibacterial properties, the prepared AgNPs could be utilized as sterilizing agents, coatings for medical equipment, and dental materials in various medical applications. Prospects of this research include scaling up the biosynthetic production of silver nanoparticles using Synechococcus and proving its efficacy against a wide spectrum of microbial populations. Additionally, these nanoparticles are eco-friendly and economically low cost.
Author Contributions
Conceptualization, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; methodology, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; software, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; validation, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; formal analysis, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; investigation, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; resources, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; data curation, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; writing—original draft preparation, Rania Omar; writing—review and editing, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; visualization, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; supervision, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem; funding acquisition, Reem Mohammed Alharbi, Neveen Abdel-Raouf, Rania Omar, Sherif Hassan, Khaled N.M. Elsayed and Ibraheem Borie M. Ibraheem. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the editor and the anonymous reviewers for their careful reading of our manuscript and their many insightful comments and suggestions which significantly improved our manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abdel-Raouf, N; Al-Enazi, N. M.; Ibraheem, I.B.M. Green biosynthesis of gold nanoparticles using Galaxaura elongata and characterization of their antibacterial activity. Arabian Journal of Chemistry. 2017, 10, S3029–S3039. [Google Scholar] [CrossRef]
- Alkhulaifi, M.M.; Alshehri, J.H.; Alwehaibi, M.A.; Awad, M.A.; Al-Enazi, N.M.; Aldosari, N.S.; Hatamleh, A.A.; Abdel- Raouf, N. Green synthesis of silver nanoparticles using Citrus limon peels and evaluation of their antibacterial and cytotoxic properties. Saudi Journal of Biological Sciences. 2020, 27, 3434–3441. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, I.B.M.; Abd-Elaziz, B.E.E.; Saad, W.F.; Fathy, W.A. Green biosynthesis of silver nanoparticles using marine Red Algae Acanthophora specifera and its antimicrobial activity. J Nanomed Nanotechnol. 2016, 7, 1–4. [Google Scholar]
- He, Y.; Xie, Y.; Huangm, Y.; Xia, D.; Zhang, Y.; Liu, Y.; Xiao, Y.; Shen, F.; He, J.; Luo, L. Optimization of cyanobacteria Microcystis aeruginosa extract to improve the yield of Ag nanoparticles with antibacterial property. International Biodeterioration & Biodegradation. 2022, 171, 105407. [Google Scholar]
- Al-Amoudi, O.A.; Mutawie, H.H.; Patel, A.V.; Blunden, G. Chemical composition and antioxidant activities of Jeddah corniche algae, Saudi Arabia. Saudi Journal of Biological Sciences. 2009, 16, 23–29. [Google Scholar] [CrossRef]
- Balantrapu, K.; Goia, D.V. Silver nanoparticles for printable electronics and biological applications. Journal of materials research. 2009, 24, 2828–2836. [Google Scholar] [CrossRef]
- Albrecht, M.A.; Evans, C.W.; Raston, C.L. Green chemistry and the health implications of nanoparticles. Green chemistry. 2006, 8, 417–432. [Google Scholar] [CrossRef]
- Li, H.; Carter, J.D.; LaBean, T.H. Nanofabrication by DNA self-assembly. Materials Today. 2009, 12, 24–32. [Google Scholar] [CrossRef]
- Rajendran, R.; Ganesan, N.; Balu, S.K.; Alagar, S.; Thandavamoorthy, P.; Thiruvengadam, D. Green synthesis, characterization, antimicrobial and cytotoxic effects of silver nanoparticles using Origanum heracleoticum L. leaf extract. Int J Pharmacy & Pharmaceutical Sci. 2015, 7, 288–293. [Google Scholar]
- Husain, S.; Sardar, M.; Fatma, T. Screening of cyanobacterial extracts for synthesis of silver nanoparticles. World Journal of Microbiology and Biotechnology. 2015, 31, 1279–1283. [Google Scholar] [CrossRef]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex. Langmuir. 2007, 23, 2694–2699. [Google Scholar] [CrossRef] [PubMed]
- Alavi, M. Bacteria and fungi as major bio-sources to fabricate silver nanoparticles with antibacterial activities. Expert Review of Anti-Infective Therapy. 2022, 20, 897–906. [Google Scholar] [CrossRef]
- Hamed, S.M.; Abdel-Alim, M.M.; Abdel-Raouf, N.; Ibraheem, I.B.M. Biosynthesis of silver chloride nanoparticles using the cyanobacterium Anabaena variabilis. Life Science Journal. 2017, 14, 25–30. [Google Scholar]
- Omar, R.; Ibraheem, I.B.M.; Hassan, S.; Elsayed, N.M.K. Biogenic Synthesis of Different forms of Bio-caped Silver Nanoparticles Using Microcystis sp. and its Antimicrobial Activity. Current Nanoscience. 2023, 19, 1–13. [Google Scholar] [CrossRef]
- Arya, A.; Gupta, K.; Chundawat, T.S.; Vaya, D. Biogenic synthesis of copper and silver nanoparticles using green alga Botryococcus braunii and Its antimicrobial activity. Bioinorganic Chemistry and Applications. 2018, 7879403. [Google Scholar] [CrossRef]
- Kordy, M.G.M.; Abdel-Gabbar, M.; Soliman, H.A.; Aljohani, G.; BinSabt, M.; Ahmed, I.A.; Shaban, M. Phyto-capped Ag nanoparticles: green synthesis, characterization, and catalytic and antioxidant activities. Nanomaterials. 2022, 12, 373. [Google Scholar] [CrossRef]
- Piasecka, A.; Krzeminska, I.; Tys, J. Physical methods of microalgal biomass pretreatment. International Agrophysics. 2014, 28, 341–348. [Google Scholar] [CrossRef]
- Shirai, M.; Matumaru, K.; Ohotake, A.; Takamura, Y.; Aida, T.; Nakano, M. Development of a solid medium for growth and isolation of axenic Microcystis Strains (Cyanobacteria). Applied and Environmental Microbiology. 1989, 55, 2569–2571. [Google Scholar] [CrossRef]
- Ranjan, R.; Chinnibabu, B.; Sadhu, N. Training Manual On Live feed for Marine finfish and shellfish culture. 17th–22nd March, 2016. Isolation and Purification of microalgae. 2016, 5, 46–54. [Google Scholar]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Herdman, M.; Stanier, R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Prabhu, N.; Raj, D.T.; Yamuna, G.K.; Ayisha, S.S.; Puspha, J.; Innocent, D. Synthesis of silver phyto nanoparticles and their antibacterial efficacy. Digest Journal of Nanomaterials & Biostructures (DJNB). 2010, 5, 185–189. [Google Scholar]
- Mock, J.J.; Barbic, M.; Smith, D.R.; Schultz, D.A.; Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. The Journal of Chemical Physics. 2002, 116, 6755–6759. [Google Scholar] [CrossRef]
- Thatoi, P.; Kerry, R.G.; Gouda, S.; Das, G.; Pramanik, K.; Thatoi, H.; Patra, J.K. Photo-mediated green synthesis of silver and zinc oxide nanoparticles using aqueous extracts of two mangrove plant species, Heritiera fomes and Sonneratia apetala and investigation of their biomedical applications. Journal of Photochemistry and Photobiology B: Biology. 2016, 163, 311–318. [Google Scholar] [CrossRef]
- Manosalva, N.; Tortella, G.; Cristina-Diez, M.; Schalchli, H.; Seabra, A.B.; Durán, N.; Rubilar, O. Green synthesis of silver nanoparticles: effect of synthesis reaction parameters on antimicrobial activity. World Journal of Microbiology and Biotechnology. 2019, 35, 88. [Google Scholar] [CrossRef] [PubMed]
- Arévalo-Gallegos, A.; Garcia-Perez, J.S.; Carrillo-Nieves, D.; Ramirez-Mendoza, R.; Iqbal, H.M.; Parra-Saldívar, R. Botryococcus braunii as a bioreactor for the production of nanoparticles with antimicrobial potentialities. International Journal of Nanomedicine. 2018, 13, 5591. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Wei, Y.; Khan, Z.U.H.; Tahir, K.; Khan, S.U.; Ahmad, A.; Khan, F.U.; Cheng, L.; Yuan, Q. Electrochemical and antioxidant properties of biogenic silver nanoparticles. International Journal of Electrochemical Science. 2015, 10, 7905–7916. [Google Scholar] [CrossRef]
- da Silva, M.P.; Serafini, M.Y.M.; Nádya Pesce Da, S.; Adriano, B. Development and characterization of phosphatidylcholine nanovesicles containing the antimicrobial peptide nisin. Food Research International. 2010, 43, 1198–1203. [Google Scholar] [CrossRef]
- Salehi, S.; Shandiz, S.A.S.; Ghanbar, F.; Darvish, M.R.; Ardestani, M.S.; Mirzaie, A.; Jafari, M. Phytosynthesis of silver nanoparticles using Artemisia marschalliana Sprengel aerial part extract and assessment of their antioxidant, anticancer, and antibacterial properties. International Journal of Nanomedicine. 2016, 11, 1835. [Google Scholar]
- Aiad, M.; Moursi, E.; El-Dissoky, R.; Amer, M. Response of maize crop to irrigation under different rates and doses of nitrogen fertilization in the North Nile Delta Region. Journal of Soil Sciences and Agricultural Engineering. 2014, 5, 77–92. [Google Scholar] [CrossRef]
- Ismail, G.A.; El-Sheekh, M.M.; Samy, R.M.; Gheda, S.F. Antimicrobial, antioxidant, and antiviral activities of biosynthesized silver nanoparticles by phycobiliprotein crude extract of the cyanobacteria Spirulina platensis and Nostoc linckia. BioNanoScience. 2021, 11, 355–370. [Google Scholar] [CrossRef]
- Marslin, G.; Siram, K.; Maqbool, Q.; Selvakesavan, R.K.; Kruszka, D.; Kachlicki, P.; Franklin, G. Secondary metabolites in the green synthesis of metallic nanoparticles. Materials. 2018, 11, 940. [Google Scholar] [CrossRef]
- Kuhn, D.A.; Vanhecke, D.; Michen, B.; Blank, F.; Gehr, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein journal of nanotechnology. 2014, 5, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- Malarkodi, C.; Rajeshkumar, S.; Paulkumar, K.; Jobitha, G.G.; Vanaja, M.; Annadurai, G. Biosynthesis of semiconductor nanoparticles by using sulfur reducing bacteria Serratia nematodiphila. Advances in nano research. 2013, 1, 83–91. [Google Scholar] [CrossRef]
- Garibo, D.; Borbón-Nuñez, H.A.; Díaz de León, J.N.; Mendoza, E.G.; Estrada, I.; Toledano-Magaña, Y.; Tiznado, H.; Ovalle-Marroquin, M.; Soto-Ramos, A.G.; Blanco, A.; Rodríguez, J.A.; Romo, O.A.; Chávez-Almazán, L.A.; Susarrey-Arce, A. Green synthesis of silver nanoparticles using Lysiloma acapulcensis exhibit high-antimicrobial activity. Scientific Reports. 2020, 10, 12805. [Google Scholar] [CrossRef] [PubMed]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Frontiers in Microbiology. 2016, 7, 1831. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Graphical Abstract Illustrating the Process of Biosynthesis of Silver Nanoparticles from Synechococcus sp using Three Methods: Direct Powder Method, Supernatant of Ethanolic Extract, and Pellets.
Figure 1.
Graphical Abstract Illustrating the Process of Biosynthesis of Silver Nanoparticles from Synechococcus sp using Three Methods: Direct Powder Method, Supernatant of Ethanolic Extract, and Pellets.
Figure 2.
Neighbor Joining Phylogenetic Tree of the 16S rRNA Sequence of Synechococcus Isolate Constructed Using MEGA 11.
Figure 2.
Neighbor Joining Phylogenetic Tree of the 16S rRNA Sequence of Synechococcus Isolate Constructed Using MEGA 11.
Figure 3.
Color Change over Time to Detect Biosynthesis of Silver Nanoparticles (AgNPs) by Synechococcus sp.: (1) Control Sample: AgNO3, (2) After Adding Synechococcus Powder Sample Directly, (3) After Adding Synechococcus Ethanolic Extract, (4) After Adding Synechococcus Ethanolic Extract Pellets.
Figure 3.
Color Change over Time to Detect Biosynthesis of Silver Nanoparticles (AgNPs) by Synechococcus sp.: (1) Control Sample: AgNO3, (2) After Adding Synechococcus Powder Sample Directly, (3) After Adding Synechococcus Ethanolic Extract, (4) After Adding Synechococcus Ethanolic Extract Pellets.
Figure 4.
UV-Visible Spectrum of Biosynthesized AgNPs Utilizing Synechococcus sp using Three Methods: Powder, Pellets, and Ethanolic Extract. The Spectrum Shows the Surface Plasmon Resonance Exhibited by the Biosynthesized AgNPs in the Region Around 420 nm.
Figure 4.
UV-Visible Spectrum of Biosynthesized AgNPs Utilizing Synechococcus sp using Three Methods: Powder, Pellets, and Ethanolic Extract. The Spectrum Shows the Surface Plasmon Resonance Exhibited by the Biosynthesized AgNPs in the Region Around 420 nm.
Figure 5.
Fourier Transform Infrared (FT-IR) Spectrum of Biosynthesized AgNPs Produced by the cyanobacterium Synechococcus sp. using three different methods: Powder, Pellets, and Ethanolic extract. The spectrum highlights organic biomolecule groups responsible for the bio-Reduction of Ag+.
Figure 5.
Fourier Transform Infrared (FT-IR) Spectrum of Biosynthesized AgNPs Produced by the cyanobacterium Synechococcus sp. using three different methods: Powder, Pellets, and Ethanolic extract. The spectrum highlights organic biomolecule groups responsible for the bio-Reduction of Ag+.
Figure 6.
Zeta Sizer Analysis of Biosynthesized AgNPs Produced by Synechococcus sp using Three Different Methods: Powder, Pellets, and Ethanolic Extract. The Analysis Demonstrates the Polydisperse Size Distribution of the Nanoparticles.
Figure 6.
Zeta Sizer Analysis of Biosynthesized AgNPs Produced by Synechococcus sp using Three Different Methods: Powder, Pellets, and Ethanolic Extract. The Analysis Demonstrates the Polydisperse Size Distribution of the Nanoparticles.
Figure 7.
Zeta Potential Chart of Biosynthesized AgNPs Produced by Synechococcus sp. using Three Different Methods: Powder, Pellets, and Ethanolic Extract. The Chart Illustrates the Negative Charges of Nanoparticles.
Figure 7.
Zeta Potential Chart of Biosynthesized AgNPs Produced by Synechococcus sp. using Three Different Methods: Powder, Pellets, and Ethanolic Extract. The Chart Illustrates the Negative Charges of Nanoparticles.
Figure 8.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the direct biomass powder technique. The size of nanoparticles for single particles ranges from 20 to 80 nm on average.
Figure 8.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the direct biomass powder technique. The size of nanoparticles for single particles ranges from 20 to 80 nm on average.
Figure 9.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the pellet technique. The size of nanoparticles for single spherical particles ranges from 42-89 nm on average.
Figure 9.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the pellet technique. The size of nanoparticles for single spherical particles ranges from 42-89 nm on average.
Figure 10.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the ethanolic extract technique. The diameter of rod-shaped AgNPs ranges from 40 to 140 nm, while the diameter of spherical AgNPs ranges from 40 to 90 nm.
Figure 10.
Scanning Electron Microscope (SEM) Images of AgNPs Biosynthesized by Synechococcus sp. using the ethanolic extract technique. The diameter of rod-shaped AgNPs ranges from 40 to 140 nm, while the diameter of spherical AgNPs ranges from 40 to 90 nm.
Figure 11.
The antibacterial inhibitory zones (in mm) of three types of AgNPs—R1, R2, and R3—generated from powder, pellet, and ethanolic extract of Synechococcus sp, respectively. Both negative control (ethanol) and positive control (Ampicillin), were also included. The experiment was conducted on five plates: Plate 1 (Staphylococcus aureus), Plate 2 (Staphylococcus sp.), Plate 3 (E. coli), Plate 4 (Salmonella typhi), and Plate 5 (Pseudomonas sp.).
Figure 11.
The antibacterial inhibitory zones (in mm) of three types of AgNPs—R1, R2, and R3—generated from powder, pellet, and ethanolic extract of Synechococcus sp, respectively. Both negative control (ethanol) and positive control (Ampicillin), were also included. The experiment was conducted on five plates: Plate 1 (Staphylococcus aureus), Plate 2 (Staphylococcus sp.), Plate 3 (E. coli), Plate 4 (Salmonella typhi), and Plate 5 (Pseudomonas sp.).
Table 1.
The utilized primers for Synechococcus sequencing during the molecular identification:.
Table 1.
The utilized primers for Synechococcus sequencing during the molecular identification:.
| Sample ID |
|
Primer Sequence |
| The studied cyanobacterium |
Forward |
GTCACGCCCGAAGTCGTTAC |
| Reverse |
CCTCTGTGTGCCTAGGTATC |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).